SUMMARY
Heterochromatin can be epigenetically inherited in cis, leading to stable gene silencing. However, the mechanisms underlying heterochromatin inheritance remain unclear. Here we identify Fft3, a fission yeast homolog of the mammalian SMARCAD1 SNF2 chromatin remodeler, as a factor uniquely required for heterochromatin inheritance, rather than for de novo assembly. Importantly, we find that Fft3 suppresses turnover of histones at heterochromatic loci to facilitate epigenetic transmission of heterochromatin in cycling cells. Moreover, Fft3 also precludes nucleosome turnover at several euchromatic loci to prevent R-loop formation, ensuring proper replication progression. Our analyses show that overexpression of Clr4/Suv39h, which is also required for efficient replication through these loci, suppresses phenotypes associated with the loss of Fft3. This work uncovers a conserved factor critical for epigenetic inheritance of heterochromatin, and describes a mechanism in which suppression of nucleosome turnover prevents formation of structural barriers that impede replication at fragile regions in the genome.
eTOC blurb
Taneja et al. show that a SNF2 family chromatin remodeler suppresses histone turnover to promote epigenetic transmission of heterochromatin in dividing cells, and uncover a mechanism in which suppression of nucleosome turnover at specific genomic sites facilitates proper replication
INTRODUCTION
The vast array of differentiated cell types is derived from identical genetic information. In addition to gene-specific regulatory mechanisms, a surprising degree of control is provided by epigenetic modifications; that is, heritable modifications of chromosomes that do not involve changes to the DNA sequence (Kaufman and Rando, 2010; Probst et al., 2009). Epigenetic modifications of DNA and/or chromatin proteins create “open” or “closed” domains referred to as euchromatin or heterochromatin, respectively (Grewal and Jia, 2007; Piunti and Shilatifard, 2016; Richards and Elgin, 2002), which enforce the transcriptional program in different cell types. Discovering how distinct chromatin domains are assembled and propagated is critical for understanding normal development as well as disease states.
Enzymes and structural proteins including chromatin-remodelers, histones and their posttranslational modifications create distinct chromatin domains (Jenuwein and Allis, 2001; Li et al., 2007; Rando and Winston, 2012). Euchromatin is marked by histone acetylation and histone H3 lysine 4 methylation (H3K4me), whereas heterochromatin is distinguished by histone hypoacetylation and histone H3 lysine 9 methylation (H3K9me) (Jenuwein and Allis, 2001; Litt et al., 2001; Noma et al., 2001). H3K9me serves as a “molecular anchor” for the HP1 family of chromodomain proteins, which recruit effectors involved in various chromosomal processes including genome stability and gene silencing (Grewal and Jia, 2007).
The fission yeast Schizosaccharomyces pombe genome contains constitutive heterochromatin domains coating pericentromeric regions, subtelomeres and the silent mating-type (mat) region as well as small blocks of facultative heterochromatin (Cam et al., 2005; Zofall et al., 2012). RNA processing factors including RNAi machinery, which degrades transcripts produced by target loci, recruit Clr4/Suv39h to methylate H3K9 (Bayne et al., 2010; Zofall et al., 2012), creating binding sites for the chromodomain proteins Chp1, Chp2 and Swi6 (Grewal and Jia, 2007). Interestingly, Clr4 binds to H3K9me via its own chromodomain, enabling heterochromatin to spread beyond nucleation sites (Al-Sady et al., 2013; Zhang et al., 2008). Chp1 tethers RNAi machinery across heterochromatin domains to enforce posttranscriptional silencing in cis (Noma et al., 2004; Petrie et al., 2005). Chp2 and Swi6 load factors including SHREC that contains the class II histone deacetylase (HDAC) Clr3 and the SNF2 family protein Mit1, as well as the Clr6 HDAC to enforce transcriptional silencing (Fischer et al., 2009; Motamedi et al., 2008; Sugiyama et al., 2007).
Remarkably, heterochromatin can promote its own reassembly, leading to clonal propagation of the silenced state (Hall et al., 2002). This property has been extensively studied at the mat locus. Replacement of the RNAi nucleation site cenH, which has homology to centromeric repeats (Grewal and Klar, 1997), with ura4+ causes defects in de novo heterochromatin assembly. Conversion from heterochromatin-depleted ura4-on to the heterochromatin-enriched ura4-off occurs at a low frequency, but once established, the ura4-off state is inherited in cis (Grewal and Klar, 1996; Hall et al., 2002; Nakayama et al., 2000). Indeed, when ura4-off and ura4-on epigenetic states that differ in H3K9me levels at the otherwise genetically identical mat region are combined in the same nuclear environment of diploid cells, the H3K9me pattern associated with each state is faithfully inherited through mitosis and meiosis (Hall et al., 2002).
The ability of Clr4 to both catalyze H3K9 methylation (“write”) and bind to H3K9me (“read”) is critical for the inheritance of heterochromatin (Aygun et al., 2013; Zhang et al., 2008). Thus, in addition to heterochromatin nucleation by DNA or RNA-based mechanisms (“de novo-nucleated”), the parental H3K9 methylated nucleosomes and their associated factors provide an additional epigenetic specificity for loading Clr4 (“epigenetic-templated”), leading to clonal propagation of heterochromatin in cis. Exactly how epigenetic memory is preserved and transmitted to sister chromatids through replication remains unknown.
We performed a genetic screen to identify factors that affect epigenetic inheritance, but not de novo-nucleated assembly, of heterochromatin. We identified Fft3, a SNF2-like nucleosome-remodeling factor that affects maintenance of heterochromatin, particularly in dividing cells. This class of remodelers has a conserved, but still undetermined, role in heterochromatic silencing. Fft3 suppresses turnover of nucleosomes to enable epigenetic-templated maintenance of heterochromatin. Loss of Fft3 in other parts of the genome enhances nucleosome turnover, causing formation of RNA-DNA hybrids and defects in replication progression. Strikingly, Clr4 overexpression rescues fft3Δ phenotypes, including silencing defects and R-loop accumulation. Our results implicate a conserved SNF2 family protein in the suppression of nucleosome turnover, which safeguards epigenetic stability and ensures the proper replication of the genome.
RESULTS
Genetic Screen for Factors Affecting the Maintenance of Heterochromatin
Reporter genes inserted within a 20-kb heterochromatin domain comprising the mat2 and mat3 loci and the interval between them (referred to as the K-region) are subject to heterochromatic silencing (Grewal and Klar, 1997; Thon et al., 1994). Screens for defective reporter gene silencing have identified mutations in conserved factors that are required for heterochromatin assembly at mat and other loci (Ekwall and Ruusala, 1994; Thon et al., 1994). However, de novo heterochromatin nucleation can mask the role of factors uniquely required for propagation. To identify such factors, we used a well-characterized strain in which the cenH heterochromatin nucleation center is replaced with ura4+ (KΔ::ura4+) (Grewal and Klar, 1997). KΔ::ura4+ cells display variegated ura4+ expression, owing to stochastic de novo nucleation of heterochromatin. Once the heterochromatin enriched ura4-off state is established, it is clonally propagated in a manner dependent upon pre-existing methylated H3K9, which provides the epigenetic template for loading Clr4 (Zhang et al., 2008).
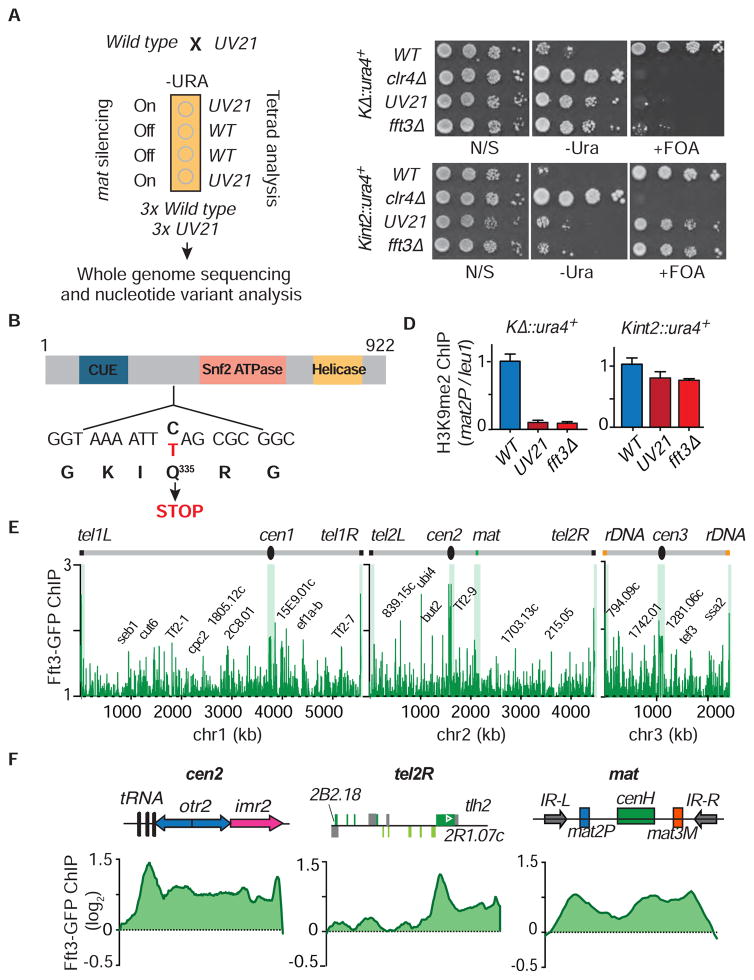
We mutagenized KΔ::ura4+ ura4-off cells and screened for mutants showing efficient conversion to the ura4-on state (Figure 1A). Next, we confirmed epigenetic maintenance of heterochromatin, but not de novo nucleation, was defective by crossing the mutants to a strain containing ura4+ inserted within cenH at the silent mat interval (Kint2::ura4+). UV19, UV21 and EMS32 showed silencing defects in the KΔ::ura4+ strain background, and little to no silencing defect (except EMS32) in the Kint2::ura4+ background, indicating that the mutants are proficient in heterochromatic silencing when de novo assembly mechanisms are operative, but fail to propagate the epigenetically-maintained silenced state (Figure 1B). We focused on UV21, which conferred a strong silencing defect in KΔ::ura4+, but had no observable effect in the Kint2::ura4+ strain background.
Figure 1. Genetic Screen for Mutations that Specifically Affect Heterochromatin Maintenance.
(A) Schematic of the mat locus showing the heterochromatic region surrounded by IR-R and IR-L boundary elements, and the strategy used to isolate mutants defective in silencing the KΔ::ura4+ locus carrying a deletion of the cenH heterochromatin nucleation element. REII and REIII indicate local silencer elements located adjacent to silent mat2P and mat3M mating-type cassettes, respectively. (B) Expression analysis of KΔ::ura4+and Kint2::ura4+ in WT and mutant backgrounds. Ten-fold serial dilutions of WT and mutants were spotted onto non-selective (N/S) media, media lacking uracil (-Ura) and counter selective FOA-containing (+FOA) media to assay for de-repression of ura4+. (C) Heterochromatin analysis in KΔ::ura4+ and Kint2::ura4+ WT and UV21 backgrounds. The H3K9me2 fold enrichment at mat2P was determined by ChIP-qPCR, relative to the control locus leu1. The normalized value of WT is set to 1. Error bars indicate + SD (n=3).
See also Figure S1.
UV21 Affects the Epigenetic Maintenance of Heterochromatin
Defective silencing in UV21 could result from changes in heterochromatin assembly. Indeed, H3K9me levels at the mat locus were severely reduced in UV21 KΔ::ura4+ cells, but only marginally affected in UV21 Kint2::ura4+ cells (Figure 1C). Thus, UV21 does not affect de novo targeting of H3K9me, but is required for epigenetic-templated maintenance of heterochromatin.
We next analyzed heterochromatin assembly at other loci. Pericentromeric regions contain dg/dh repeats that are “hot spots” for production of siRNA required for RNAi-mediated de novo assembly of heterochromatin (Cam et al., 2005). Heterochromatin at subtelomere regions is nucleated at specific sites and spreads in a manner dependent on nucleosome stabilizing factors (such as the HDAC Clr3), similar to the mat locus. We observed no major change in H3K9me levels at pericentromeric repeats in UV21, however, H3K9me at subtelomeric regions was affected, particularly in the regions adjoining the nucleation sites (such as tlh1 or tlh2) (Figure S1A). Therefore, UV21 shows specific defects in the epigenetic maintenance of heterochromatin.
Interestingly, further examination revealed that the H3K9me reduction in UV21, linked to the KΔ::ura4+ ura4-off to ura4-on conversion, occurs broadly across the entire silent mat domain (Figure S1B), rather than being restricted to a specific region.
UV21 Contains a Mutation in the SNF2 Family Member Protein Fft3
To identify the locus harboring the UV21 mutation, we backcrossed UV21 to the non-mutagenized parental strain and performed tetrad analyses. Silencing of the ura4+ reporter was used to monitor the segregation of the mutant allele (Figure 2A). We sequenced three wild-type 6 (WT) and three UV21 mutant segregants from the final cross and found one substitution mutation (C-to-T) uniquely present in the mutants. This mutation mapped to fft3, which encodes one of the S. pombe homologs of the highly conserved SNF2 family chromatin remodeler, called SMARCAD1 in humans and Fun30 in budding yeast (Neves-Costa et al., 2009; Rowbotham et al., 2011; Steglich et al., 2015; Stralfors et al., 2011). Conventional DNA sequencing confirmed UV21 is a nonsense mutation that introduces a premature stop codon upstream of the ATPase domain in Fft3 (Figure 2B).
Figure 2. The UV21 Mutant Is Associated with the SNF2 Family Chromatin Remodeler Fft3.
(A) Identification of the UV21 mutation by whole genome sequencing. The UV21 mutant strain was backcrossed to WT and the phenotype was tracked by monitoring for de-repression of ura4+ inserted at the mat locus. Segregants were subjected to whole genome sequencing and nucleotide variant analysis. (B) Schematic representation of Fft3. The position of the nonsense mutation associated with UV21 is indicated. (C) Expression analysis of KΔ::ura4+and Kint2::ura4+ in UV21 and fft3Δ. Serial dilutions of WT and mutants were spotted onto non-selective (N/S) media, media lacking uracil (-Ura) and counter selective FOA-containing (+FOA) media to assay for de-repression of ura4+. (D) Heterochromatin analysis in KΔ::ura4+ and Kint2::ura4+ WT and mutant backgrounds. The H3K9me2 fold enrichment at mat2P was determined by ChIP-qPCR, relative to the control locus leu1. The normalized value of WT is set to 1. Error bars stand for + SD (n=3). (E) Genome wide localization of Fft3. Fft3-GFP distribution along the S. pombe chromosomes was determined by ChIP-chip. (F) Fft3 associates with heterochromatin domains. Fft3-GFP was determined by ChIP-chip.
See also Figure S2.
fft3Δ Mimics the Heterochromatin Defects in UV21
To confirm that the effects on heterochromatin are functionally linked to a mutation in fft3+, we crossed UV21 with fft3Δ. Tetrad analysis showed that UV21 is tightly linked to fft3 (i.e. all four segregants showed the mutant phenotype in 40 analyzed tetrads, suggesting that UV21 is likely a mutation in fft3). Indeed, deletion of fft3 in the KΔ::ura4+ ura4-off background caused a majority of cells to convert to ura4-on, whereas fft3Δ Kint2::ura4+ showed little or no conversion to ura4-on (Figure 2C). Consistently, H3K9me was severely reduced in fft3Δ KΔ::ura4+, and only marginally affected in Kint2::ura4+ (Figure 2D). Thus, analogous to UV21, fft3Δ affects epigenetic-templated, but not de novo-nucleated assembly of heterochromatin at the mat locus.
In cells lacking Fft3, H3K9me levels were maintained at pericentromeric regions, but reduced at subtelomeric regions, particularly at sequences adjoining the heterochromatin nucleation sites (Figure S2A). Moreover, H3K9me was abolished throughout the silent mat interval of fft3Δ KΔ::ura4+ cells that are defective in de novo-nucleation, but not in the WT counterpart (Figure S2B). Together, these results show that the phenotypic effects of fft3Δ mimic those of UV21.
Fft3 Localizes to Heterochromatin Domains and to Several Euchromatic Loci
ChIP-chip analysis revealed significant peaks of Fft3 at all major heterochromatic loci (Figure 2E). The localization pattern of Fft3 closely resembled the distribution of heterochromatin at pericentromeric regions and the silent mat interval surrounded by IR boundary elements (Figure 2F). Fft3 was also enriched at subtelomeric regions (Figure 2F). These results suggest that Fft3 acts directly and may cooperate with heterochromatin factors to maintain silenced chromatin. 7 Notably, our previous purification of Swi6, which affects the inheritance of heterochromatin in cis (Hall et al., 2002; Nakayama et al., 2000), contained Fft3 peptides (Fischer et al., 2009). Indeed, Fft3 co-immunoprecipitated with Swi6 (Figure S2C), supporting a close connection to heterochromatin factors.
We also noted Fft3 peaks distributed across all three chromosomes (Figure 2E). Consistent with earlier findings (Steglich et al., 2015; Stralfors et al., 2011), Fft3 was enriched at 55% of solo LTRs, 68% of tRNAs and 67 % of loci encoding snRNAs (Figure S2D). Fft3 was also highly enriched throughout Tf2 retrotransposons and their related wtf elements, and at several non-coding RNA loci (Figure S2D). Surprisingly, Fft3 was preferentially enriched at highly transcribed genes and genes containing short internal repeats within their ORFs, where it was distributed across the transcribed regions (Figure S2D). Therefore, Fft3 may perform additional functions in other parts of the genome.
Loss of Fft3 Causes a Variegated Expression Pattern
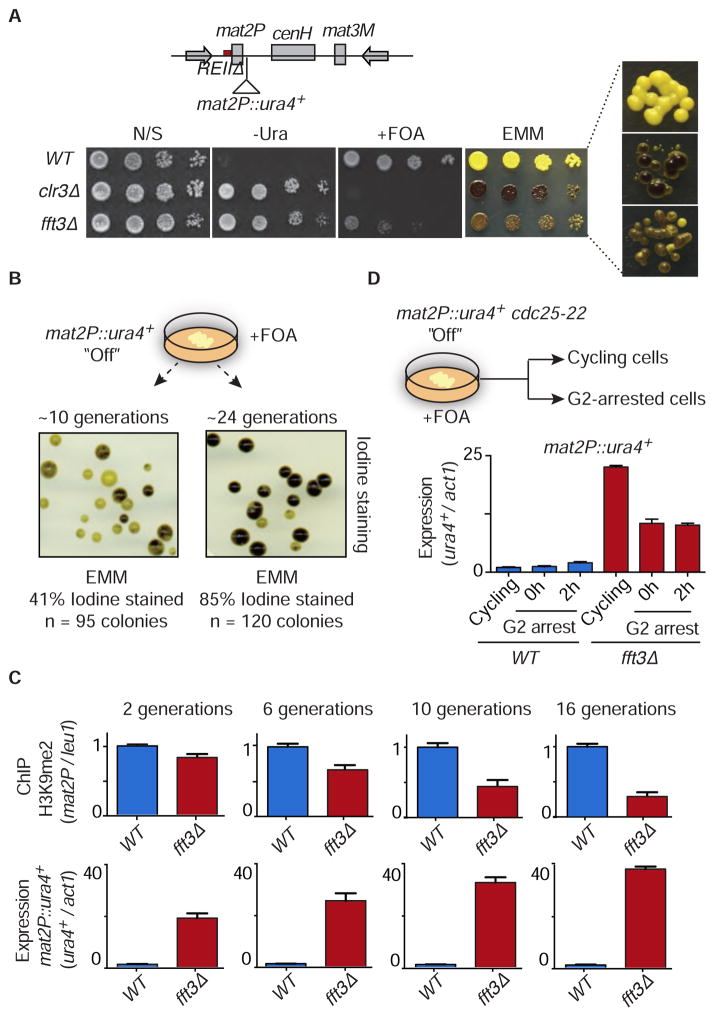
We next focused on the role of Fft3 in heterochromatin maintenance. Because of the rapid loss of heterochromatin in KΔ::ura4+ strains, we employed an alternative sensitized system using cells that carry a deletion of a locally acting silencer element (termed REII) adjacent to mat2P. Deletion of REII noticeably affects mat2P silencing only in cells impaired in the assembly of heterochromatin (Thon et al., 1994). In non-switchable mat1-M cells, the expression of mat2P leads to haploid meiosis and the formation of a starch compound detectable by iodine staining (Thon et al., 1994). Black coloration indicates a severe loss of silencing whereas a yellow color (unstained) indicates a silenced mat2P region. Interestingly, loss of Fft3 yielded a mixture of colonies, that ranged from highly stained (black) to unstained (yellow), and caused variegated expression of ura4+ inserted adjacent to mat2P (mat2P::ura4+) (Figure 3A). This phenotype is distinct from the uniform de-repression of mat2P and the ura4+ reporter in cells lacking core heterochromatin silencing machinery such as Clr3 and Clr4 (Figure 3A) (Thon et al., 1994). Together, our analyses support a role for Fft3 in maintaining the repressive heterochromatic state.
Figure 3. Variegated Loss of Heterochromatin at the REIIΔ Site in fft3Δ.
(A) Schematic of the mat locus in the REIIΔ strain, showing the location of the ura4+ insertion next to mat2P (top). Serial dilutions were spotted onto the indicated media. A panel showing iodine staining at high magnification is displayed (right). Note that iodine staining of fft3Δ yields a mixture of yellow and black colonies, in contrast to WT (all yellow) and clr3Δ (all black). (B) Gradual loss of heterochromatin in fft3Δ cells. fft3Δ cells selected for transcriptionally silenced mat2P::ura4+ (“Off”) on FOA-containing media were grown in non-selective medium for either ~10 generations or ~24 generations. Colonies were grown on an Edinburgh Minimal Medium (EMM) plate and subjected to iodine staining. The number of stained colonies was scored for each generation length. (C) WT and fft3Δ selected for mat2P::ura4+ “Off” were grown in N/S media for 2, 6, 10 and 16 generations. The H3K9me2 fold enrichment was determined by ChIP-qPCR (top) and ura4+ silencing was determined by RT-qPCR (bottom). The fold enrichment of H3K9me2 at mat2P relative to the control leu1 locus is shown as the mean + SD (n=3). The normalized value of WT is set to 1. Similarly, expression of ura4+ relative to the control locus act1 is shown as the mean + SD (n=3). The normalized value of ura4+ expression in WT is set to 1. (D) Loss of silencing in cycling fft3Δ cells. WT or fft3Δ cells containing the cdc25-22 allele were selected for mat2P::ura4+ “Off” on FOA containing media, and were grown at 26°C or arrested in G2 by shifting to 36°C. Cells were collected either immediately after the arrest (0h G2 arrest) or after an additional 2 hours of arrest (2h G2 arrest). Transcript levels of ura4+ relative to act1 in WT (blue bars) and fft3Δ (red bars) were determined by RT-qPCR and are shown as the mean + SD (n=3).
See also Figure S3.
Fft3 Facilitates the Propagation of Heterochromatin in Cycling Cells
The variegated phenotype of fft3Δ might reflect defects in the fidelity of heterochromatin inheritance during cell division. To test this, we grew fft3Δ cells carrying REIIΔ on counter-selective FOA medium to select for mat2P::ura4+ silencing. These cells were allowed to multiply on a non-selective medium before assaying for mat2P expression. Iodine staining revealed a mixture of yellow and black stained colonies. The percentage of black stained colonies, indicating defects in mat2P silencing and reduction in H3K9me (Figures 3A and S3A), increased with the number of cell divisions (Figure 3B). H3K9me in fft3Δ steadily decreased as the number of cell divisions increased (Figure 3C, top panels), correlating with loss of mat2P::ura4+ silencing (Figure 3C, bottom panels). Thus, maintaining the heterochromatic state during successive cell divisions requires Fft3, and its loss results in a variegated expression pattern due to frequent conversion to the expressed state.
We next compared mat2P::ura4+ expression in WT and fft3Δ ura4-off cells, which were either allowed to cycle or were arrested at the G2/M boundary using the temperature-sensitive (ts) cdc25-22 allele. Expression of mat2P::ura4+ was assayed immediately (0 hour) or 2 hours after the arrest. Notably, de-repression of mat2P::ura4+ occurred in both cycling and G2 arrested fft3Δ cells. However, de-repression was considerably higher in cycling cells (Figure 3D), indicating that Fft3 is particularly important for maintenance of heterochromatin in cycling cells.
Fft3 Interacts with Replication Factor C
The propagation of heterochromatin in cis presumably requires collaboration between chromatin assembly factors and DNA replication machinery. We identified all subunits of the Replication Factor C complex (RFC) in our purification of GFP-tagged Fft3 (Fft3-GFP) from a synchronized S phase population (Figure S3B). RFC is a five subunit DNA polymerase (Pol) δ and ε accessory factor required for loading the essential processivity factor PCNA onto nascent DNA strands (Tsurimoto and Stillman, 1989). Flag-tagged RFC subunits co-immunoprecipitated with Fft3-GFP (Figure S3C). Thus, Fft3 interacts with replication fork associated proteins, similar to human SMARCAD1 (Rowbotham et al., 2011), which might link DNA replication with maintenance of epigenetic chromatin states (Franco and Kaufman, 2004).
Fft3 Facilitates Heterochromatin Maintenance by Suppressing Histone Turnover
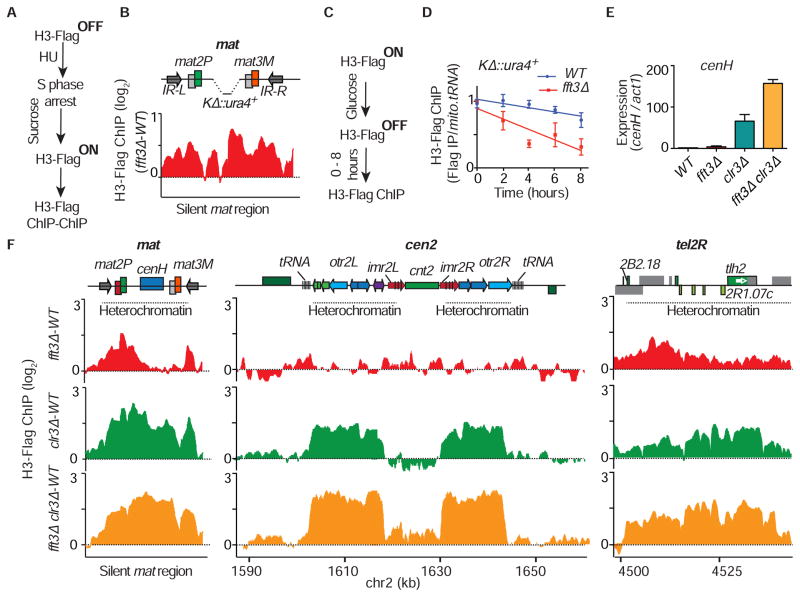
Pre-existing methylated nucleosomes are believed to recruit Clr4 for clonal propagation of heterochromatin (Zhang et al., 2008). Interestingly, loss of the anti-silencing factor Epe1, which promotes turnover of histones (Aygun et al., 2013), rescued heterochromatin maintenance defects in fft3Δ (Figure S4A), suggesting Fft3 may affect nucleosome stability. To explore this, we performed a nucleosome exchange assay (Aygun et al., 2013) in which carboxy-terminal Flag-tagged histone H3 (H3-Flag) is rapidly induced by shifting the carbon source from glucose to sucrose (Figure S4B). We induced H3-Flag expression in KΔ::ura4+ cells after blocking DNA replication with hydroxyurea (HU) and determined the level of H3-Flag incorporation (Figure 4A). Remarkably, elevated levels of H3-Flag occurred throughout the silent mat region in fft3Δ (Figure 4B), suggesting that Fft3 suppresses nucleosome exchange.
Figure 4. Fft3 Suppresses Nucleosome Turnover at Heterochromatic Regions.
(A) Experimental design used to measure histone H3-Flag incorporation. (B) H3-Flag incorporation in fft3Δ was determined by microarray analysis following the schematic in (A) and is shown for the KΔ::ura4+ mat locus. The signal is normalized to WT. (C) Experimental design to analyze nucleosome retention at KΔ::ura4+. (D) The H3-Flag fold enrichment relative to mitochondrial tRNA was determined by ChIP-qPCR for cycling WT and fft3Δ cells at the indicated time points following the schematic in (C) and is shown as the mean ± SD (n=3). The 0 hour value was set to 1 for comparison between different time points. (E) Loss of Clr3 and Fft3 causes cumulative defects in cenH silencing. The relative expression of cenH transcripts over act1 was determined by RT-qPCR in the indicated strains and are shown as the mean + SD (n=3). (F) H3-Flag incorporation in fft3Δ, clr3Δ and the double mutant was determined by ChIP-chip following the schematic in (A) and is shown for the indicated heterochromatic regions. Signals were normalized to WT.
See also Figure S4.
Since Fft3 is particularly important in cycling cells, we wondered whether the “dilution” of parental histones coupled to DNA replication occurs more rapidly in fft3Δ. To address this, we modified the nucleosome exchange assay by pre-loading chromatin with H3-Flag nucleosomes and monitoring their loss in cycling cells. Cells were grown overnight in sucrose-containing medium to express Flag-tagged H3 and then were shifted to glucose medium to turn off H3-Flag expression (Figures 4C and S4C). Time course analyses of H3-Flag dilution at the mat locus in KΔ::ura4+ cells showed that loss of H3-Flag occurred more rapidly in fft3Δ as compared to WT (Figure 4D). This difference was observed at both the transcribed ura4+ reporter and a transcriptionally inert region (Figures 4D and S4D), indicating that increased transcription cannot be solely responsible for the loss of histones in fft3Δ. This result further supports the role of Fft3 in the suppression of histone turnover to facilitate epigenetic-templated assembly of heterochromatin in cycling cells.
Fft3 and Clr3 Perform Overlapping Functions to Preclude Histone Turnover
The HDAC Clr3 is distributed across all major heterochromatin domains (Sugiyama et al., 2007), and also suppresses nucleosome turnover (Aygun et al., 2013). Cells lacking Fft3 and Clr3 showed a cumulative increase in centromeric repeat (cenH) transcripts (Figure 4E), suggesting overlapping functions. We therefore analyzed histone turnover in clr3Δ, fft3Δ or clr3Δ fft3Δ strains blocked using HU treatment. We observed an elevated histone turnover throughout heterochromatin domains in clr3Δ (Figure 4F), and a similar, albeit weaker, increase in histone turnover in fft3Δ (Figure 4F). This is consistent with a variegated expression pattern in fft3Δ that contrasts with the uniform de-repression in clr3Δ (Figure 3A). Remarkably, the clr3Δ fft3Δ double mutant showed a cumulative increase in histone turnover throughout heterochromatin domains (Figure 4F), which was most evident at pericentromeric regions and the central centromere core (Figure 4F). Together with cumulative derepression of centromeric repeats, these results support a causal relationship between nucleosome stability and heterochromatic silencing, and suggest Fft3 and Clr3 perform distinct functions to preclude histone turnover.
Fft3 Localized at Euchromatic Loci Promotes Replication Progression
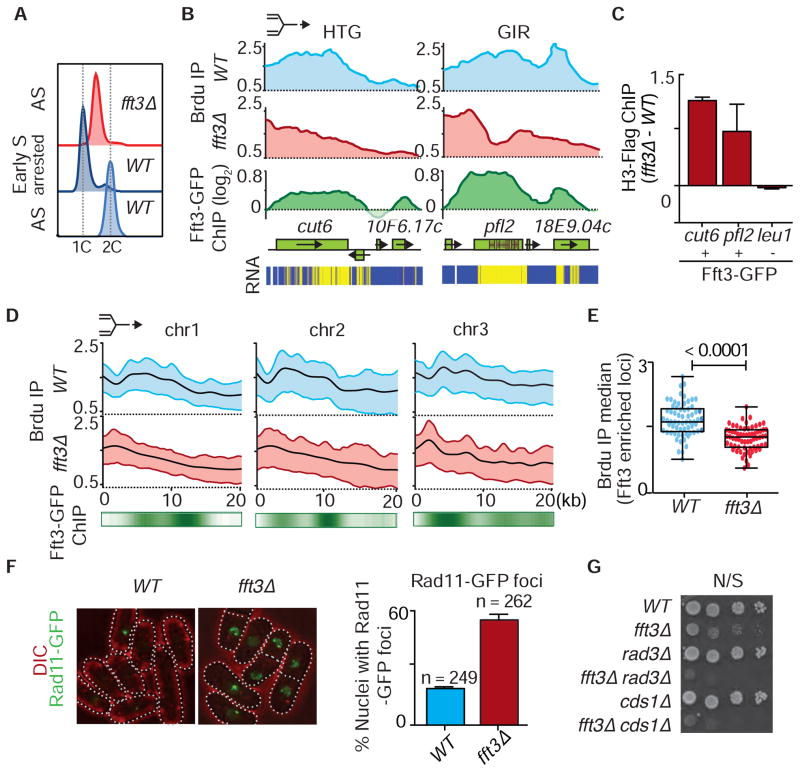
Since Fft3 associates with RFC and localizes to specific euchromatic loci, such as natural replication fork barriers including tRNAs, retroelements and their remnants, and genes containing short internal repeats (Figures 2E and S2D) (Steglich et al., 2015; Stralfors et al., 2011), we asked whether fft3Δ is defective in replication progression. Notably, fft3Δ cells show a growth defect at low temperature (Figure S5A), and are sensitive to genotoxins (see below). Interestingly, asynchronous fft3Δ cells and fft3Δ cells released from the HU block showed a prolonged S phase (Figures 5A and S5B), indicative of possible defects in DNA replication.
Figure 5. fft3Δ Cells Display Replication Defects Across Various Euchromatic Loci.
(A) FACS profiles of asynchronous (AS) fft3Δ, asynchronous WT and HU blocked WT cells. (B) Fork progression at a highly transcribed gene (HTG) cut6+ and a gene containing short internal repeats (GIR) pfl2+, as determined by ChIP-chip analysis of BrdU incorporation in WT and fft3Δ. The replication fork symbol on top indicates the direction of fork progression. Fft3-GFP distribution is shown below. The locations of internal repeats are marked within the ORF. Expression analysis (RNA) of the indicated loci is shown as a heat map (bottom panel, yellow: high level of cDNA; blue: transcriptionally cold region). (C) ChIP-qPCR analysis of H3-Flag incorporation at loci with or without Fft3-GFP enrichment in fft3Δ normalized to WT. (D) Cumulative analysis of fork progression through a 20kb region for 67 loci enriched with Fft3 (see Table S1) as determined by ChIP-chip analysis of BrdU incorporation in WT and fft3Δ. The heat map below shows the distribution of Fft3-GFP. (E) Box plot of the median of total BrdU incorporation at each of 67 loci (see Table S1) enriched with Fft3, determined using ChIP-chip. The corresponding differences between the two indicated strains are highly significant as determined by t-test (p<0.0001). (F) Cells lacking Fft3 accumulate single-stranded DNA. Rad11-GFP foci were scored in WT and fft3Δ cells. (G) Cells lacking Fft3 require replication checkpoint proteins for survival. Serial dilutions of the indicated strains were spotted onto non-selective media (N/S) for growth analysis.
See also Figures S5, S6 and Table S1.
To directly investigate replication, we arrested cells at the G2/M boundary and monitored bromo-2-deoxyuridine (BrdU) incorporation into DNA (Hayashi et al., 2007). Despite efficient origin firing, BrdU incorporation was low at specific locations in fft3Δ (Figures 5B, S5C and S5D). We identified ~67 Fft3 enriched locations showing defects in replication progression (Table S1). Strikingly, most contained highly transcribed genes or genes containing short internal repeats that show Fft3 enrichment (Figure 2E and S2D). Similar but less pronounced results were obtained with G2/M arrested cells released in the absence of HU, presumably due to faster progression of the replication program (Figures S5D and S5E). Fft3 suppressed nucleosome turnover at the affected loci, similar to its effect at heterochromatin (Figures 5C, S5F and S5G). Cumulative BrdU profiles confirmed these defects at affected loci on all three chromosomes (Figure 5D). Statistical analyses revealed defective replication specifically at genomic locations showing Fft3 localization (Figures 5E and S5H), with no major changes in the global replication pattern (Figure S5I). We conclude that Fft3 precludes histone turnover and promotes progression of replication forks at specific euchromatic loci.
Requirement for Replication Checkpoint Machinery in fft3Δ Cells
Replication defects in fft3Δ might cause accumulation of single stranded DNA marked by Rad11, a component of the single-stranded DNA binding complex, replication protein A (RPA) (Parker et al., 1997). Indeed, 55% of fft3Δ nuclei accumulated Rad11 foci, compared to 19% of WT nuclei (Figure 5F). We also quantified nuclei with foci containing Rad52, a protein that is recruited to DNA double-strand break sites (Kim et al., 2000). Only 6% of WT nuclei contained Rad52-GFP foci, in comparison to 32% of fft3Δ nuclei (Figure S5J). To further explore replication stress in fft3Δ cells, we looked for growth defects when fft3 was deleted in cells lacking the replication checkpoint proteins Rad3 (homolog of ATR) or Cds1 (homolog of Chk2) (Cimprich and Cortez, 2008). The severe growth defects of the double mutants fft3Δ rad3Δ and fft3Δ cds1Δ (Figure 5G) indicate that replication checkpoint proteins are required for the survival of fft3Δ cells and supports the involvement of Fft3 in replication fork progression.
We wondered whether silencing defects could mainly be attributed to defects in replication progression in fft3Δ. To address this, we isolated a conditional mutant allele of fft3 (fft3-ts) that displays heterochromatin silencing defects when grown at 33°C but not at 26°C (Figure S6A). fft3-ts contains a point mutation in the ATPase domain (Fft3-R602K, Figure S6B) that does not affect steady state Fft3 protein levels (Figures S6C). fft3-ts and WT KΔ::ura4-off cells were arrested in G1 at permissive temperature by nitrogen starvation, and then shifted to non-permissive temperature for 4 hours. Notably, defective heterochromatic silencing could be observed in fft3-ts cells without progression through S phase (Figure S6D).
Clr4 overexpression Rescues Defects Caused by Loss of Fft3
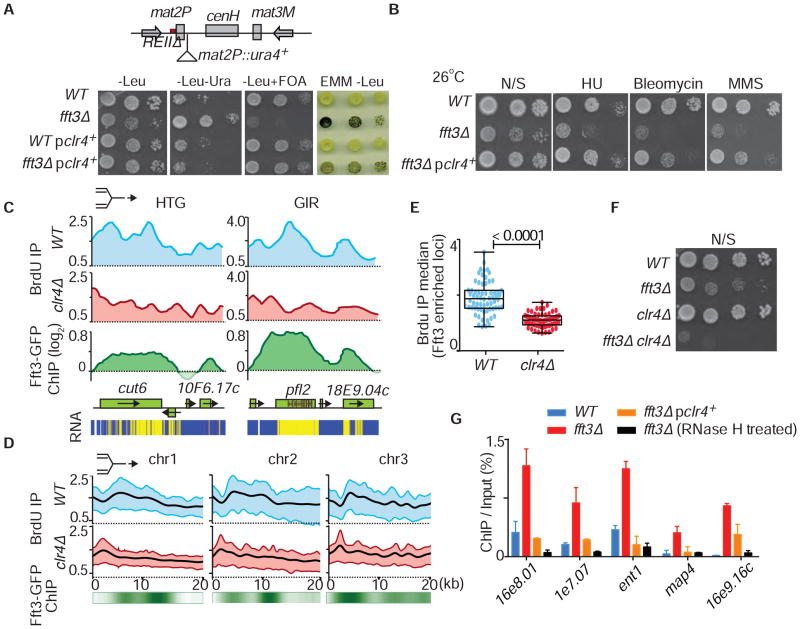
We first attempted to identify UV21 by cloning the gene from a genomic DNA library. Although we failed, we did identify complementing clones containing clr4+. Indeed, multiple copies of clr4+can suppress the silencing defect in UV21. Consistently, Clr4 overexpression partially restored silencing in fft3Δ REIIΔ mat2P::ura4+ cells (Figure 6A), and suppressed the sensitivity of fft3Δ to genotoxins (Figure 6B). These results suggested that Fft3 and Clr4 might perform overlapping functions. Interestingly, defects in BrdU incorporation occurred at the same genomic regions in fft3Δ and clr4Δ cells (Figure S7A), showing slower fork progression through highly transcribed genes and genes containing internal repeats (Figure 6C). Cumulative BrdU profiles confirmed these effects on replication progression (Figure 6D). Loss of Clr4 specifically affected BrdU incorporation at Fft3 enriched loci (Figures 6E and S7B), further indicating that both Fft3 12 and Clr4 facilitate replication at these regions. Notably, fft3Δ clr4Δ showed a severe growth defect (Figure 6F). Taken together, these results uncover functional interplay between Fft3 and Clr4 in promoting heterochromatic silencing and replication progression at specific regions of the genome.
Figure 6. Over-expression of Clr4 Rescues Defects Caused by Loss of Fft3.
(A) Serial dilutions and iodine staining of the indicated strains are shown. (B) Growth defects and genotoxin sensitivity of fft3Δ are suppressed by Clr4 overexpression. Plates were incubated at 26°C for 5–6 days. (C) Analysis of fork progression as determined by ChIP-chip analysis of BrdU incorporation is shown for the highly transcribed gene (HTG) cut6+ and the pfl2+ gene containing short internal repeats (GIR) in WT and clr4Δ. The replication fork symbol on top indicates the direction of fork progression. Fft3-GFP distribution at the indicated loci is shown below. Expression analysis of the indicated loci is shown as a heat map (bottom panel, high level of cDNA; blue: transcriptionally cold region). BrdU microarray data for clr4Δ and WT were generated previously (Zofall et al., 2016). (D) Cumulative analysis of fork progression through a 20kb region at 67 loci enriched with Fft3-GFP (see Table S1) as determined by ChIP-chip analysis of BrdU incorporation in WT and clr4Δ. Fft3-GFP distribution is shown below as a heat map. (E) Box plot of the median of total BrdU incorporation at each of 67 loci (see Table S1) enriched with Fft3 as determined using ChIP-chip. The corresponding differences between the two indicated strains are highly significant as determined by t-test (p<0.0001). (F) Cells lacking Fft3 and Clr4 show severe growth defects. Serial dilutions of the indicated strains were spotted onto non-selective medium. (G) Rescue of R-loop formation in fft3Δ cells by over-expression of Clr4. Formation of R loops in WT (blue bars), fft3Δ (red bars), fft3Δ overexpressing Clr4 (orange bars) and fft3Δ with RNase H treatment (black bars) was determined by DRIP assay using S9.6 antibody, followed by qPCR. Prior to ChIP with S9.6 antibody, chromatin fractions were treated with RNase A. ChIP data are quantified as the percentage of input DNA. Data are shown as the mean + SD (n=2).
Fft3 and Clr4 Prevent R-loop Formation at Loci showing Replication Defects
We next investigated the underlying cause of replication defects in fft3Δ. Unstable nucleosomes at transcribed regions in fft3Δ might provide an opportunity for nascent RNA to hybridize with DNA and form R-loops that obstruct replication forks (Richard and Manley, 2016; Santos-Pereira and Aguilera, 2015; Skourti-Stathaki and Proudfoot, 2014). We used S9.6 antibody (Hu et al., 2006) to isolate RNA-DNA hybrids (Ginno et al., 2012) from cells synchronized in S phase. Loci showing replication arrest in fft3Δ accumulated R-loops at a significantly higher level than in WT (Figures 6G and S7A), with no significant change in their transcript levels (Figure S7C). Treatment with RNase H reduced the signal, confirming our detection of bona fide R-loop structures (Figure 6G). Moreover, overexpression of RNase H suppressed the growth defect and the genotoxin sensitivity of fft3Δ (Figure S7D), supporting the conclusion that R-loop formation underlies the phenotypes associated with the loss of Fft3.
Since overexpression of clr4+ suppressed the genome instability and silencing defects in fft3Δ, we tested whether R-loop formation was also mitigated. Indeed, we observed a reduction in R-loops at regions with replication defects (Figure 6G). These results reveal that both Fft3 and Clr4 suppress R-loop formation specifically at sites that show replication defects in their absence, further highlighting the significance of the genetic interaction between Clr4 and Fft3.
DISCUSSION
Heterochromatin is critical for gene regulation and the integrity of eukaryotic genomes (Grewal and Jia, 2007). De novo assembly pathways have been described, however, little is known about the factors involved in epigenetic inheritance of heterochromatin. Here we identify a conserved SNF2 family protein, Fft3, as a factor dispensable for de novo assembly but required for epigenetic-templated maintenance of heterochromatin, wherein parental methylated H3K9 facilitates recruitment of Clr4 to promote clonal propagation of heterochromatin (Hall et al., 2002; Zhang et al., 2008). Fft3 suppresses turnover of histones to ensure faithful propagation of heterochromatin during cell division. Fft3 also precludes histone turnover at euchromatic regions that are difficult to replicate, which prevents the formation of aberrant structures that can obstruct replication machinery.
Fft3 Suppresses Histone Turnover and Promotes Heterochromatin Inheritance
Our unbiased genetic screen revealed an important role for Fft3 in the epigenetic inheritance of heterochromatin. Fft3 likely directly promotes propagation of heterochromatin. Supporting this, Fft3 is preferentially enriched at all major heterochromatin domains. Moreover, Fft3 interacts with Swi6/HP1, previously implicated in the propagation of heterochromatin in cis (Hall et al., 2002; Nakayama et al., 2000). Fft3 may act with Swi6 to ensure the stable transmission of epigenetic imprints critical for heterochromatin inheritance.
Fft3 homologs have been implicated in heterochromatin silencing from yeast to mammals, suggesting their conserved role in this process (Byeon et al., 2013; Neves-Costa et al., 2009; Rowbotham et al., 2011; Stralfors et al., 2011). Here we find that Fft3 suppresses histone turnover to promote epigenetic stability. SHREC, which contains the SNF2 chromatin remodeler Mit1 in addition to the HDAC Clr3 (Creamer et al., 2014; Sugiyama et al., 2007), also precludes turnover of nucleosomes (Aygun et al., 2013). However, Fft3 and SHREC likely perform distinct functions. First, Fft3 and Clr3 act redundantly to suppress histone turnover across heterochromatin domains. Second, loss of these factors causes cumulative de-repression of heterochromatic repeat elements. Third, loss of Clr3 causes uniform de-repression across heterochromatin domains, whereas loss of Fft3 compromises the fidelity of heterochromatin inheritance leading to a variegated expression pattern. We envision that SHREC enforces heterochromatin silencing during interphase in the major G2 phase of the S. pombe cell cycle. Fft3 rather has a more prominent role in ensuring epigenetic transmission of heterochromatin in dividing cells. Notably, Fft3 is the first factor shown to suppress histone turnover to transmit epigenetic memory and maintain heterochromatin in dividing cells.
The suppression of histone turnover by Fft3 has direct implications for the epigenetic-templated inheritance of heterochromatin (Figure 7). The cis inheritance of heterochromatin (Audergon et al., 2015; Hall et al., 2002; Nakayama et al., 2000; Ragunathan et al., 2015) requires the retention of methylated H3K9 for recruitment of Clr4 through its chromodomain 14 (Zhang et al., 2008) to modify newly assembled nucleosomes and replicate the parental histone modification pattern. How parental histones, which are transferred to daughter DNA strands (Alabert and Groth, 2012; Jackson and Chalkley, 1985; Xu et al., 2010), are maintained during DNA replication is a key question. Fft3 and its mammalian homolog SMARCAD1, which associate with replication machinery (this study) (Rowbotham et al., 2011), might help in displacing nucleosomes during the passage of polymerases and then re-depositing them. As per this model, Fft3 might allow events such as replication and transcription to occur without loss of epigenetic information in the form of modified histones. Supporting this, “old” histones are lost more rapidly in fft3Δ than in WT (Figure 4D). Histone chaperones Asf1 and CAF-1 (Alabert and Groth, 2012; Kaufman and Rando, 2010; Probst et al., 2009; Zhang et al., 2000), which associate with Swi6/HP1 implicated in heterochromatin inheritance may also participate in this process (Dohke et al., 2008; Yamane et al., 2011).
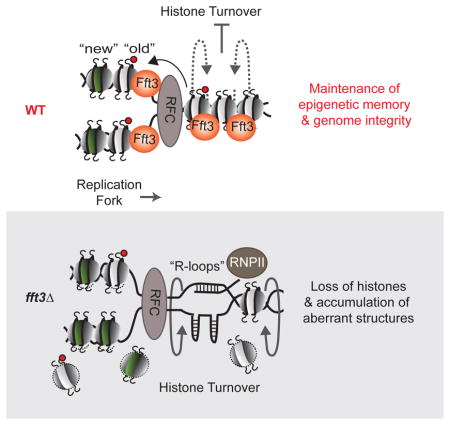
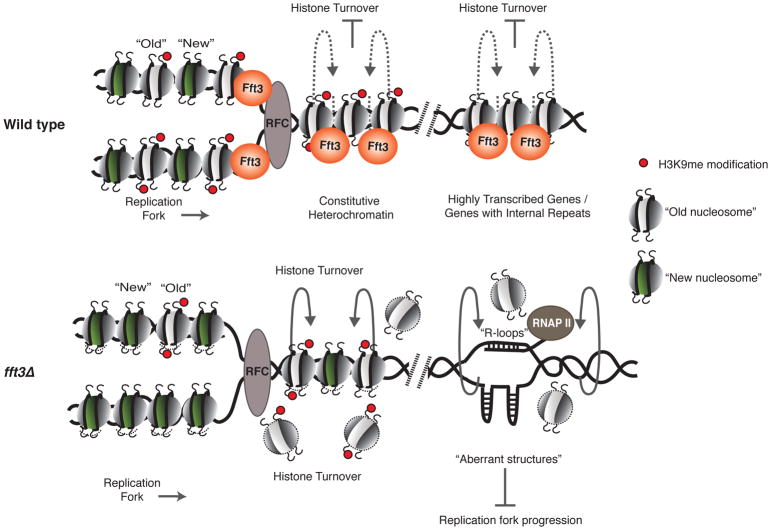
Figure 7. Model Illustrating the Mechanism of Action of the Fft3 Chromatin Remodeler.
Fft3 precludes the exchange of nucleosomes over heterochromatic as well as euchromatic loci such as highly transcribed genes and genes containing short internal repeats. Loss of Fft3 leads to unstable nucleosomes and a gradual loss of old H3K9me nucleosomes at heterochromatic regions, causing defects in the propagation of heterochromatin. At euchromatic loci, loss of Fft3 leads to an open chromatin structure and formation of R-loops and/or aberrant structures, which further destabilize nucleosomes and impede replication fork progression, causing replication defects.
A previous study noted the appearance of euchromatin marks at subtelomeric regions and at the centromere central core in fft3Δ (Stralfors et al., 2011), which could result from increased histone turnover and incorporation of newly synthesized histones with distinct modifications (Alabert and Groth, 2012). As observed for heterochromatic regions, loss of Clr3 and Fft3 has a cumulative effect on histone turnover at the central core. Given that Clr3 and Fft3 have overlapping functions and co-localize at several loci, it is conceivable that Fft3 may act more broadly to suppress histone turnover, but its effects are masked by other factors acting in parallel.
Fft3 as a Chromatin Organizer for Replication Progression
Cells lacking Fft3 are defective in replication progression and accumulate single-stranded DNA. Consistent with DNA damage, fft3Δ cells accumulate Rad52 repair foci and are sensitive to genotoxic agents and the loss of the replication checkpoint proteins Rad3 and Cds1. Indeed, replication checkpoint activation may explain the prolonged S phase in fft3Δ. Importantly, loss of Fft3 does not affect the global replication program, but only impairs progression at specific loci including highly transcribed genes or genes containing short internal repeats in the ORF. This suggests that transcription-replication collisions or formation of secondary structures might impede replication forks.
How could Fft3 facilitate the passage of replication forks? Fft3 and its mammalian counterpart interact with replication machinery (Rowbotham et al., 2011) (this study), and could directly resolve replication barriers, similar to Isw2 and Ino80 remodelers in budding yeast (Vincent et al., 2008), and/or facilitate DNA repair (Chen and Symington, 2013). Notably, we find accumulation of R-loops known to impede replication machinery in fft3Δ, leading to the formation of recombinogenic DNA breaks (Richard and Manley, 2016; Santos-Pereira and Aguilera, 2015; Skourti-Stathaki and Proudfoot, 2014). Fft3 suppresses turnover of histones at highly transcribed regions, which likely protects the DNA fiber from invading RNA and prevents R-loop formation (Figure 7). Similarly, nucleosomes at loci containing short internal repeats may prevent single stranded DNA from forming secondary structures. In this model, enhanced turnover of histones in fft3Δ creates open chromatin, allowing formation of R-loops and/or secondary DNA structures, which could further aggravate histone turnover and lead to stalled replication forks and DNA damage (Figure 7). By preventing such aberrant structures from forming, factors such as Fft3 ensure the unimpeded passage of replication forks.
Interestingly, Fft3 and Clr4 perform overlapping functions. Overexpression of Clr4 can rescue fft3Δ phenotypes including heterochromatin defects, R-loop formation and sensitivity to genotoxic agents. Indeed, cells lacking both Clr4 and Fft3 show severe growth defects, and clr4Δ cells show replication defects at loci that are similarly affected in fft3Δ. Consistent with the idea that clr4Δ might also accumulate R-loops, loss of H3K9me correlates with R-loop accumulation in C. elegans, but the exact role of heterochromatin in this process is unknown (Zeller et al., 2016). Notably, Clr4-dependent heterochromatin assembly precludes histone turnover across domains showing RNA-DNA hybrid formation (Aygun et al., 2013; Nakama et al., 2012). Thus, increased histone turnover in clr4Δ could allow nascent RNA to gain access to DNA to form R-loops. Although heterochromatin modifications are not detected at genes showing replication fork stalling, it is possible that heterochromatin assembly occurs transiently.
Our work has important implications for understanding how eukaryotic cells ensure stable propagation of gene expression patterns and protect genomic integrity through suppression of nucleosome turnover. In higher eukaryotes, low nucleosome turnover is observed at repressed regions such as Polycomb-associated sites (Deal et al., 2010), and it is possible that factors suppressing histone turnover promote propagation of the silenced chromatin state. Genomic regions that show defects in replication fork progression caused by R-loops or collision with secondary DNA structures are a major source of genome instability, and are often referred to as fragile sites (Helmrich et al., 2013). Importantly, mutations in SMARCAD1, the human homolog of Fft3, have been linked to genetic instability and various human diseases including cancer. Since modulating the DNA replication and repair process is a common tool for treatment of various cancers, a deeper understanding of the Fft3 family of chromatin remodelers may reveal therapeutic targets and provide a unique perspective on the roles of these factors in various chromosomal processes.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shiv Grewal (grewals@mail.nih.gov).
Experimental Model and Subject Details
Schizosaccharomyces pombe strains used in this study are derivatives of the standard laboratory strain 972 and are listed in Table S2
Method Details
Strains, Media and Plasmid Construction
Yeast strains used in this study are listed in Table S2. Standard techniques and media were used to culture S. pombe. Deletion strains and epitope-tagged strains were constructed using a standard PCR-based methodology. To construct the pclr4+ plasmid that is maintained at 3–4 copies per cell, a DNA fragment containing the full-length clr4+ gene under the control of its native promoter was cloned in the LEU2-marked pWH5 plasmid at the HindIII site. p-nmt1-rnh1+ was constructed by cloning the rnh1+ gene into the pREP3 plasmid carrying the nmt1 inducible promoter. The resulting plasmids were used to transform strains with leucine auxotrophy and colonies containing the plasmid were selected on leucine-depleted plates. PMG-Leu medium lacking thiamine was used to express rnh1+ under the control of nmt1 promoter. For the genotoxin sensitivity assay, cells were grown to mid-log phase in medium lacking leucine and then spotted onto non-selective medium or medium supplemented with 10mM hydroxyurea (HU), 2.5 mU/ml bleomycin or 0.01% methyl methanesulfonate (MMS). Plates were incubated at 26°C for 5–6 days before imaging.
Genetic Screen
Exponentially growing cells were mutagenized with EMS or UV (200J/m2) using a UV cross linker (Thermo Fisher Scientific). Colonies were screened for loss of silencing of the ura4+ reporter inserted at the mat locus lacking the cenH nucleation region (KΔ::ura4+). Mutants affecting the silencing of KΔ::ura4+ but not of Kint2::ura4+ were isolated and backcrossed three times to a non-mutagenized strain. Genomic DNA isolated from three wild type and three mutant segregants from the final cross was subjected to whole genome sequencing using the HiSeq platform (Illumina). The mutations were identified using bioinformatic analysis. The mutations were further verified using conventional PCR-based Sanger sequencing.
ChIP and ChIP-chip
For H3K9me ChIP experiments, exponentially growing cells were fixed at room temperature with 3% paraformaldehyde. For ChIP analysis of Fft3-GFP protein, exponentially growing cells were shifted to 18°C for 6 h prior to fixation with 3% paraformaldehyde for 30 min, followed by cross-linking with 10 mM dimethyl adipimidate (Sigma-Aldrich) at room temperature for 45 min. Cells were resuspended in ChIP lysis buffer (50 mM HEPES/KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) supplemented with Roche Complete protease inhibitors and were disrupted by glass bead lysis (Sigma) using a Biospec Mini-Beadbeater-16. The whole cell crude extract was sonicated to shear the chromatin into 500–1000 bp fragments. Chromatin was cleared by centrifugation at 1500 x g for 10 min at 4°C. ChIPs were performed using anti-GFP (ab290, Abcam) or anti-H3K9me2 (Abcam) antibody (1–3 μg). Antibodies were recovered using Protein A: Protein G (1:1) bead slurry. Chromatin bound beads were washed with lysis buffer followed by two washes with high-salt lysis buffer (50 mM HEPES/KOH, pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate), and once each with wash buffer (10 mM Tris/HCl pH 8, 250 mM LiCl, 0.5% NP40, 0.5% sodium deoxycholate and 1 mM EDTA) and TE buffer. Immunoprecipitated DNA was eluted with TE supplemented with 1% SDS buffer and reverse cross-linked at 65°C overnight. The recovered immunoprecipitated chromatin fraction was purified using a QIAquick PCR purification kit (QIAGEN) after treatment with RNase A (5 μg) and Proteinase K (40 μg) for 2 h each at 37°C. Immunoprecipitated DNA and input DNA were analyzed by performing real-time PCR using iTaq™ Universal SYBR® Green Supermix (BioRad), or processed for microarray analysis (ChIP-chip). Immunoprecipitated DNA and input DNA were labeled by amminoallyl-dUTP in random-primed PCR. Aminoallyl-labeled DNA was conjugated with Cy5(immunoprecipitated DNA) or Cy3(input DNA) and equal amounts of Cy5- and Cy3-labeled DNA were hybridized to a custom Agilent 4x44K 60-mer oligonucleotide array spanning the whole genome at 300 bp intervals. Hybridization, wash and array scan followed Agilent’s recommended procedure. After scanning on an Agilent SureScan Microarray scanner, the Cy3 and Cy5 signals were extracted by the Agilent Feature Extraction protocol. Ratios of Lowes normalized signals were used to calculate enrichment values for each 60-mer. Enrichment values above 2 were replaced by average enrichments of its two neighbors unless at least one neighboring probe exceeded value of 2.
Purification and Co-Immunoprecipitation
Cells expressing untagged or GFP tagged Fft3 (Fft3-GFP) were synchronized in S phase using the temperature-sensitive cdc25-22 allele. Protein purification was performed as previously reported (Sugiyama et al., 2007) with some modifications. Briefly, cells harvested from 4 L cultures were flash frozen in liquid nitrogen and then crude cell lysates were prepared using a house-hold blender. The cell lysates were cleared by centrifugation, and subjected to affinity-purification on anti-GFP agarose beads, GFP-Trap® A (antibodies-online) or anti-Flag M2 affinity gel (Sigma). Beads were extensively washed, and purified proteins were eluted three times with 0.2 M glycine (pH 2.0). Eluted proteins were precipitated with trichloroacetic acid and resolved on a 4–12% Bis-Tris gel (Thermo Fisher Scientific). Proteins were visualized using SimplyBlue™ SafeStain (Thermo Fisher Scientific). The protein search was performed against the UnitPro Schizosaccharomyces pombe database from the European Bioinformatics Institute.
RT-PCR
Total RNA was isolated using the MasterPure™ Yeast RNA Purification Kit (Epicentre) according to the manufacturer’s instructions. Gene-specific and strand-specific cDNA was synthesized from DNase treated total RNA using the SuperScript-III Reverse Transcriptase (Invitrogen) and expression was analyzed by performing real-time PCR using iTaq™ Universal SYBR® Green Supermix (BioRad).
Nucleosome Turnover Assay
The nucleosome turnover assay was performed as previously reported (Aygun et al., 2013) except that sonication was used to fragment DNA instead of MNase treatment. Briefly, cells were grown to OD595 0.1–0.15 in EMM-Leu + 8% glucose media and were synchronized by adding 15 mM HU for 4 h. Cells were pelleted and washed twice with EMM-Leu-glucose + 4% sucrose media containing 15mM HU and then allowed to grow for 2 h to induce the expression of H3-Flag. Cells were cross-linked with 1% formaldehyde at room temperature for 20 min. Cells were lysed by bead beating in ChIP lysis buffer (50 mM HEPES/KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% DOC) and the chromatin fraction was recovered and fragmented to 500–1,000 bp by sonication. Chromatin was cleared by centrifugation at 1500 x g, 10 min at 4°C and immunoprecipitated using a 20 μl bed volume of pre-washed anti-FLAG M2 agarose beads (Sigma) for 4 h at 4°C with slow rotation. Beads containing the chromatin fraction were washed extensively and the immunoprecipitated DNA was recovered by reverse cross-linking at 65°C, as described for the ChIP procedure. Input and immunoprecipitated DNA were purified using the QIAquick-PCR purification kit (QIAGEN). H3-Flag immunoprecipitated DNA and input DNA were processed as in the ChIP-chip procedure and analyzed on a custom Agilent 4x44K array tiling a large portion of chromosome 2 (chr2:1,000,000–3,049820 and chr2:4,491,831–4,541,531) at 50 bp resolution. Enrichments were processed as in ChIP-chip except the values were subjected to a sliding window filter which replaced every enrichment value exceeding more than twice or falling below 50% of average enrichment of its two neighbors by the average enrichment of the neighboring positions. Additionally, the enrichments were subjected to a 500 bp averaging window. When appropriate positions corresponding to deleted regions of the mating type locus (REIIΔ or KΔ) were excluded from the analysis, as were positions corresponding to sequence shared by mat1M and mat3M loci.
Flow Cytometry
Mid-log phase cells were fixed in 70% (v/v) cold ethanol. Cells were arrested in early S phase by treatment with 12 mM HU for 4 h, washed twice with 50 mM sodium citrate buffer and treated with 0.1 mg/ml RNase A at 37°C for 2 h. Cells were stained with 1μM SYTOX Green (Invitrogen) at room temperature and the DNA content was measured using a FACS Calibur flow cytometer (BD Biosciences).
Microscopy and Image Analysis
To quantify Rad11-GFP and Rad52-GFP foci, mid-log phase cells were collected and imaged on a glass slide using DeltaVision Elite microscope (GE Healthcare) with a 100x 1.4NA Plan Super Apochromat oil lens. Multiple z sections 0.3μm thickness were acquired and subsequently deconvoluted using SoftWoRX 6.0 (GE Healthcare).
BrdU Incorporation Assay and Analysis
The BrdU incorporation assay was performed as previously described (Hayashi et al., 2007; Zofall et al., 2016). Cells carrying the thymidine kinase expression module Pnmt1-TK and the nucleoside transporter module were synchronized using the cdc25-22 temperature sensitive allele. Cells were grown in EMM media to OD595 0.1–0.2 and were blocked at the G2/M boundary by shifting to 36°C for 4 h 15 min. Cells were released from the block by lowering the temperature to 26°C in the presence of 200 μM 5-bromo-2′-deoxyuridine (BrdU) and 10 mM HU. Cells grown for 120 min in the presence of BrdU and HU were collected and fixed with 0.1% sodium azide (w/v). After washing with PBS, cells were lysed using glass beads in lysis buffer (10 mM Tris/HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 2% Triton X-100, and 1% SDS) combined with phenol-chloroform. DNA was recovered from the aqueouS phase by ethanol precipitation and was fragmented by sonication and treated with 2 μg RNase A. DNA was purified using phenol-chloroform and then subjected to overnight immunoprecipitation at 4°C with mouse anti-BrdU antibody (BD Pharmingen) coupled to Dyna1 anti-mouse IgG magnetic beads (Invitrogen). Immunoprecipitated DNA was washed and recovered as described in the ChIP procedure. Immunoprecipitated and input DNA were random-prime PCR amplified and analyzed on Agilent 4x44K microarrays as described for ChIP-chip. A sliding window filter was applied for noise reduction, and enrichment data were smoothed using the unweighted sliding average method over intervals corresponding to the DNA fragment size after sonication (~1kb). BrdU incorporation was analyzed for a total of 67 Fft3 enriched loci in wild type and mutant strains. The nearest annotated origin of replication upstream (relative to the direction of replication) of the local maximum of the BrdU trace within a target region was identified. BrdU and Fft3 signal from the region running from the position 2000 base pairs upstream to 20,000 base pairs downstream of this origin was then used for composite trace construction. A spline curve (R function smooth.spline) (https://www.r-project.org/) was fit to each individual BrdU incorporation trace and each Fft3-GFP occupancy trace to allow for interpolation of a signal for every trace at every base. An average spline was subsequently fit to these interpolated values to produce the composite trace. The resulting composite BrdU traces were plotted as curves with 1 standard deviation envelopes while the Fft3-GFP traces are represented below as heatmap strips. For each strain, separate composite traces were computed for chromosomes I, II, and III.
DRIP Assay
DNA/RNA hybrid immunoprecipitation (DRIP) was performed as previously described (Ginno et al., 2012) with some modifications. cdc25-22 cells were synchronized by blocking at the G2/M boundary by shifting to 36°C for 4 h 15 min and were released by shifting to 26°C. Cells corresponding to maximum septation were collected and treated with 0.1% sodium azide (w/v). Cells were lysed by 2.5 mg/ml Zymolyase-100T treatment at 37°C for 40 min in PEMS buffer (100 mM PIPES/KOH pH 6.9, 1 mM EGTA, 1 mM MgSO4 and 1.2 M sorbitol). The spheroplasts were resuspended in TE buffer supplemented with 1% SDS and incubated at 65°C for 10 min. The chromatin fraction was isolated by adding 175 μl of 5 M potassium acetate followed by a 5 min incubation on ice and centrifugation at 16 k x g for 10 min at 4°C. Chromatin was precipitated with an equal volume of isopropanol and the pellet was resuspended in TE and treated with 2 μg of RNase A for 30 min followed by 100 μg of Proteinase K for 30 min at 65°C. DNA was gently purified with phenol:chloroform (1:1) extraction followed by ethanol precipitation in the presence of 30 μl of 3 M sodium acetate. Purified DNA dissolved in TE was digested with 50 U of HindIII, EcoRI, BsrGI, XbaI and SspI with 2.5 μg BSA at 37°C overnight. As a negative control, half of the mix was treated with 3 μl of RNase H (NEB) overnight at 37°C. DNA was phenol/chloroform extracted and immunoprecipitated overnight with 10 μl of S9.6 antibody (1 mg/ml) in 500 μl of binding buffer (10 mM Na2HPO4, 140 mM NaCl, 0.05% Triton X-100) at 4°C. The immunoprecipitated DNA was recovered using Dynabeads Protein A (Invitrogen) and washed three times with binding buffer. DNA was eluted in 100 μl of elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 0.5% SDS) followed by treatment with 7 μl of Proteinase K (20 mg/ml) for 45 min at 55°C. Immunoprecipitated DNA was purified using the QIAquick-MinElute PCR Purification kit (QIAGEN).
Quantification and Statistical Analysis
Statistical parameters are reported in the Figures and Figure Legends.
Data and Software Availability
Accession number for genome-wide datasets deposited in NCBI GEO under the reference number GEO: GSE88904. Accession link for original datasets deposited in Mendeley Data: http://dx.doi.org/10.17632/srw6sb93by.1
Supplementary Material
Highlights.
SMARCAD1 homolog Fft3 suppresses histone turnover to promote epigenetic inheritance
Fft3 facilitates epigenetic transmission of heterochromatin in dividing cells
Stabilization of nucleosomes at genes prevents R-loop replication barriers
Clr4/Suv39h suppresses R-loop accumulation and silencing defects in fft3Δ cells
Acknowledgments
We thank Hisao Masukata and Julie Cooper for sharing strains and Stephen Leppla for sharing S9.6 antibody. We also thank Jothy Dhakshnamoorthy, H. Diego Folco and Takeshi Mizuguchi for help with experiments and strain/plasmid constructions. We thank Jemima Barrowman for editing the manuscript, and members of the S.I.S.G. lab for helpful suggestions. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
AUTHOR CONTRIBUTIONS
N.T. and S.I.S.G. designed the experiments; N.T., V.B., T.S. Mi.Z. performed the experiments; Ma.Z., G.T. and D.W. performed analyses of genome-wide datasets; all authors contributed to data analysis; and N.T. and S.I.S.G. wrote the manuscript.
Supplemental Information including seven figures and three tables can be found with this article online
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Sady B, Madhani HD, Narlikar GJ. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol Cell. 2013;51:80–91. doi: 10.1016/j.molcel.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012;13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- Audergon PN, Catania S, Kagansky A, Tong P, Shukla M, Pidoux AL, Allshire RC. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science. 2015;348:132–135. doi: 10.1126/science.1260638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygun O, Mehta S, Grewal SI. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat Struct Mol Biol. 2013;20:547–554. doi: 10.1038/nsmb.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon B, Wang W, Barski A, Ranallo RT, Bao K, Schones DE, Zhao K, Wu C, Wu WH. The ATP-dependent chromatin remodeling enzyme Fun30 represses transcription by sliding promoter-proximal nucleosomes. J Biol Chem. 2013;288:23182–23193. doi: 10.1074/jbc.M113.471979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- Chen H, Symington LS. Overcoming the chromatin barrier to end resection. Cell Res. 2013;23:317–319. doi: 10.1038/cr.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer KM, Job G, Shanker S, Neale GA, Lin YC, Bartholomew B, Partridge JF. The Mi-2 homolog Mit1 actively positions nucleosomes within heterochromatin to suppress transcription. Mol Cell Biol. 2014;34:2046–2061. doi: 10.1128/MCB.01609-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohke K, Miyazaki S, Tanaka K, Urano T, Grewal SI, Murakami Y. Fission yeast chromatin assembly factor 1 assists in the replication-coupled maintenance of heterochromatin. Genes Cells. 2008;13:1027–1043. doi: 10.1111/j.1365-2443.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Ruusala T. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics. 1994;136:53–64. doi: 10.1093/genetics/136.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SI. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci U S A. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco AA, Kaufman PD. Histone deposition proteins: links between the DNA replication machinery and epigenetic gene silencing. Cold Spring Harb Symp Quant Biol. 2004;69:201–208. doi: 10.1101/sqb.2004.69.201. [DOI] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Klar AJ. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Klar AJ. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics. 1997;146:1221–1238. doi: 10.1093/genetics/146.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Katou Y, Itoh T, Tazumi A, Yamada Y, Takahashi T, Nakagawa T, Shirahige K, Masukata H. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J. 2007;26:1327–1339. doi: 10.1038/sj.emboj.7601585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Nudler E, Tora L. Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol. 2013;20:412–418. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH. An antibody-based microarray assay for small RNA detection. Nucleic Acids Res. 2006;34:e52. doi: 10.1093/nar/gkl142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V, Chalkley R. Histone segregation on replicating chromatin. Biochemistry. 1985;24:6930–6938. doi: 10.1021/bi00345a027. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Rando OJ. Chromatin as a potential carrier of heritable information. Curr Opin Cell Biol. 2010;22:284–290. doi: 10.1016/j.ceb.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WJ, Lee S, Park MS, Jang YK, Kim JB, Park SD. Rad22 protein, a rad52 homologue in Schizosaccharomyces pombe, binds to DNA double-strand breaks. J Biol Chem. 2000;275:35607–35611. doi: 10.1074/jbc.M007060200. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells. 2012;17:218–233. doi: 10.1111/j.1365-2443.2012.01583.x. [DOI] [PubMed] [Google Scholar]
- Nakayama JII, Klar AJS, Grewal SIS. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- Neves-Costa A, Will WR, Vetter AT, Miller JR, Varga-Weisz P. The SNF2-family member Fun30 promotes gene silencing in heterochromatic loci. PLoS One. 2009;4:e8111. doi: 10.1371/journal.pone.0008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Parker AE, Clyne RK, Carr AM, Kelly TJ. The Schizosaccharomyces pombe rad11+ gene encodes the large subunit of replication protein A. Mol Cell Biol. 1997;17:2381–2390. doi: 10.1128/mcb.17.5.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie VJ, Wuitschick JD, Givens CD, Kosinski AM, Partridge JF. RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast. Mol Cell Biol. 2005;25:2331–2346. doi: 10.1128/MCB.25.6.2331-2346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352:aad9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- Ragunathan K, Jih G, Moazed D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2015;348:1258699. doi: 10.1126/science.1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Winston F. Chromatin and transcription in yeast. Genetics. 2012;190:351–387. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. R loops and links to human disease. J Mol Biol epub. 2016 doi: 10.1016/j.jmb.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Rowbotham SP, Barki L, Neves-Costa A, Santos F, Dean W, Hawkes N, Choudhary P, Will WR, Webster J, Oxley D, et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell. 2011;42:285–296. doi: 10.1016/j.molcel.2011.02.036. [DOI] [PubMed] [Google Scholar]
- Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich B, Stralfors A, Khorosjutina O, Persson J, Smialowska A, Javerzat JP, Ekwall K. The Fun30 chromatin remodeler Fft3 controls nuclear organization and chromatin structure of insulators and subtelomeres in fission yeast. PLoS Genet. 2015;11:e1005101. doi: 10.1371/journal.pgen.1005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stralfors A, Walfridsson J, Bhuiyan H, Ekwall K. The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet. 2011;7:e1001334. doi: 10.1371/journal.pgen.1001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Thon G, Cohen A, Klar AJ. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics. 1994;138:29–38. doi: 10.1093/genetics/138.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol Cell Biol. 1989;9:609–619. doi: 10.1128/mcb.9.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat Struct Mol Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- Yamane K, Mizuguchi T, Cui B, Zofall M, Noma K, Grewal SI. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell. 2011;41:56–66. doi: 10.1016/j.molcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P, Padeken J, van Schendel R, Kalck V, Tijsterman M, Gasser SM. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat Genet. 2016;48:1385–1395. doi: 10.1038/ng.3672. [DOI] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SIS. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- Zofall M, Smith DR, Mizuguchi T, Dhakshnamoorthy J, Grewal SI. Taz1-Shelterin promotes facultative heterochromatin assembly at chromosome-internal sites containing late replication origins. Mol Cell. 2016;62:862–874. doi: 10.1016/j.molcel.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.