Abstract
Polycomb group proteins form two main complexes, PRC2 and PRC1, which generally coregulate their target genes. Here, we show that PRC1 components act as neoplastic tumor suppressors independently of PRC2 function. By mapping the distribution of PRC1 components and the histone H3K27me3 mark, we identify a large set of genes that acquire PRC1 in the absence of H3K27me3 in Drosophila larval tissues. These genes massively outnumber canonical targets and they are preeminently involved in the regulation of cell proliferation, signaling and polarity. Mutation in PRC1 components specifically deregulates this set of genes, whereas canonical targets are derepressed in both PRC1 and PRC2 mutants. In human ES cells, PRC1 components colocalize with H3K27me3 like in Drosophila embryos, whereas in differentiated cell types they are selectively recruited to a large set of proliferation and signaling-associated genes that are H3K27me3 negative, showing that the redeployment of PRC1 components during development is evolutionarily conserved.
Polycomb group (PcG) proteins form two main classes of evolutionarily conserved complexes called PRC2 and PRC1. In Drosophila, PRC2 contains the enzymatic E(Z) subunit that deposits the H3K27me3 mark, along with SU(Z)12, ESC and p55 1–4. A second class of PcG complex, PRC1, was shown to contain PH, PC, PSC and the SCE subunits 5. These two complexes are recruited to their target sites by a set of DNA binding proteins, notably PHO 6. They colocalize almost perfectly in embryogenesis7, and their embryonic phenotypes are similar, with posterior homeotic transformations due to misexpression of HOX genes 8. Later in development, mutations in PcG components induce cancer 9,10, suggesting that PcG proteins may be dynamically recruited to new target genes. Since previous work suggested that larval mutations in different PcG components can induce phenotypes of different severity 11, we analyzed the effect of PRC1 and PRC2 mutations side by side, by using the FLP/cell-lethal system 12 to generate eye discs composed predominantly of mutant cells.
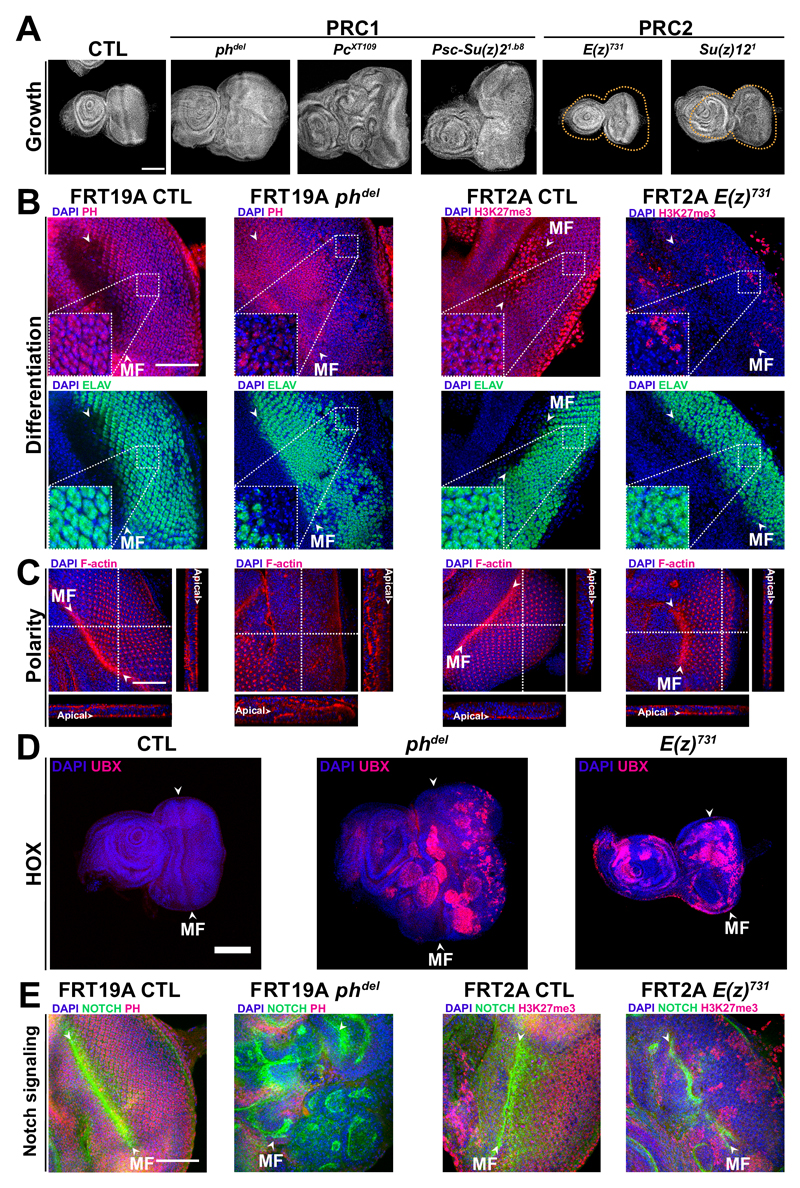
Different PRC1 complexes exist. All of them contain a catalytic subunit which, in Drosophila, is encoded by the Sce gene. In addition, canonical PRC1 (cPRC1) complexes contain PC, PH and PSC proteins. Null mutations in the Psc and ph loci 9,10, as well as in Pc, induce tumors in a highly penetrant manner (Fig. 1A, Supplementary Fig. 1A, 2A and B), showing that canonical PRC1 is required for tumor suppression. In contrast, mutations in the PRC2 components E(z) and Su(z)12 generate eye discs with normal appearance and smaller size than wild type controls (Fig. 1A, Supplementary Fig. 1 A and B, see also 13,14). ph knock out (KO) cells are also negative for the Elav neuronal differentiation marker, whereas E(z) KO cells are Elav positive even in the absence of the PRC2-dependent H3K27me3 mark (Fig. 1B and Supplementary Fig. 1C). ph mutant cells show massive overproliferation in the posterior part of the eye disc. In contrast, E(z) KO cells hypoproliferate (Supplementary Fig. 1C and D). Furthermore, cell polarity is normal in E(z) and Su(z)12 PRC2 mutants, whereas it is dysregulated in ph, Psc and Pc KO cells, with multilayered cell growth and disruption of the apical localization of F-actin (Fig. 1C, Supplementary Fig. 1E and Supplementary Fig. 2A and B). The contrast between tumor induction by cPRC1 mutation and hypoproliferation in PRC2 mutants does not depend on Hox genes, since they are strongly derepressed in both classes of mutants (Fig. 1D, Supplementary Fig. 1C and Supplementary Fig. 2C). While ph mutation induces expression of Notch (N) in a cell-autonomous manner 10, no overexpression of N is observed in E(z) KO, H3K27me3 negative cells (Fig. 1E and Supplementary Fig. 1C). qChIP at the N locus shows that PC and PH bind to N in eye discs in the absence of H3K27me3 (Supplementary Fig. 3).
Fig. 1. Differential effects of PRC1 and PRC2 mutations on tumorigenesis.
A- DAPI staining shows that PRC1 mutants (phdel, PcXT109 and Psc-Su(z)21.b8) overgrow while PRC2 mutants (E(z)731 and Su(z)121) exhibit small eye phenotype. B- Double staining of PH or H3K27me3 and of the ELAV neuronal differentiation marker. Dashed squares indicate higher magnification. C- Staining of the F-actin polarity marker. White dashed lines indicate the positions of the XZ and YZ cross-sections shown respectively below and to the right of each panel. D- Staining of the Hox UBX protein in PRC1 and PRC2 mutants, as indicated. E- Staining of PH or H3K27me3 and of N in PRC1 and PRC2 mutants, as indicated. Scale bars are 50 µm in B, C, E and 100 µm in A, D. The morphogenetic furrow (MF) is indicated by arrowheads and discs are oriented with the posterior on the right side.
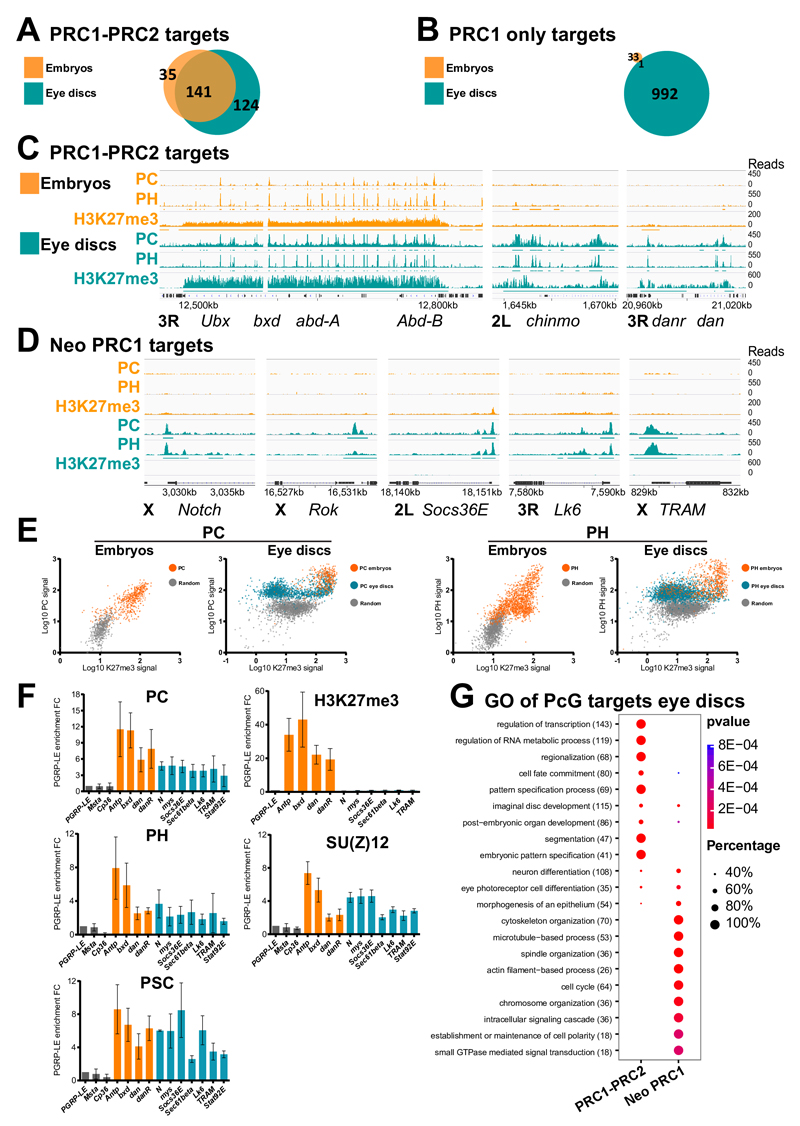
In order to test whether other genes might acquire PRC1 binding in the absence of H3K27me3, we mapped the PRC1 subunits PH, PC as well as the H3K27me3 mark in embryos 6 and in the two larval eye and wing imaginal discs by ChIP-Seq (See Supplemental materials and methods). PRC1-bound genes (co- bound by PC and PH) in embryos are usually marked by H3K27me3 7. Most of the 176 canonical embryonic target genes marked by the two complexes maintain PRC1 and H3K27me3 in larval tissues (Figs. 2A, 2C, Supplementary Figs. 4A, 5A and Supplementary Table 1). H3K27me3 increases during development on these canonical targets (Supplementary Fig. 5A and 6). PcG-mediated silencing is, however, dynamic, as revealed by Immuno-FISH experiments (Supplementary Fig. 7). In particular, the hedgehog (hh) gene is expressed in the latest phases of eye disc development, posteriorly to the morphogenetic furrow 15. hh is strongly localized within nuclear PH foci in anterior cells, before the passage of the furrow. However, it is released from PH in most of the cells after the passage of the furrow (Supplementary Fig. 7B), suggesting that, similar to mammalian systems 16, PcG-mediated cellular memory is dynamically regulated during Drosophila development 17,18. Dynamic Polycomb binding also occurs on many PRC1-PRC2 targets (Supplementary Fig. 5A) where PcG binding strongly increases between embryonic and larval stages as illustrated for danr, dan and chinmo (Fig. 2C).
Fig. 2. Massive redeployment of PRC1 from canonical to neo PRC1 gene targets during development.
A- Venn diagram showing the overlap of PcG PRC1-PRC2 target genes in embryos and in eye imaginal discs. B- Venn diagram representing the overlap of genes marked only by PC/PH in the absence of the H3K27me3 mark (PRC1 only targets) in wild type embryos and in third instar eye imaginal discs. C- ChIP-Seq for PC, PH and H3K27me3 in embryos and eye imaginal discs for PRC1-PRC2 targets illustrated at the Bithorax complex (BXC), at chinmo, danr and dan. Significantly enriched regions are shown as horizontal bars under each ChIP-Seq track. D- Distribution of PC/PH and of H3K27me3 mark in embryos and eye imaginal discs at neo PRC1 target genes. E- Scatter plots showing the ChIP-Seq signal levels of PC or PH versus H3K27me3 on their targets in embryos and larval eye discs. Regions that were already enriched in embryos are shown in orange, whereas green indicates targeting in larval stage only. A control random set of H3K27me3 negative regions is represented in grey. F- qChIP for PC, PH and PSC from the PRC1 complex, SU(Z)12 from the PRC2 complex and for H3K27me3. Results are normalized using the PGRP-LE gene (negative control). Error bars represent the s.d of three experiments. G- Comparative Gene Ontology (GO) analysis of the maintained versus neo PcG targets in eye imaginal discs. The number of PcG target genes in each GO category is indicated in brackets. The fraction of bound genes and p-values are indicated by the size and color of each spot.
In addition, we found a large set of genes that become bound by cPRC1 proteins in larval tissues in the absence of H3K27me3 (Fig. 2B, 2D, 2E and Supplementary Table 1). This category, named as “neo PRC1”, includes 894 genes in eye discs, of which 654 are in common with wing discs (Supplementary Fig. 4B). Neo PRC1 targets are strongly enriched in genes regulating the cell cycle, cell polarity and cytoskeletal organization, as well as signaling and signal transduction pathways (Fig. 2G, Supplementary Tables 2 and 3). These data show that, after a first wave of deployment during embryogenesis, a second developmental wave massively recruits PRC1 components to novel genes during larval development.
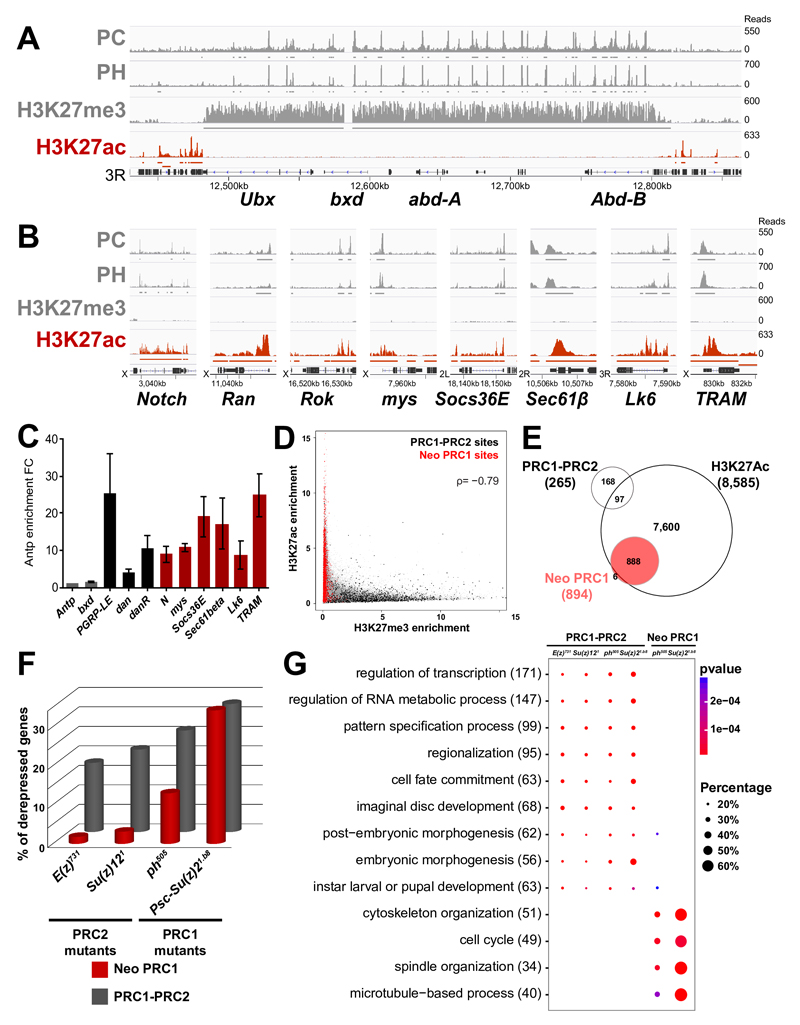
We analyzed the neo PRC1 category in more detail. First, we verified PRC1-specific targeting to six of these genes by qChIP in eye discs (Fig. 2F) and H3K27me3 is absent from all of them, while it is clearly detected in PRC1-PRC2 targets. Interestingly however, the SU(Z)12 and E(Z) subunits of the PRC2 are present on neo PRC1 sites (Fig. 2F) and, in genome wide mapping of SU(Z)12 19, 66.5% of the neo PRC1 genes are bound by SU(Z)12 (Supplementary Fig. 8). Therefore, we compared PC binding and H3K27me3 in ph and in E(z) null mutant eye disc tissue. At PRC1-PRC2 target genes, H3K27me3 decreases upon E(z) mutation. As expected, PC binding also decreases in both ph and in E(z) mutants (Supplementary Fig. 9A). H3K27me3 also decreases slightly upon mutation of ph at PRC1-PRC2 target genes (Supplementary Fig. 9A), suggesting that PRC1 may have an effect in stabilizing PRC2 function. Although PC and PH binding at neo PRC1 is generally weaker than at PRC1-PRC2 targets (Fig. 2 E and F, Supplementary Fig. 5), PC levels decrease at neo PRC1 target genes in ph tissue. However, PC levels are not affected in E(z) mutant tissue, showing that these peaks reflect specific binding and that PC is recruited on neo PRC1 targets independently from PRC2 (Supplementary Fig. 9B). We then analyzed the mark of PRC1, namely H2AK118Ub. Mutant discs for the Drosophila PRC1 catalytic subunit Sce show no growth defects (See 20, Supplementary Fig. 10 A) and no polarity defects (Supplementary Fig. 10 B and C), suggesting that PRC1 acts on neo PRC1 targets independently of its associated histone mark. qChIP experiments show that the H2AK118Ub mark is not significantly enriched on neo PRC1 targets (Supplementary Fig. 10 D). We showed earlier that the DNA binding protein PHO, a known recruiter of PcG components, also binds at low levels to many other target loci in fly embryos. Genome-wide comparisons and qChIP shows that many neo PRC1 peaks colocalize with PHO binding sites, suggesting that neo PRC1 genes correspond to progressive assembly of PRC1 to weaker PHO sites during development (see Supplementary Fig. 11 and 7). In contrast to PRC1-PRC2 targets, neo PRC1 genes are robustly transcribed (Supplementary Figs. 12 and 13). Binding does not reflect the presence of open chromatin however, since genes that are similarly or more transcribed than the neo PRC1 group are mostly devoid of PC or PH proteins (Supplementary Fig. 12). Since H3K27Ac is frequently associated with active genes and since neo PRC1 targets lack the counteracting H3K27me3 mark, we mapped H3K27Ac in eye discs. While this mark is mostly absent from canonical PcG target genes (Figs. 3A and 3E), we observed strong H3K27Ac at neo PRC1 target genes (Fig. 3B, 3C and 3E), slightly downstream to the TSS (Supplementary Fig. 14). The presence of H3K27me3 and H3K27Ac are clearly anti-correlated, with genes bound by both PRC1 and PRC2 carrying the former and the neo PRC1 targets carrying the latter (Fig. 3D). Therefore, the level of expression and the mark at H3K27 define PcG target genes categories (Fig. 3E and Supplementary Fig. 12).
Fig. 3. PRC1 represses transcriptionally active neo PRC1 targets.
A- ChIP-Seq profiles for PC, PH, H3K27me3 and H3K27Ac in the BXC locus in eye imaginal discs. Enriched regions are shown under each ChIP-Seq track. B- ChIP-Seq profiles as above in a set of neo PRC1 target loci. C- qChIP against H3K27Ac on a set of canonical and neo PRC1 target genes. Results are normalized using the Antp gene (negative control). Error bars represent the s.d of three experiments. D- Scatter plot showing the ChIP-Seq distribution of H3K27me3 and H3K27Ac. PRC1-PRC2 target sites (in black) mainly carry H3K27me3 while neo PRC1 target sites (in red) carry H3K27Ac. The H3K27Ac and H3K27me3 enrichments are anti-correlated with a spearman’s rank correlation coefficient ρ of –0,79. E- Venn diagram showing the overlap of the different classes of PcG target genes with the H3K27Ac mark, based on genome-wide ChIP-Seq data analysis in eye imaginal discs. F- Percentage of upregulated genes in RNA-seq of PRC1 (ph505 and Psc1.b8) or PRC2 (E(z)731 and Su(z)121) mutant eye discs. G- Comparative Gene Ontology (GO) analysis of genes upregulated in PRC1 (ph505 and Psc1.b8) or PRC2 (E(z)731 and Su(z)121 ) mutants, stratified by categories of PcG ChIP-Seq targets. The total number of upregulated genes in each GO category is indicated in brackets. The fraction of bound genes that are upregulated in each category of PcG targets and the p-values are indicated by the size and the color of each spot.
Although the finding of PcG protein binding to active genes is not unprecedented 21–24, the massive recruitment of PRC1 components to genes involved in cell proliferation, signaling and polarity was notable. We thus performed RNA-Seq of eye discs mutated in PRC1 (ph505, Psc/Su(z)21.b8) or PRC2 (E(z)731 and Su(z)121) components (Supplementary Table 4). Mutations in different components of the same PcG complex regulate similar sets of genes (72% of the genes upregulated in ph mutants are also induced upon Psc/Su(z)2 mutation and 68% of the genes upregulated in E(z) mutants are also induced in Su(z)12 null tissues) (Supplementary Table 4, Supplementary Fig. 15A). Hox and other canonical PcG targets are derepressed in PRC1 and PRC2 mutants (Fig. 3F, Supplementary Fig. 15B and Supplementary Table 4). Some PRC1-PRC2 target genes are more strongly derepressed in PRC1 mutants (Supplementary Fig. 15B). This includes genes of the JAK/STAT pathway that were previously shown to be linked to Polycomb-dependent tumorigenesis 9, 25. A significant fraction of neo PRC1 genes is only upregulated in PRC1 mutants, whereas they are unaffected by PRC2 mutations (Fig. 3F and Supplementary Fig. 15C). Upregulated neo PRC1 genes, representing putative direct targets of PRC1, are strongly enriched in cancer-related ontologies such as cell cycle, cytoskeleton organization and tissue polarity (Fig. 3G, Supplementary Fig. 16 and Supplementary Table 5). Many of these genes play key roles in signaling pathways promoting cell proliferation and involved in tumorigenesis, such as the Notch pathway (including N, shaggy, Nedd4 and Apc 10,26,27), the JAK/STAT pathway (including domeless, stat92E 9,28) and the Rho/JNK pathway (including Ran, Rho1 and puckered 29). Two of these, N and stat92E, were previously shown to mediate tumorigenesis upon loss of ph or of the Psc locus 9,10. Therefore, Drosophila PRC1 components might suppress tumor formation by dampening the expression of a set of cell proliferation, cell signaling and polarity genes independently of PRC2.
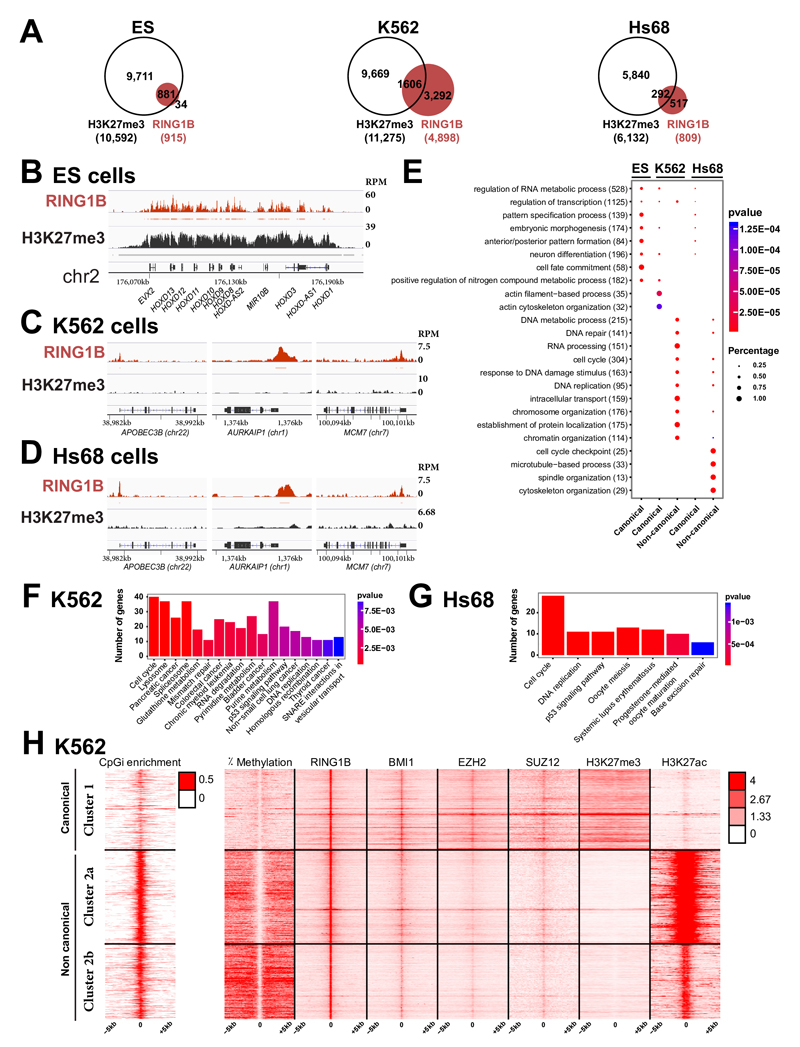
While PcG proteins and many of their target genes are strongly conserved in evolution 30,31, the prevalent view is that mammalian PcG components act as oncogenes, both through INK4A-ARF-dependent and independent mechanisms 32,33. However, recent evidence suggests that several PcG members can act as tumor suppressors 32 and that PRC1 may be present at a large subset of sites devoid of H3K27me3 34. We therefore analyzed genome-wide maps of H3K27me3 and the key PRC1 RING1B subunit in human embryonic stem (ES) cells, in the myelogenous leukemia cell line (K562) and in normal fibroblasts (Hs68) (Figs. 4A to D). As expected, a large fraction (96%) of genes bound by RING1B in ES cells show colocalization of RING1B with H3K27me3. Strikingly, this fraction decreases in the two differentiated cell types, with only 33% and 36% of RING1B target genes marked by H3K27me3 in K562 and Hs68 fibroblast cells (Supplementary Table 6). While canonical targets encode for transcriptional regulators involved in developmental pathways (Fig. 4E), non-canonical targets (bound by RING1B without H3K27me3) show a difference in gene ontologies, with preeminent functions in cell cycle regulation, cytoskeleton organization, DNA repair and signaling pathways (Figs. 4E, F and G) and including strongly expressed genes in both differentiated cell types (Supplementary Fig. 17A and B). Mammalian cells contain a variety of PRC1-like complexes, all of which contain RING1B 34. Moreover, RING1B is involved in the formation of non-PRC1 complexes 35. In order to test whether other PRC1 subunits distribute similarly with respect to H3K27me3, we analyzed ChIP-Seq profiles in K562 cells. For cPRC1, we selected BMI1. For PRC2, we analyzed EZH2 and SUZ12, and we added H3K27Ac as well as DNA methylation. The data show that the majority of the targets containing both RING1B and BMI1 does not colocalize with H3K27me3 (Supplementary Fig. 18A and B). Although EZH2 and SUZ12 are significantly bound to them, these PRC1 targets are strongly marked by H3K27Ac. Most of them correspond to DNA-unmethylated CpG islands and are located close to TSSs of highly expressed genes (Fig. 4H, Supplementary Figs. 17C, 18C and 19), highly reminiscent of the situation in Drosophila.
Fig. 4. PcG proteins localize to a large set of DNA-unmethylated CpG islands in the absence of H3K27me3 in differentiated human cells.
A- Venn diagram showing the overlap of RING1B target genes (in red) and genes carrying H3K27me3 (in white) in human ES cells (ES), K562 myelogenous leukemia cells and foreskin fibroblasts (Hs68). B- ChIP-Seq profiles for RING1B and H3K27me3 in ES at the HOXD complex. Significantly enriched regions are shown as horizontal bars under each ChIP-Seq track C- ChIP-Seq profiles for RING1B and H3K27me3 at a set of PRC1 loci in K562 cells. D- ChIP-Seq profiles for RING1B and H3K27me3 at a set of PRC1 loci in Hs68 cells. E- Comparative Gene Ontology (GO) analysis of canonical versus non-canonical PcG targets in ES, K562 and Hs68 cells. The total number of PcG target genes in each GO category is indicated in brackets. The fraction of bound genes and the p-values are indicated by the size and the color of each spot, respectively. F- KEGG pathways enriched in the non-canonical targets in K562 cells. G- KEGG pathways enriched in the non-canonical targets in Hs68 cells. H- Heat maps and profiles of percentage of DNA methylation, occupancies of RING1B, BMI1, EZH2, SUZ12, H3K27me3 and H3K27Ac within -/+ 5kb from RING1B peaks in K562 cells.
The present work uncovers a substantial redeployment of PRC1 during development. PRC1 components prevent tumorigenesis both by silencing signaling pathway genes in conjunction with PRC2 as well as by limiting the expression of neo PRC1 genes coordinately regulating cell cycle, signaling and cell polarity. Therefore, PRC1-mediated control of cell proliferation and tissue polarity is at least equally important as its canonical developmental patterning role that involves silencing of transcription factor genes. Furthermore, the analysis of PcG targeting in human cells suggests a parallel with Drosophila. In ES cells, while non canonical PRC1 complexes can bind to active genes in the absence of H3K27me3 36, 37, canonical PRC1 and PRC2 components colocalize, similar to fly embryogenesis. In contrast, other normal differentiated cells or cancer cells23,34,38 may feature widespread association of PRC1 components to a large set of target genes in the absence of H3K27me3. Of note, the Mel18 subunit of cPRC1 was suggested to be also capable of activating a subset of its target genes. A similar function was reported for part of the PRC1 targets in Drosophila 24, suggesting possible conserved activation function for Polycomb components. Finally, it will be important to analyze whether the tumor suppressor function that was recently identified for several PRC1 components32,39 involves regulation of neo PcG-target genes, similar to the Drosophila case.
Online Methods
Genetics
Flies were raised in standard corn meal yeast extract medium at 25°C. The Oregon-R w1118 line (referred to as wild type) was obtained from R. Paro (ZMBH, University of Heidelberg, Germany). Mosaic imaginal discs were generated using the eye-FLP-cell lethal clonal method as described in 12 using UAS-flp under the control of ey-GAL4 to induce recombination. Discs are composed predominantly of mutant cells (referred to in the text as mutant discs). The strong or null mutants used in this study are: ph-p505; phdel, Psc-Su(z)21b8, E(z)731; Su(z)121, SceKO.
Genotypes of control and PcG-mutant clones
FRT19A/ FRT19A GMR-hid; ey-GAL4 UAS-FLP1/+
UAS-GFP/ ey-GAL4 UAS-FLP1; FRT2A/ FRT2A GMR-hid
FRT42D/ FRT42D GMR-hid; UAS-GFP/ ey-GAL4 UAS-FLP1
Ey-GAL4 UAS-FLP/+; FRT82B/ FRT82B GMR-hid
ph505, FRT19A/ FRT19A GMR-hid; ey-GAL4 UAS-FLP1/+
phdel, FRT19A/ FRT19A GMR-hid; ey-GAL4 UAS-FLP1/+
Su(z)21.b8, FRT42D/ FRT42D GMR-hid; UAS-GFP/ ey-GAL4 UAS-FLP1
UAS-GFP/ ey-GAL4 UAS-FLP1; E(z)731, FRT2A/ FRT2A GMR-hid
UAS-GFP/ ey-GAL4 UAS-FLP1; Su(z)121, FRT2A/ FRT2A GMR-hid
Ey-GAL4 UAS-FLP/+; SceKO FRT82B/ FRT82B GMR-hid
Staining procedures
Eye-antennal imaginal discs were dissected in PBS from L3 wandering larvae and fixed in 4% paraformaldehyde/PBS for 20 min at R.T. Discs were permeabilized for 1 hr at R.T in 0,5% PBTr (PBS + 0,5% Triton X-100) and blocked for 1 h at room temperature with 3% BSA in 0,025% PBTr (PBS + 0,025% Triton X-100). Discs were then incubated O/N at 4°C on a rotating wheel with the primary antibodies in 0,025% PBTr (PBS + 0,025% PBTr) and 1% BSA. The following antibodies were used: goat anti-PH ((Grimaud et al., 2006), 1:500), rabbit anti-H3K27me3 (Diagenode, C15410195, 1:1000), mouse anti-ELAV (DSHB, 9F8A9-c, 1:500), mouse anti-UBX (DSHB, FP3.38-s, 1/20), mouse anti-ABD-B (DSHB, 1A2E9-s, 1/10), chicken anti-GFP (LifeTechnologies, A10262, 1/200), rabbit anti-H2AK119Ub (Cell Signaling Technology, D27C4, 1:500), mouse anti-NOTCH (DSHB, C17.9C6, 1:1000). The second day, discs were washed 3 times with 0,025% PBTr and incubated for 2h at R.T on a rotating wheel with secondary antibodies diluted in 0,025% PBTr + 1% BSA. Following secondary antibodies were used: donkey anti-rabbit Alexa Fluor 488® (LifeTechnologies, A-21206, 1:200), donkey anti-rabbit Alexa Fluor 555® (LifeTechnologies, A-31572, 1:200), donkey anti-rabbit Alexa Fluor 647® (LifeTechnologies, A-31573, 1:200), donkey anti-goat Alexa Fluor 555® (LifeTechnologies, A-21432, 1:200), donkey anti-mouse Alexa Fluor 488® (LifeTechnologies, A-21202, 1:200), donkey anti-mouse Alexa Fluor 555® (LifeTechnologies, A-31570, 1:200), donkey anti-goat Alexa Fluor 647® (LifeTechnologies, A-31571, 1:200) and donkey anti-Chicken Alexa Fluor 488® (LifeTechnologies, A-11039, 1:200). F-actin was stained by adding Rhodamin Phalloidin Alexa Fluor 555® (LifeTechnologies, R415, 1:200) to secondary antibodies. Eye-antennal discs were then washed and stained with DAPI diluted in 0,025% PBTr (1µg/mL final) for 20 min at R.T. Discs were rinsed in 0,025% PBTr and put in PBS. Discs were mounted in Vectashield medium (Vector Laboratories) and visualized on Leica SP8-UV confocal microscope. EDU incorporation experiments were realized using EDU Click-iT kit, following manufacturer’s recommendations (Alexa Fluor 488®, Invitrogen, C10337). Eye discs sizes were measured by manually defining the limits of the eye discs using ImageJ.
Chromatin Immunoprecipitation (ChIP) Experiments on whole Drosophila embryos or larval imaginal discs
ChIP of whole embryos (16-18h) was essentially performed as previously described40. Briefly, cross-linking was performed for 15 min in the presence of 1.8% formaldehyde during tissue homogenization. Chromatin extracts of embryos were sonicated using a Bioruptor (Diagenode) for 15 min (settings 30 sec on, 30 sec off, high power).
ChIP in Drosophila imaginal discs was carried out on third instar larval eye-antennal and wing imaginal discs as previously described in 7 with the following modifications: after dissection and fixation of the imaginal discs the chromatin was sonicated using a Bioruptor (Diagenode) for 15 min (settings 30 s on, 60 s off, high power) in A2 buffer at 1% SDS. The size of the sheared chromatin fragments ranges from 500 to 1000 bp. After sonication SDS concentration was brought back to 0.1%. For ChIP-Seq experiments, IPs were carried out in a total volume of 250µL using the following antibodies (diluted 1:100): PC and PH antibodies described in7, H3K27ac antibody from Abcam (#4729), H3K27me3 antibody from Upstate Biotechnology (#07–449) and Pleiohomeotic (PHO) antibody provided by Judith A. Kassis and described in41.
Sequencing of the ChIP samples was performed by the sequencing platform MGX (see URL section). To obtain the recommended quantity of DNA several Ips were prepared (using 50 eye-antennal discs per IP), pooled and resuspended in a volume of 20µl (12 to 14 Ips for Pc or Ph, 3 to 4 Ips for H3K27me3 and 3 Ips for H3K27ac). Sample preparation was done with Illumina kit (ref. IP-102-1001) following manufacturer’s instructions.
ChIP followed by qPCR (qChIPs) were carried out with the same protocol, using 500 eye-antennal discs per IP. The following antibodies were used: rabbit anti-PC, anti-PH (described in 7, 1:100), rabbit anti-PSC (described in42, 1:200), rabbit anti-H3K27Ac and anti-H3K27me3 (Active Motif, #39134 and #39155, 1:100), rabbit anti-H2AK119Ub (Cell Signaling Technology, D27C4, 1:100), rabbit anti-SU(Z)12 (cf material and methods, Supplementary Fig. 8B, 1:100), rabbit anti-E(Z) (Santa Cruz Biotechnology, #98265, lot A0109, 1:50). Primers for qPCR reactions are listed in Supplementary Table 7. For ChIP experiments performed in PcG mutant eye-antennal discs, the antenna was removed to work with as many mutant cells as possible.
Drosophila ChIP-Seq analysis
ChIP experiments have been performed in duplicates and DNA samples have been sequenced on HiSeq2000, filtered and aligned with CASAVA from Illumina. The number of reads and the correlations between replicates are provided in Supplementary Tables 8 and 9, respectively.
PC, PH and H3K27ac ChIP-Seq data have been analyzed using MACS43 version 1.3.7 with standard parameters except for the genome size with a value of 120Mb and the tag size with a value of 36 nucleotides. Only the peaks from MACS with a minimum enrichment of 2 fold and a maximum FDR of 10% are considered as enriched.
The H3K27me3 ChIP-Seq data have been analyzed using SICER44 with input as control library, a redundancy threshold of 4, a window size of 500bp, an effective genome fraction of 0.7, a gap size of 2kb and a threshold of 10 %. To define highly confident targets, the 500bp windows with 2 fold enrichment have been considered. However, for assignment of neo PRC1 target genes, H3K27me3 enriched regions detected using default settings have been used.
For each condition, the final list of peaks was obtained as the intersection between the peaks of both replicates.
SU(Z)12 ChIP-Seq data are available with accession number GSE36039 and were re-analyzed with the same settings as the other ChIP-Seq analyses performed for this manuscript. PHO enriched regions were obtained from ChIP-on-chip data, which are available with accession number E- MEXP-1708.
To Visualize and present ChIPseq data, we used IGV (Integrative Genomics Viewer), see URL section45, 46.
For the scatter plots showing PC (or PH) versus H3K27me3 enrichments at PC (or PH) sites and a random set of H3K27me3 negative sites in embryos and in eye discs, ChIP-Seq signals were quantified using EaSeq version 1.01, see URL section47. Enrichment values were then plotted using GraphPadPrism 6. The scatter plot showing H3K27Ac and H3K27me3 enrichments were calculated through 500bp-enriched windows (enrichment >=2) sliding along the genome. Density plots showing the average enrichment of ChIP-Seq tracks around the TSS of target genes were obtained using the Bioconductor package “seqplots”, with default parameters.
Assignment of genes to enriched regions
The method is based on the one used by ModEncode when they map regulatory elements in Drosophila48. A gene is considered as marked if an enriched region is located between 1kb upstream of one of the TSS of the gene and a distance downstream to the TSS, equal to the length of the longest transcript, with a maximum of 2Kb.
The annotations that have been used were computed with dedicated scripts using the API EnsEMBL Core, with EnsEMBL v73 database that corresponds to release 5.46 in Flybase.
Assignment of genes to the different categories of Polycomb targets
Larval PcG targets have been analysed using the following scheme: PRC1-PRC2 target genes are enriched for PRC1 (PC and PH) and for H3K27me3 using a stringent 2 fold enrichment cut-off. Neo PRC1 target genes are strictly defined as enriched for PRC1 (PC and PH) but not enriched for H3K27me3 when using default settings of SICER. Genes already bound in embryos either by PC and/or PH and/or H3K27me3 using SICER default settings are not considered as neo PRC1 targeted during larval stages.
Immuno-DNA-FISH (Fluorescence in situ hybridization) in Drosophila imaginal discs
All details for the FISH protocol are available at the URL provided in the URL section. Briefly, WT eye-antennal and wing third instar imaginal discs were dissected and fixed in PBT (PBS-0.1% Tween) 4% paraformaldehyde. Hybridization of 10ng of probes was done overnight on FHB (Fish hybridization buffer). For the immunostaining, after posthybridization washes, discs were blocked in PBSTr-10% Normal Goat Serum (NGS) for 2h at RT, and incubated overnight at 4°C with an anti-PH antibody7 at a dilution of 1:700 in PBSTr-10% NGS. Discs were washed several times in PBSTr, blocked again in PBSTr-10% NGS for 1h at RT, and incubated for 1h at RT with an anti-rabbit-Cy5 (Jackson Laboratories) at a dilution of 1:200 in PBSTr-10% NGS. Discs were finally stained with DAPI (0.1µg/mL in PBT for 10min) and mounted in ProLong antifade (Molecular Probes).
Fluorescence high resolution wide-field image acquisition was performed on a Leica DMRXA equipped with a micromax YHS1300 CCD camera (Roper Scientific), a 100x/NA 1.40 oil immersion objective (Leica Microsystems).
Generation of Fluorescent probes
For each gene, the FISH probes cover a region of 12 kb significantly enriched for PRC1 proteins (PC, PH) together with H3K27me3 histone modification. FISH probes were generated using 4 to 6 genomic PCR fragments of approximately 1.5 kb. Primer sequences to generate these fragments are listed in Supplementary Table 10. Probes were labeled using FISH Tag™ kits (Invitrogen life technologies) following manufacturer’s instructions. The specificity of each probe has been systematically tested by FISH on polytene chromosomes.
To calculate the percentage of colocalization, the gene and the PH foci were considered colocalized when FISH signal overlapped or was juxtaposed to the immunostaining signal in a nucleus. For each condition, 3D stacks were collected from 3–4 different tissues (optical sections were collected at 0.5 µm intervals along the z-axis), and 50 to 100 nuclei were observed in 3D stack using Metamorph software (Universal Imaging Corp.) to obtain the percentage of colocalization.
Drosophila RNA-seq data
RNA-seq data from mutant conditions have been compared to their respective control genotypes i.e neutral clones generated in the same genetic background (see Online Methods). The expression levels were calculated based on 2 biological replicates for each conditions.
RNA-Seq libraries were constructed with the TruSeq RNA sample preparation (Low throughput protocol) kit from Illumina (performed by Montpellier MGX, Montpellier, France; part number 15008136). One µg of total RNA was used for the construction of the libraries. The RNA is fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments are copied into first strand cDNA using SuperScript II reverse transcriptase and random hexamer primers. The Second strand cDNA is synthesized. These cDNA fragments then go through an end repair process, the addition of a single 'A' base and the subsequent ligation of the adapter. The products are then purified and enriched with 15 cycles of PCR. The final cDNA libraries are validated with a DNA 1000 Labchip on a Bioanalyzer (Agilent) and quantified with a KAPA qPCR kit.
For one sequencing lane of a flowcell V3, three libraries were pooled in equal proportions, denatured with NaOH and diluted to 7 pM in hybridization buffer. Cluster formation, primer hybridization and single read 50 cycles sequencing were performed on cBot and HiSeq2000 (Illumina, San Diego, CA) respectively.
The RNA-Seq data have been aligned only on the transcripts by using TopHat49 v2.0.8b and Bowtie 2.1.0.0 with standard parameters. For each gene, a RPKM score (Reads per kilobase per Million mapped reads) has been computed.
Image analysis and base-calling were performed using the HiSeq Control Software and Real-Time Analysis component. The quality of the data was assessed using fastqc from the Babraham Institute and the Illumina software SAV (Sequence Analysis Viewer). Demultiplexing, alignment and RNA counting was performed using CASAVA 1.8.2 (Illumina). Alignment was made with eland_rna on the dm3 version of Drosophila melanogaster genome and on several contaminants (the ribosomal RNA sequences, the mitochondrial chromosome, the PhiX genome and the Illumina adaptors). The transcript annotation was retrieved from UCSC (refFlat file), version dated August 14, 2012. GeneIDs are from the NCBI (gene2refseq.gz; 2012-08-14). Before statistical analysis, genes with less than 10 reads (cumulating all the analysed samples) were filtered and thus removed.
Differentially expressed genes were identified using the Bioconductor50 package edgeR 2.6.251. Data were normalized in parallel using the Trimmed Mean of M values (TMM) 52. Genes with adjusted p-value less than 5% (according to the FDR method from Benjamini-Hochberg) were considered differentially expressed.
Wild type RNA-seq from eye discs and wing discs were obtained from GEO (Accession number GSE43341).
RNA interference in S2 cells
Double-stranded RNA against Su(z)12 and GFP were synthesized by in-vitro transcription (Ambion Megascript T7 kit) of PCR products amplified from w1118 genomic DNA or from GFP coding sequence from pAWG (The Drosophila Gateway Vector Collection, Carnegie Institute of Science), using gene-specific primers, each of which included T7 promoter sequences at their 5’ends, as follows: T7-Su(z)12-R CATCCCAAAGAGCTGGACAT, T7-Su(z)12- S GTCTTGTAGACCGTCTCGGC (Su(z)12); T7-GFP-R GACGTAAACGGCCACAAGTT, T7-GFP-S TGCTCAGGTAGTGGTTGTCG (GFP). S2 cells were cultured at a concentration of 106 cells/ml and incubated with 6µg/ml of dsRNA for 4 days before harvesting.
Production of a rabbit anti-Drosophila SU(Z)12 antibody
A rabbit anti- SU(Z)12 antiserum was raised against a peptide containing the residues 624 to 900 from the Drosophila SU(Z)12 (Covalab, Inc.).
Western-Blot
To assess the specificity of SU(Z)12 rabbit antibody, equal amounts of proteins from nuclear extracts were boiled for 5min in Cracking Buffer (125mM Tris-Hcl ph7, 5% B-mercapto-ethanol, 2% SDS, 4M Urea) and separated on 4-20% gradient SDS-PAGE (TGX Stain-free precast Gels, Bio-Rad). After activation of Stain-free Gel by UV-light and transfer onto nitrocellulose membrane using trans- blot Turbo (Bio-Rad), the membrane was imaged under UV-light for loading control. The membrane was blocked in 5% milk, 1X PBS, 0,1% Tween, and incubated overnight with anti- SU(Z)12 (1:1000; antibody described in this paper), followed by incubation for 2h at room temperature with anti-rabbit secondary antibody (1:5000) coupled to HRP (Sigma-Aldrich). Detection was performed using Clarity Western ECL substrate (Bio-Rad) using a ChemiDoc XRS+ Imager (Bio-Rad).
Gene Ontology (GO) analysis
The ontologies have been obtained using DAVID53,54 database through DAVID web interface. We applied the “functional Annotation” function considering only “Biological Process” ontologies (BP). Gene Ontologies are visualized using the R package “clusterProfiler”55 with “row.percentage” settings.
Human ChIP-Seq analysis
The ChIP-Seq datasets for RING1B in human ES cells were obtained from Ku et al., 200856 (GSE13084); and H3K27me3 datasets in ES cells from Gifford et al., 201345 (GSM772750). The ChIP-Seq datasets for RING2 (RING1B) and H3K27me3 in K562 cells were from the Encode consortium (ENCSR138FUZ and ENCSR000AKQ) and in fibroblasts (Hs68) from Pemberton et al., 201423 (GSE40740). All of the datasets were analyzed according to the following methodology: sequences were aligned to the human genome (UCSC build hg38) with Bowtie2 using default parameters. PCR duplicates and reads with low mapping quality (MAPQ<30) were removed and only sequences that mapped uniquely to the genome were used for further analysis. Individual replicates were pooled and peaks were called using MACS2.1 with default parameters for RING1B and with the -broad option on for H3K27me3 to identify domains, using whole-cell extract input from the corresponding cell line as control. RING1B peaks were associated with a gene using the same criteria as for Drosophila.
The data for BMI1, EZH2, SUZ12, H3K27Ac and DNA methylation in K562 cells are from the ENCODE consortium (ENCSR782WRO, ENCSR000AQE, ENCSR000AUC, ENCSR000AKP and ENCSR765JPC). The definition for CpG islands was obtained from the UCSC genome browser (genome version hg38).
For the clustering analysis, the normalized enrichment values were extracted from the ChIP-Seq tracks in a window of 10kb centered either on the RING1B peaks or on CpG islands in K562 cells. These values were then used for k-mean based clustering with 3 clusters. The data was visualized using java TreeView. The average enrichment per cluster was plotted using R.
Human RNA-seq analysis
Stranded RNA-seq data from K562 cells (encode consortium - ENCSR000AEM) and Hs68 cells23 was mapped to the human genome (UCSC version hg38) using STAR (version 2.4) with the following parameters: --outFilterMultimapNmax 20 --alignSJoverhangMin 8 -- alignSJDBoverhangMin 1 --outFilterMismatchNmax 999 --outFilterMismatchNoverLmax 0.04 --alignIntronMin 20 --alignIntronMax 1000000 --alignMatesGapMax 1000000 -- outFilterIntronMotifs RemoveNoncanonical. Gene expression was quantified using cufflinks (version 2.2.1) against the reference gencode v22 transcriptome and normalized across all datasets using cuffnorm (version 2.2.1).
Supplementary Material
One Sentence Summary.
PRC1 components bind to a large set of genes in the absence of H3K27me3 in human and Drosophila and suppress the formation of larval tumors in a PRC2-independent manner.
Acknowledgments
We thank Frédéric Bantignies for his contribution on Immuno-DNA-FISH experiments, Solène Roux and Caroline Jacquier for producing and testing anti-SU(Z)12 rabbit antibodies. We thank the Montpellier RIO imaging facility for assistance with microscopy and the MGX facility for genomic data production and first-level processing. We are grateful to Judith Kassis for anti-PHO antibodies, Vincenzo Pirrotta and Rick Jones for anti-E(z) antibodies, Jürg Müller for Sce mutant fly lines. We thank the Bloomington Stock Center (Indiana University) and the Developmental Studies Hybridoma Bank (University of Iowa) for fly stocks and antibodies. We thank the laboratory of Jian Wang for the phdel stock. We thank Silvia Perez- Lluch and Montserrat Corominas for providing the RNA-seq data on wild-type eye and wing imaginal discs prior to publication. We are grateful to the Genotoul bioinformatics platform Toulouse Midi-Pyrenees for providing computing and storage resources. Research in the laboratory of G.C. was supported by grants from the European Research Council (ERC-2008-AdG No 232947), the CNRS, the European Network of Excellence EpiGeneSys, the European H2020 EINFRA MuG grant, the Agence Nationale de la Recherche, the Laboratory of Excellence EpiGenMed, and the Fondation ARC pour la Recherche sur le Cancer. A-M.M. was supported by a grant of the Canceropole Grand Sud-Ouest N°2012-E10. B.B. is supported by the Sir Henry Wellcome Postdoctoral Fellowship (WT100136MA). V.L is supported by a doctoral fellowship from the labex EpiGenMed. B.S. is supported by INSERM. S.S is supported by a post-doctoral fellowship from the labex EpiGenMed.
Footnotes
URLs
ChIP followed by DNA sequencing (ChIP-Seq) was performed by the sequencing platform MGX (Montpellier GenomiX: http://www.mgx.cnrs.fr/).
To Visualize and present ChIPseq data, we used IGV (Integrative Genomics Viewer) 45,46 (https://www.broadinstitute.org/igv/).
ChIP-Seq signals were quantified using EaSeq version 1.01 (http://easeq.net).
Density plots showing the average enrichment of ChIP-Seq tracks were obtained using Bioconductor package “seqplots” (http://github.com/przemol/seqplots ).
The FISH protocol is available at http://www.epigenesys.eu/en/protocols/fluorescence-microscopy/182-two-colour-fluorescent-in-situ-dna-hybridization-on-whole-mount-drosophila-embryos-and-larval-imaginal-discs.
Accession codes
Drosophila ChIP-Seq and RNA-Seq data are deposited to the Gene Expression Omnibus (GEO) repository under the accession number GSE74080.
Author Contributions
V.L. and A.D. performed developmental genetics, molecular biology, microscopy and genomics experiments. A.T. and B.B. performed bioinformatics analysis, B.S and S.S. performed molecular biology experiments, AM.M. performed developmental genetics and molecular biology experiments, AM.M, V.L, A.D. and G.C. wrote the manuscript and all authors commented and corrected the manuscript.
Competing Financial Interests Statement
The authors declare that they have no competing financial interests.
References and Notes
- 1.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–96. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 2.Muller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 3.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 4.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 6.Schuettengruber B, et al. Cooperativity, Specificity, and Evolutionary Stability of Polycomb Targeting in Drosophila. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.08.072. [DOI] [PubMed] [Google Scholar]
- 7.Schuettengruber B, et al. Functional Anatomy of Polycomb and Trithorax Chromatin Landscapes in Drosophila Embryos. PLoS Biol. 2009;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–75. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 9.Classen AK, Bunker BD, Harvey KF, Vaccari T, Bilder D. A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK-STAT signaling. Nat Genet. 2009;41:1150–5. doi: 10.1038/ng.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez AM, et al. Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nat Genet. 2009;41:1076–82. doi: 10.1038/ng.414. [DOI] [PubMed] [Google Scholar]
- 11.Beuchle D, Struhl G, Muller J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development. 2001;128:993–1004. doi: 10.1242/dev.128.6.993. [DOI] [PubMed] [Google Scholar]
- 12.Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–9. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurzhals RL, Tie F, Stratton CA, Harte PJ. Drosophila ESC-like can substitute for ESC and becomes required for Polycomb silencing if ESC is absent. Dev Biol. 2008;313:293–306. doi: 10.1016/j.ydbio.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips MD, Shearn A. Mutations in polycombeotic, a Drosophila polycomb-group gene, cause a wide range of maternal and zygotic phenotypes. Genetics. 1990;125:91–101. doi: 10.1093/genetics/125.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- 16.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–66. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Oktaba K, et al. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell. 2008;15:877–89. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Kwong C, et al. Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet. 2008;4:e1000178. doi: 10.1371/journal.pgen.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herz HM, et al. Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila. Mol Cell Biol. 2012;32:1683–93. doi: 10.1128/MCB.06503-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez L, et al. The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development. 2012;139:117–27. doi: 10.1242/dev.074450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol Cell. 2012;45:255–62. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pemberton H, et al. Genome-wide co-localization of Polycomb orthologs and their effects on gene expression in human fibroblasts. Genome Biol. 2014;15:R23. doi: 10.1186/gb-2014-15-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaaf CA, et al. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet. 2013;9:e1003560. doi: 10.1371/journal.pgen.1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bunker BD, Nellimoottil TT, Boileau RM, Classen AK, Bilder D. The transcriptional response to tumorigenic polarity loss in Drosophila. Elife. 2015;4 doi: 10.7554/eLife.03189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mummery-Widmer JL, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–92. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munoz-Descalzo S, Tkocz K, Balayo T, Arias AM. Modulation of the ligand-independent traffic of Notch by Axin and Apc contributes to the activation of Armadillo in Drosophila. Development. 2011;138:1501–6. doi: 10.1242/dev.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher KH, Wright VM, Taylor A, Zeidler MP, Brown S. Advances in genome-wide RNAi cellular screens: a case study using the Drosophila JAK/STAT pathway. BMC Genomics. 2012;13:506. doi: 10.1186/1471-2164-13-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregory SL, et al. A Drosophila overexpression screen for modifiers of Rho signalling in cytokinesis. Fly (Austin) 2007;1:13–22. doi: 10.4161/fly.3806. [DOI] [PubMed] [Google Scholar]
- 30.Grossniklaus U, Paro R. Transcriptional Silencing by Polycomb-Group Proteins. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–42. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 32.Martin-Subero JI, Koppens M, van Lohuizen M. Context-dependent actions of Polycomb repressors in cancer. Oncogene. 2015 doi: 10.1038/onc.2015.195. [DOI] [PubMed] [Google Scholar]
- 33.Piunti A, et al. Polycomb proteins control proliferation and transformation independently of cell cycle checkpoints by regulating DNA replication. Nat Commun. 2014;5:3649. doi: 10.1038/ncomms4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–56. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vidal M. Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. Int J Dev Biol. 2009;53:355–70. doi: 10.1387/ijdb.082690mv. [DOI] [PubMed] [Google Scholar]
- 36.Morey L, Aloia L, Cozzuto L, Benitah SA, Di Croce L. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep. 2013;3:60–9. doi: 10.1016/j.celrep.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 37.Farcas AM, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloet SL, et al. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat Struct Mol Biol. 2016;23:682–90. doi: 10.1038/nsmb.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oguro H, et al. Lethal myelofibrosis induced by Bmi1-deficient hematopoietic cells unveils a tumor suppressor function of the polycomb group genes. J Exp Med. 2012;209:445–54. doi: 10.1084/jem.20111709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Negre N, et al. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown JL, Fritsch C, Mueller J, Kassis JA. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;130:285–94. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- 42.Strutt H, Paro R. The polycomb group protein complex of Drosophila melanogaster has different compositions at different target genes. Mol Cell Biol. 1997;17:6773–83. doi: 10.1128/mcb.17.12.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–8. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol. 2016;23:349–57. doi: 10.1038/nsmb.3180. [DOI] [PubMed] [Google Scholar]
- 48.Negre N, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–31. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 54.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ku M, et al. Genomewide analysis of PRCl and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:el000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.