SUMMARY
Synthetic biology is increasingly used to develop sophisticated living devices for basic and applied research. Many of these genetic devices are engineered using multi-copy plasmids, but as the field progresses from proof-of-principle demonstrations to practical applications, it is important to develop single-copy synthetic modules that minimize consumption of cellular resources and can be stably maintained as genomic integrants. Here we use empirical design, mathematical modeling and iterative construction and testing to build single-copy, bistable toggle switches with improved performance and reduced metabolic load that can be stably integrated into the host genome. Deterministic and stochastic models led us to focus on basal transcription to optimize circuit performance and helped to explain the resulting circuit robustness across a large range of component expression levels. The design parameters developed here provide important guidance for future efforts to convert functional multi-copy gene circuits into optimized single-copy circuits for practical, real-world use.
INTRODUCTION
Multi-copy plasmids are the workhorses of genetic engineering and are commonly used by both microbes in the environment and biologists in the lab to express and exchange genetic information. Microbes use plasmids to acquire genetic components including virulence factors and antibiotic resistance cassettes that provide an evolutionary advantage in a particular environmental niche (Davies and Davies, 2010; Norman et al., 2009). These plasmids serve as a common vehicle for horizontal gene transfer to surrounding microbes, but genetic components that are particularly useful are often integrated into the genome to increase their stability and reduce the metabolic burden of plasmid maintenance (Bergstrom et al., 2000; Davison, 1999; Ochman et al., 2000; Rankin et al., 2011). In a broadly similar approach, basic and applied biologists rely on multi-copy plasmids to construct and test genetic components for a wide range of applications, from simple gene expression in a target organism to complex gene circuit design for industrial or therapeutic use. Synthetic biologists and metabolic engineers commonly use an iterative “design-build-test” approach that relies on the ease of multi-copy plasmid construction and purification to generate complex genetic circuits (Cameron et al., 2014; Keasling, 1999; Khalil and Collins, 2010; Lee et al., 2012). As these applied-biology fields progress from proof-of-principle demonstrations to practical applications, however, these circuits must also be converted into single-copy synthetic modules that minimize resource consumption and can be stably integrated into the genome to minimize the possibility of horizontal gene transfer.
As both natural and applied systems must transfer genetic elements from multi-copy plasmids to single-copy genomic integrants, it is important to understand the design parameters that enable proper regulation and expression of these genetic components during this process. In this study, we use the conversion of a multi-copy genetic toggle switch to an optimized single-copy circuit as a case study to examine the design principles that affect circuit performance. We use empirical design and iterative construction and testing to build a bistable, genome-integrated toggle switch with reduced growth burden. To identify regulatory elements that are important for circuit function, we test a series of control mechanisms including promoter-level transcriptional control (repressor binding strength and operator site location and number), post-transcriptional control (5′ untranslated region (UTR)), and translational control (ribosome binding site (RBS) strength). In parallel to this empirical approach, we use deterministic and stochastic models to identify important design parameters that affect circuit function and stability, and we use these models to explain the robustness of the optimized toggle switch to translation-based perturbations. Finally, to demonstrate the practical application of the optimized circuit, we use the single-copy toggle switch to build a genome-integrated “kill switch” that provides exogenous control of cell viability with minimal metabolic load that is highly stable. This case study, and the design parameters that we identify, offer fundamental guidance for future efforts to convert multi-copy gene circuits into functional single-copy circuits, and provide a window into the intricate adaptations that microbes must make as they incorporate plasmid-borne genes into their genome.
RESULTS AND DISCUSSION
Conversion of the multi-copy toggle switch to single-copy
To identify the design parameters important for single-copy genetic circuit conversion, we chose to study a multi-copy genetic toggle switch that uses reciprocal regulation of the transcriptional repressors LacI and TetR to generate a bistable system (Cameron and Collins, 2014; Gardner et al., 2000; Kobayashi et al., 2004; Litcofsky et al., 2012) (Figure 1A). The toggle remains in its designated state in the absence of any exogenous input, but can be switched to the opposite state with the small molecule inducers anhydrotetracycline (ATc) or isopropyl β-D-1-thiogalactopyranoside (IPTG), which regulate TetR and LacI, respectively. To clearly identify each toggle state in the single-copy circuit, we first modified the RBS of both the GFP and mCherry reporters to increase their induced expression levels (Figures 1B and S1). Using control cells to define the fluorescence threshold for each toggle state (Figure S1), we quantified circuit stability in both the LacI+ and TetR+ states and found that the circuit was only stable in the TetR+ state (red state) (Figure 1C). Upon removal of the ATc used to induce the LacI+ state (green state), LacI repression of TetR and mCherry diminished rapidly, allowing TetR to quickly repress LacI and GFP to switch the toggle state back to the TetR+ state (Figure 1C). Earlier work by Ying et al. to create a single-copy toggle switch also resulted in circuit monostability in the absence of chemical inducers (Ying et al., 2010), further demonstrating the need for specific design principles to produce functional, single-copy synthetic circuits.
Figure 1. Initial conversion of the multi-copy toggle switch to single-copy.
(A) Schematic illustration of the toggle switch. Reciprocal transcriptional repressions by LacI and TetR form coupled feedback loops that enable bistability.
(B) FACS histograms of E. coli with no plasmid (null), the single-copy toggle switch (SC), the single-copy toggle switch with enhanced GFP and mCherry expression (SC enhanced), and the multi-copy toggle switch (MC). Cells were induced with ATc (Green state, left) or IPTG (Red state, right).
(C) Time course of toggle stability. The multi-copy toggle (MC) and single-copy toggle (SC enhanced) were pushed into the green or red states with ATc or IPTG, respectively, and the percentage of cells that remained in that state was measured by FACS at each time point. The multi-copy toggle is bistable, while the single-copy toggle is only stable in the red state. Data represent the mean ± SD of three replicates.
(D) Nullcline analysis of the mathematical model reveals that the toggle switch should be bistable if ε < ε* where ε* is given in Supplemental procedure 3. The heat map represents this critical value ε*, so that a toggle with ε values below the threshold is bistable with given parameters of μ and k .
(E–F) Reduction of the plasmid copy number from n = 50 (E) to n = 1 (F) lowers μ, reshaping the region of bistability (shaded region). Notably, the regions of bistability in (E) and (F) are slices from the heat map that correspond to two different μ values in (D). At the indicated parameter combination (dot), the toggle switch is predicted to lose bistability when transferred to a single-copy plasmid. See also Figure S1 and Table S3.
Mathematical modeling to recover toggle switch bistability
To better understand single-copy toggle switches and recover circuit bistability, we developed a mechanistic model of toggle switch dynamics (see Experimental procedures and Supplemental procedures 1–2). This model is derived from a biochemical rate equation formulation of gene expression with parameters including the plasmid copy number n, the dissociation constants k of the repressors to their respective promoters, the lumped constant α encompassing the transcription and translation rate constants, the decay rate constant δ of the repressors, and ε characterizing the leakage of the promoters (ε = 0 when the promoter is tightly repressed, and ε = 1 if the promoter is not repressed at all by its repressor) (see Experimental procedures and Supplemental procedures 1–2). In this model, the equilibria of the toggle switch can be characterized in a high-dimensional parameter space using only numerical methods, and as a result, this approach provides little guidance to recover bistability (see Supplemental procedure 3). To enable the analytical characterization of bistability, we constrained the model to symmetric toggle switches, thereby limiting the parameter space to three dimensions (k, μ, ε), where μ = nα/δ is the steady-state concentration of the unrepressed proteins (see Supplemental procedure 3). In particular, the toggle switch is bistable if and only if
Further analysis yields that the above condition holds true if and only if the leakage ε is smaller than the critical threshold ε*, plotted in Figure 1D (the expression of ε* is given in Supplemental procedure 3). The value of μ decreases with plasmid copy number in this model, reshaping the region of bistability to render the single-copy toggle switch monostable (Figures 1D–F, detailed in Supplemental procedure 4). As a result, while the pair (k, ε) lies in the region of bistability for the multi-copy toggle (Figure 1E), it lies outside in the single-copy case (Figure 1F). Furthermore, the model predicts that bistability can be recovered by increasing TetR and LacI expression to restore μ to its original value and by decreasing the promoter leakage ε (see Supplemental procedure 4).
Rebalanced regulator expression in the single-copy toggle
To enhance LacI and TetR expression on the single-copy plasmid, we mutagenized the RBS for each gene and used C-terminal mCherry fusions to LacI and TetR to measure their expression. This approach enabled us to bring induced TetR expression levels up to that seen in the multi-copy plasmid (Figure 2A and Supplemental procedure 5), but additional mutagenesis of the LacI promoter and 5′ UTR was required to achieve LacI expression levels equivalent to the multi-copy plasmid (Figures 2B, S2 and Supplemental procedure 5). To determine if increased LacI and TetR expression, and the resulting restoration of μ in our mechanistic model, were sufficient to recreate the bistability seen in the multi-copy plasmid, several combinations of LacI and TetR expression variants were tested on the single-copy plasmid. A majority of the circuits remained monostable in the red state, and the high LacI expression variant (L5) shifted the monostability to the green state, but none of the combinations displayed the desired bistability (Figures 2C–D and Table S1). In particular, the LacI and TetR expression variants that most closely recapitulated the multi-copy expression level (L5-T3) did not produce a bistable circuit (Figure 2E).
Figure 2. Rebalanced regulator expression in the single-copy toggle.
(A) Increased TetR was achieved by RBS modification using rational design (T1, T2, and T4) and random mutagenesis (T3). TM shows TetR expression in the original multi-copy plasmid, and TS shows its expression when moved to the single-copy plasmid.
(B) Increased LacI expression was achieved by stepwise modification of its RBS (L1 and L3), promoter (L2), and 5′ untranslated region (L4-L6). LM and LS indicate the unmodified TetR expression in the multi-copy and single-copy plasmids, respectively.
(C) TetR and LacI expression variants from (A) and (B) were paired and tested for bistability in the single-copy toggle. Toggle stability was measured 24 hours after removal of IPTG or ATc for the red or green state, respectively. The box indicates the variant that most closely recapitulated the multi-copy expression level (L5-T3).
(D) Enhanced LacI expression in the L5-TS toggle changed its stability to monostable in the green state. LS-TS indicates the unmodified single-copy toggle.
(E) LacI and TetR expression levels that recapitulate their multi-copy expression (L5-T3) do not provide bistability to the single-copy toggle. The circuit is monostable in the red state. LM-TM indicates the unmodified multi-copy toggle. Data represent the mean ± SD of three replicates. See also Figure S2 and Tables S1 and S3.
Tightened transcriptional repression to recover circuit bistability
Our mechanistic model of the circuit predicts that reduced leakage ε will increase toggle switch bistability, yielding greater robustness to parameter variations (Supplemental procedure 6), and a complementary stochastic model of circuit function similarly predicts that tight transcriptional control plays a fundamental role in providing robustness to noise in single-copy circuits (Supplemental procedure 7). We therefore focused our attention on reducing the basal expression of LacI and TetR in their repressed state. We first sought to minimize leaky transcription from the PTrc2 promoter driving TetR expression since unwanted TetR expression was the likely cause of LacI+/GFP+ instability in nearly all of the single-copy toggle variants. Mutations in LacI that increase affinity to its native O1 operator site did not improve toggle bistability (Figure S3), so we next sought to reduce basal transcription by replacing the O1 operator site with the palindromic operator site Oid that binds LacI more strongly (Sadler et al., 1983) (Figure 3A). As seen in Figure 3B, real-time PCR (RT-qPCR) analysis of tetR transcription showed that the Oid operator significantly lowered basal tetR transcription, and inclusion of additional Oid sites upstream of the PTrc2 −35 element caused a further reduction in tetR mRNA levels. To determine if the lowered tetR mRNA levels would affect circuit stability, we cloned the improved promoters PT2, PT3 and PT4 into the L6-T4 single-copy toggle and measured circuit bistability. As seen in Figure 3C, toggles containing PT2, PT3 and PT4 promoters displayed strong bistability for over 72 hours. As PT3 provided equivalent stability to PT4, we chose PT3 for continued circuit development.
Figure 3. Tightened transcriptional repression provides bistability to the single-copy toggle.
(A) The O1 LacI operator in PTrc2 was replaced with the Oid operator to create the PT1 promoter. Additional Oid operators were included to create PT2, PT3 and PT4. Open circles indicate the −35 and −10 elements.
(B) RT-qPCR was used to measure tetR transcription from PTrc2 and PT1-PT4 in the L6-T4 single-copy plasmid. The promoters were tested in the presence of ATc and 4 hours after its removal.
(C) Promoters PT2, PT3 and PT4 were cloned into the L6-T4 single-copy toggle and tested for bistability over 72 hours.
(D) A range of LacI and TetR expression variants were tested for bistability in the single-copy toggle containing PT3. A wide-range of expression variants showed bistability (compare to Figure 2C).
(E) Genomic integration of the L6-PT3T4 toggle switch improved its stability compared to the single-copy plasmid. The flow cytometry distribution of GFP and mCherry at 72 hours is shown on the right.
(F) Growth analysis of cells containing the genomically integrated L6-PT3T4 toggle. Cells containing the original multi-copy toggle switch (MC) or no plasmid are shown as controls, and growth was measured for cells held in the ATc-induced green state (left) or IPTG-induced red state (right). Data represent the mean ± SD of three replicates. See also Figures S3–S4 and Tables S2–S3.
With the clear role of transcriptional leakage ε in toggle bistability, we reexamined the range of LacI and TetR expression levels that maintain bistability in single-copy toggles with low promoter leakage. In contrast to toggle variants containing the original PTrc2 promoter (Figure 2C), ones containing the PT3 promoter achieved bistability across a wide range of unrepressed LacI and TetR expression levels (Figures 3D and S3B and Table S2), and long-term growth experiments confirmed that these toggles were bistable over 72 hours (Figure S3C). These experimental results confirm the mathematical analysis that decreased promoter leakage ε would move the toggle switch towards bistability, yielding greater robustness to parameter variations (deterministic model, Supplemental procedure 6) and greater robustness to noise (stochastic model, Figure S4 and Supplemental procedure 7). Interestingly, the model also predicts that increased repressor expression does not always increase bistability (Supplemental procedure 6), in agreement with experimental data in which increased LacI expression reduces circuit bistability when paired with low TetR expression (Figure 3D).
Genomic integration of the toggle switch and metabolic load analyses
Genomic integration allows removal of plasmid selection and replication systems that can place a high metabolic load on the host cell and contribute to horizontal gene transfer and the potential spread of antibiotic resistance (Bush and Fisher, 2011; Woodford et al., 2011). To determine if the single-copy toggle switch remained functional upon genomic integration, we tested toggle switch function and stability following integration in the lacZ locus in E. coli. As shown in Figure 3E, the integrated L6-PT3T4 toggle maintained state stability for more than 72 hours without antibiotic selection, as did several other single-copy toggle variants (Figure S3D). Furthermore, flow cytometry analysis of these cell populations showed monomodal distributions of cells in each state, confirming the population-wide stability of the circuits. Interestingly, the integrated single-copy toggle exhibited a narrowed distribution of fluorescence reporter expression compared to the single-copy plasmid, resulting in a measurable increase in circuit stability (Figure 3E). This is likely due to non-synchronized replication of the single-copy plasmid and the bacterial genome, a well-studied phenomenon that results in variable copy number of the plasmid in a subset of cells (Helmstetter et al., 1997).
As the field of synthetic biology moves from proof-of-concept construction to practical implementation, it is increasingly important to consider the physiological context of the engineered genetic circuits. To determine if the integrated toggle has a reduced physiological burden on the cell, we compared the growth profiles of cells containing the integrated L6-PT3T4 toggle switch to cells containing the parental multi-copy toggle switch. In contrast to the multi-copy plasmid that caused a clear growth defect, cells containing the integrated toggle displayed an identical growth rate to cells without the plasmid (Figure 3F).
Genetic kill switches using the integrated single-copy toggle
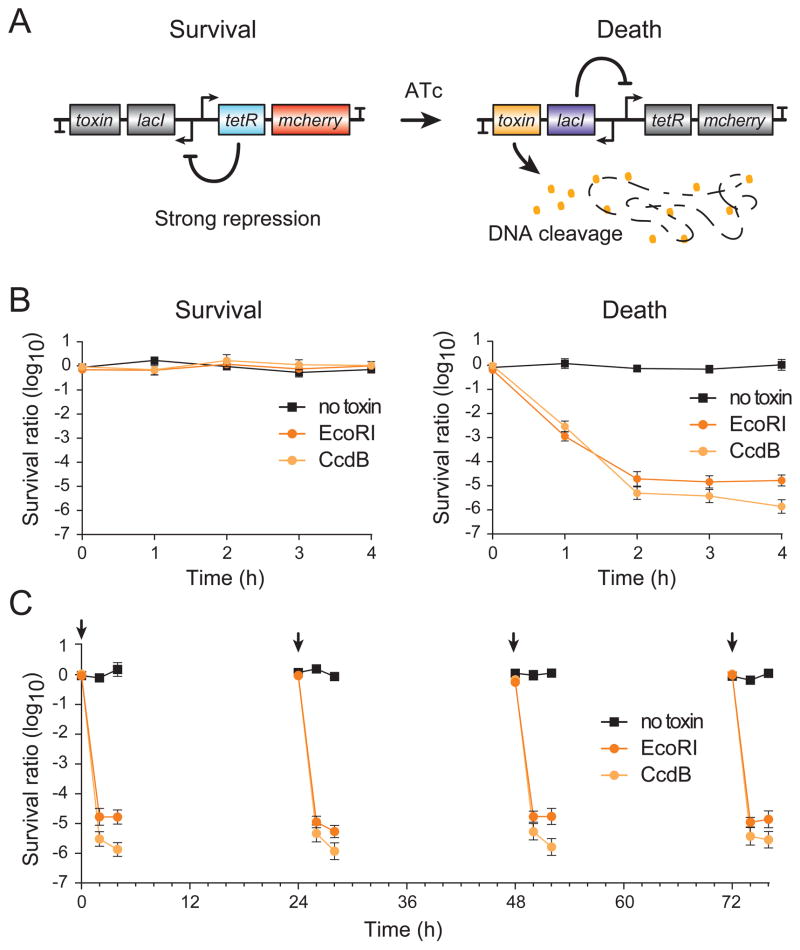
To demonstrate the stability and functionality of the genome-integrated toggle switch, we used the bistable toggle to develop a genetic kill switch in which toxin expression from the LacI+ state creates a ‘death’ state that kills the cell and the TetR+ state provides a ‘survival’ state in which the toxin is strongly repressed (Figure 4A). In this circuit design, cell viability provides a direct measure of circuit function, and the escapee rate provides a sensitive measure of circuit stability, as any leaky expression of toxins from the survival state provides a strong selective pressure for inactivating mutations (Chan et al., 2016). We cloned the DNA gyrase inhibitor CcdB (Smith and Maxwell, 2006) or the type II endonuclease EcoRI (Cheng et al., 1984), into the LacI+ state, and cells were exposed to IPTG to push them into the TetR+ survival state. Circuit bistability should allow these cells to remain viable following removal of IPTG, but ATc induction should push cells into the LacI+ death state where toxin expression causes cell death. As predicted, cells containing CcdB- or EcoRI-encoded toggle switches remained fully viable in the absence of IPTG (Figure 4B), but displayed > 4-log drop in viability within 2 hours after exposure to ATc (Figure 4C). To measure long-term circuit stability and robustness, we maintained the circuits in their survival state without inducer for 72 hours and periodically measured killing efficiency following ATc induction. The kill switches remained fully functional at each time point, displaying killing activity at 72 hours that was comparable with the killing activity at 0 hours (Figure 4C).
Figure 4. Development of genetic kill switches using the integrated single-copy toggle.
(A) Schematic of the kill switch in the ‘survival’ and ‘death’ states. TetR blocks LacI and toxin expression in the survival state, while LacI repression of TetR enables toxin expression and cell death in the death state.
(B) Kill switch stability and function was measured by colony forming units (CFUs) in the survival state following removal of IPTG (left) and in the death state following ATc induction (right). Kill switches containing the EcoRI or CcdB toxins do not affect cell viability in the survival state, but cause cell viability to drop to < 1×10−4 in the death state.
(C) Long-term robustness test for the CcdB and EcoRI kill switches. Cells containing the indicated kill switches were passaged in the survival state for 72 hours, and sub-populations of these cells were periodically switched to the death state with ATc. Cells passaged for 72 hours displayed comparable kill kinetics to those tested at 0 hours. Data are represented as mean ± SD of three replicates.
Concluding remarks
In this case study for synthetic circuit development, we used rational design and empirical analysis to convert a multi-copy genetic toggle switch into a single-copy circuit that displays increased bistability and places minimal metabolic burden on the host cell. Deterministic and stochastic models and experimental tests revealed the importance of transcriptional leakage ε in low-copy number circuits, as reduced basal TetR transcription improved the circuit’s robustness to parameter variation and noise. Conversion of existing multi-copy circuits into single-copy genetic networks will enable their integration into complex, multi-layered systems with minimized effect on the host cell’s viability and metabolic capacity. Additionally, the empirical strategies we developed to improve gene expression and reduce transcriptional leakage provide valuable design criteria for single-copy circuit construction. Strong agreement between the mathematical theory and the empirical results suggests that this forward-engineering approach will aid the design and construction of single-copy, genome-integrated genetic circuits.
EXPERIMENTAL PROCEDURES
Media and chemicals
Toggle switch tests were performed in M9 minimal media (Fisher Scientific; 6.8 g/L Na2PO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl) with 5 g/L glucose (Sigma) and 5 g/L casamino acids (Sigma), using appropriate antibiotics and inducers at the following concentrations: ampicillin (50 μg/ml), chloramphenicol (10 μg/mL), kanamycin (50 μg/mL), IPTG (1 mM), ATc (50 ng/mL), and arabinose (1 mg/mL). Luria-Bertani (LB) media (Fisher Scientific; 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) was used for cell growth of E. coli cloning strains. All cloning was conducted using E. coli EPI300 (Epicentre; F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ (ara, leu)7697 galU galK λ− rpsL (StrR) nupG trfA dhfr), and E. coli MG1655 ΔlacI was used for all toggle tests and expression level quantification.
Cell growth
Cells were inoculated from single colonies on LB agar plates and grown overnight in LB medium at 37°C and 300 rpm. Overnight cultures were diluted 200-fold into 96-well plates containing M9 medium as defined above with either IPTG or ATc and cultured for 12 h at 37°C and 900 rpm. Cells were washed once with M9 media and then serially passaged in M9 media without IPTG or ATc by 200-fold dilution every 12 h. Cell growth was monitored by measuring optical density at 600 nm (OD600), and all measurements were performed in 200 μl in 96-well, flat-bottom plates.
Single-copy plasmid construction
All plasmids were constructed using established molecular cloning techniques (Maniatis et al., 1982) and Gibson assembly (Gibson et al., 2009). To construct the single-copy toggle switches, toggle elements in pKDL071 (Litcofsky et al., 2012) were cloned into the conditionally amplifiable, single-copy plasmid pBAC/OriV (Wild et al., 2002) to make pBAC-SC and pBAC-SCenhanced (GenBank accession numbers KX264176 and KX264177). Plasmid pBAC-TS (GenBank accession number KX264178) was then generated to enable further modification to the promoter and RBS regions. Changes to genetic parts in this plasmid are listed in Table S3. Automated translation rate estimation (Salis, 2011; Salis et al., 2009) and site-saturation mutagenesis were used to enhance LacI, TetR, GFP, and mCherry expression in the single-copy toggle switch. In particular, LacI expression was improved by modifying its PLtetO promoter (Kincade and deHaseth, 1991; Lutz and Bujard, 1997) to include point mutations in the −10 site and the insertion of 5–10 random nucleotides around the 5′ UTR (Table S3). To minimize basal tetR expression from the PTrc2 promoter, the lacI-binding operator O1 was replaced by an Oid operator (Sadler et al., 1983). Further minimization of the basal level expression was achieved by including up to four Oid operators as listed in Table S3. Restriction enzymes, T4 DNA Ligase, Phusion PCR kits and Gibson Assembly Mix from New England BioLabs were used for cloning, and custom oligo DNAs were synthesized by Integrated DNA Technologies (Coralville, IA). Genewiz (Cambridge, MA) and Quintara Biosciences (Boston, MA) were used for sequencing analysis.
Quantification of LacI and TetR in vivo
C-terminal mCherry fusions to LacI and TetR were used to quantify LacI and TetR expression levels (pBAC-LC and pBAC-TC, respectively; GenBank accession numbers KT893256 and KT893257) (Chan et al., 2016). These backbone vectors were used to measure the expression of all LacI and TetR expression variants as listed in Table S3. A SpectraMax M5 microplate reader (Molecular Devices) was used to measure GFP and mCherry fluorescence with excitation/emission wavelengths of 488 nm/520 nm and 587 nm/610 nm and an emission filter cutoff at 515 nm and 610 nm, respectively.
Stability tests using flow cytometer measurements
The fluorescence distribution of each sample was measured using a BD FACSAriaII flow cytometer (BD Biosciences). For all FACS samples, cells were fixed in PBS containing 2% paraformaldehyde before analysis. At least 30,000 events were collected for each measurement, and FloJo (Treestar) was used for data analysis. Threshold values for GFP and mCherry were set based on the background fluorescence intensity of MG1655ΔlacI harboring a plasmid without GFP or mCherry. The stability of the toggle switch was then defined by calculating the percentage of cells with fluorescence values above the threshold value (Figure S1B).
RT-qPCR
To quantify basal tetR or lacI expression, we sampled cells in exponential growth (OD600 ~ 0.3) in the presence of IPTG or ATc. Cultures were stabilized with RNAprotect Bacteria Reagent (Qiagen), and total RNA was extracted using RNeasy Mini Kit (Qiagen). An RNase-free DNase Kit (Qiagen) was used to prevent possible DNA contamination, and total RNA was quantified using an ND-1000 NanoDrop spectrophotometer. Standard PCR was conducted using qPCR primers to identify any DNA contamination in the RNA samples, and the RNA samples were stored at −80°C. cDNA was synthesized from RNA samples using a Superscript III First Strand Synthesis kit (Invitrogen) and stored at −20°C. qPCR primers were designed using Prism3Plus software (Untergasser et al., 2007; Vega et al., 2012), and qPCR reactions were prepared according to the manufacturer’s instructions (LightCycler 480 SYBR Green I Master Kit, Roche Applied Science). A qPCR reaction without cDNA was used as a negative control, and five concentrations of lacI and tetR cDNA synthesized through in vitro transcription were included for each primer set in all qPCR operations. In vitro transcription was performed according to the manufacturer’s instructions (MEGAscript T7 Transcription Kit, Ambion Inc). The in vitro transcription cassette containing T7 promoter was constructed in pECJ3 (Cameron and Collins, 2014) by inserting the T7 promoter upstream of tetR or lacI, and the transcription cassettes for tetR or lacI were then amplified by PCR with P1 and P2 primers as follows: P1-tetR: AAACGTGGCTGGCCTGGTTCAC; P2-tetR: TTGATTCTCTCGAGACGGTCC; P1-lacI: TACAATGTAGGCTGCTCTAC; and P2-lacI: TCCAATTTGTGTCCAAGAATGT. These amplified DNA fragments were used as templates for in vitro transcription. qPCR reactions with primers P3 and P4 were prepared as follows: pre-incubation (95°C for 5 minutes) and 45 cycles of three-stage amplification (95°C for 10 sec, 58°C for 10 sec, and 72°C for 10 sec). The primer sequences for P3 and P4 are as follows: P3-tetR: ATTTAGGTACACGGCCTACA; P4-tetR: CTTGATGCTCTTGATCTTCC; P3-lacI: GACATCTCGGTAGTGGGATA; and P4-lacI: ACAGTTGATTGCCCTTCAC. After the thermal cycling reaction, a melting curve reaction (65°C ramped to 98°C, 10 sec at each 1°C interval) was conducted to detect non-specific amplification or primer-dimer formation. To calculate the fg value per 1 ng total RNA, standard lacI and tetR cDNA from in vitro transcription (5000, 1000, 200, 40, 8, 1.6, and 1 fg RNA per reaction) were analyzed using Roche LightCycler 480 software with default parameters for absolute quantification analysis. qPCR was performed using 4–6 biological replicates for each reported measurement.
Genomic integration
The conditional replicative plasmid pWM91 (Metcalf et al., 1996) was used for the genomic integration of the genetic toggle switches. This plasmid contains the R6Kγ origin, which only replicates in the presence of pir gene expression. E. coli SM10λ pir+ strain was used to replicate pWM91 and its derivatives. Toggle L6-PT3T4 (pBAC-L6-PT3T4; GenBank accession number KX264179) was inserted into a pWM91 derivative containing homologous regions to lacZ (pWM91-L6-PT3T4 (GenBank accession number KX264180)) and inserted into the lacZ locus according to previously described methods (Metcalf et al., 1996). Toggles L6-PT2T4 and L6-PT4T4 were inserted into the E. coli genome in the same manner using plasmids pWM91-L6-PT2T4 and pWM91-L6-PT4T4, respectively. To integrate kill switch variants of the toggle switch into the E. coli genome, ecoRI or ccdB along with cat, attHK022 and FRT sites were first cloned into pBAC-L6-PT3T4 to make pBAC-L6-PT3T4Ei or pBAC-L6-PT3T4Ci, respectively (GenBank accession numbers KX264181 and KX264182). These plasmids were then amplified by PCR to remove the replication origin using the following primers: P5: CTGGTGTCCCTGTTGATACC; P6: AGGCCAGAAAGCATAACCCGGGATCCTGGCCTGAATATTCTCTCTGG. After BamHI digestion, the DNA fragments were recircularized and transformed into E. coli MG1655ΔlacI harboring pAH69 using previously described methods (Haldimann and Wanner, 2001). Successful single integrations were verified by PCR.
Kill switch test
Cells were grown overnight with IPTG and then transferred into fresh LB medium containing 50 ng/mL ATc, 1 mM IPTG or no inducer to measure the death state, IPTG survival state, or post-IPTG survival state, respectively. Cell viability was assayed by colony forming unit (CFU) measurement in LB medium. Samples were serially diluted in PBS over a 7-log range, and 5 μL of each dilution were spotted onto square Petri dishes containing LB agar (1.5% agar) with IPTG and then incubated at 37°C overnight. CFU/mL and the survival ratio were calculated as follows: CFU/mL = [(number of colonies) × (dilution factor)]/0.005 mL, survival ratio = (CFU/mL ATc-treated or post-IPTG)/(CFU/mL IPTG-treated), and survival ratio for long-term stability = (CFU/mL ATc-treated)/(CFU/mL post-IPTG)).
Mathematical model
Let L and T denote LacI and TetR, respectively. Furthermore, introduce pL and pT to denote the promoter of LacI and TetR, where cL and cT denote the repressed promoters of LacI and TetR by TetR and LacI, respectively, so that
where ML = 4 and MT = 2 (LacI and TetR form tetramers and dimers, respectively). Let αL and αL denote the production rate of LacI when its promoter is free of TetR and when it is repressed by TetR, respectively, so that
Introduce αT and αT similarly for TetR, yielding
Furthermore, define εL = αL/αL so that it captures the leakiness of the LacI promoter: if εL = 0 the promoter is tightly repressed by TetR, whereas εL = 1 corresponds to the case when TetR does not repress the production of LacI at all. Similarly, εT = αT/αT captures the leakiness of the TetR promoter. Finally, let δL and δT denote the decay rates of LacI and TetR, respectively, so that
To address robustness to noise, we consider a bistable toggle in one of the two stable equilibria, say the LacI+ state, so that the concentration of TetR is low and that of LacI is high. Despite its promoter being repressed by LacI, TetR may be expressed due to noise or leakiness. If the concentration of TetR reaches some threshold X, the toggle then switches into the TetR+ equilibria. We are interested in the expected first passage time (FPT) for the repressed protein concentration to reach this threshold. To analyze the effect of plasmid copy number, we consider the stochastic model of gene expression in which protein expression occurs in bursts (Kaern et al., 2005). In particular, we assume that mRNAs degrades instantaneously after expressing a burst of protein molecules x (TetR in the above example), modeled by the reactions
where the burst size B is distributed geometrically with parameter b, and the time between bursts from any given copy of plasmid is an exponential random variable with mean τ = 1/β, which decreases as promoter leakage increases. The effects of the plasmid copy number n can be incorporated into the reaction parameters as β = nβ0 and b = b0/n where β0 and b0 are fixed constants, and they denote the values of β and b in the single-copy plasmid case (n = 1). The resulting mathematical model and its analysis are detailed in the Supplemental Information.
Supplementary Material
Acknowledgments
We are grateful to Sriram Chandrasekaran and Eduardo D. Sontag for helpful discussions. This work was supported by funding from the Defense Threat Reduction Agency Grant HDTRA1-14-1-006, Air Force Office of Scientific Research grant FA9550-14-1-0060, Defense Advanced Research Projects Agency Grant N66001-11-C-4203, and the ONR MURI grant N000141110725. J.W.L. was also supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2012R1A6A3A03039833), and A.G. was also supported by the NIH grant P50 GM098792.
Footnotes
ACCESSION NUMBERS
DNA sequences have been submitted to GenBank under the following accession numbers: KX264176, KX264177, KX264178, KX264179, KX264180, KX264181 and KX264182.
Supplemental Information includes experimental procedures, four figures and three tables and can be found with this article online at TBD.
AUTHOR CONTRIBUTIONS
J.W.L., A.G., D.E.C., and J.J.C. designed the study. J.W.L., A.G., D.E.C., and J.J.C. analyzed data and wrote the paper. J.W.L., A.G., N.P. and K.R.C., performed the experiments. J.W.L., A.G., D.E.C., J.C.W., P.A.S., D.D.V., and J.J.C. discussed the results and commented on the manuscript.
References
- Bergstrom CT, Lipsitch M, Levin BR. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics. 2000;155:1505–1519. doi: 10.1093/genetics/155.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K, Fisher JF. Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from gram-negative bacteria. Annu Rev Microbiol. 2011;65:455–478. doi: 10.1146/annurev-micro-090110-102911. [DOI] [PubMed] [Google Scholar]
- Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nat Rev Microbiol. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- Cameron DE, Collins JJ. Tunable protein degradation in bacteria. Nat Biotechnol. 2014;32:1276–1281. doi: 10.1038/nbt.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Lee JW, Cameron DE, Bashor CJ, Collins JJ. ‘Deadman’ and ‘Passcode’ microbial kill switches for bacterial containment. Nat Chem Biol. 2016;12:82–86. doi: 10.1038/nchembio.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SC, Kim R, King K, Kim SH, Modrich P. Isolation of gram quantities of EcoRI restriction and modification enzymes from an overproducing strain. J Biol Chem. 1984;259:11571–11575. [PubMed] [Google Scholar]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J. Genetic exchange between bacteria in the environment. Plasmid. 1999;42:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstetter CE, Thornton M, Zhou P, Bogan JA, Leonard AC, Grimwade JE. Replication and segregation of a miniF plasmid during the division cycle of Escherichia coli. J Bacteriol. 1997;179:1393–1399. doi: 10.1128/jb.179.4.1393-1399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Keasling JD. Gene-expression tools for the metabolic engineering of bacteria. Trends Biotechnol. 1999;17:452–460. doi: 10.1016/s0167-7799(99)01376-1. [DOI] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincade JM, deHaseth PL. Bacteriophage lambda promoters pL and pR: sequence determinants of in vivo activity and of sensitivity to the DNA gyrase inhibitor, coumermycin. Gene. 1991;97:7–12. doi: 10.1016/0378-1119(91)90003-t. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol. 2012;8:536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- Litcofsky KD, Afeyan RB, Krom RJ, Khalil AS, Collins JJ. Iterative plug-and-play methodology for constructing and modifying synthetic gene networks. Nat Methods. 2012;9:1077–1080. doi: 10.1038/nmeth.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- Norman A, Hansen LH, Sorensen SJ. Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci. 2009;364:2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Rankin DJ, Rocha EP, Brown SP. What traits are carried on mobile genetic elements, and why? Heredity (Edinb) 2011;106:1–10. doi: 10.1038/hdy.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler JR, Sasmor H, Betz JL. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc Natl Acad Sci U S A. 1983;80:6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis HM. The ribosome binding site calculator. Methods Enzymol. 2011;498:19–42. doi: 10.1016/B978-0-12-385120-8.00002-4. [DOI] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AB, Maxwell A. A strand-passage conformation of DNA gyrase is required to allow the bacterial toxin, CcdB, to access its binding site. Nucleic Acids Res. 2006;34:4667–4676. doi: 10.1093/nar/gkl636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nat Chem Biol. 2012;8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild J, Hradecna Z, Szybalski W. Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Res. 2002;12:1434–1444. doi: 10.1101/gr.130502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- Ying BW, Ito Y, Shimizu Y, Yomo T. Refined method for the genomic integration of complex synthetic circuits. J Biosci Bioeng. 2010;110:529–536. doi: 10.1016/j.jbiosc.2010.05.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.