Abstract
Using the lanthanide ion praseodymium, Pr(III), metallated ion formation and electron transfer dissociation (ETD) were studied for 25 biological and model acidic peptides. For chain lengths of seven or more residues, even highly acidic peptides that can be difficult to protonate by electrospray ionization will metallate and undergo abundant ETD fragmentation. Peptides composed of predominantly acidic residues form only the deprotonated ion, [M + Pr − H]2+; this ion yields near complete ETD sequence coverage for larger peptides. Peptides with a mixture of acidic and neutral residues, generate [M + Pr]3+, which cleaves between every residue for many peptides. Acidic peptides that contain at least one residue with a basic side chain also produce the protonated ion, [M + Pr + H]4+; this ion undergoes the most extensive sequence coverage by ETD. Primarily metallated and non-metallated c- and z-ions form for all peptides investigated. Metal adducted product ions are only present when at least half of the peptide sequence can be incorporated into the ion; this suggests that the metal ion simultaneously attaches to more than one acidic site. The only site consistently lacking dissociation is at the N-terminal side of a proline residue. Increasing peptide chain length generates more backbone cleavage for metal-peptide complexes with the same charge state. For acidic peptides with the same length, increasing the precursor ion charge state from 2+ to 3+ also leads to more cleavage. The results of this study indicate that highly acidic peptides can be sequenced by ETD of complexes formed with Pr(III).
Keywords: Acidic peptide, electron transfer dissociation, lanthanide metal adduction, sequence effects, peptide size effects, praseodymium ions
Introduction
Electron transfer dissociation (ETD) and electron capture dissociation (ECD) are important tandem mass spectrometry (MS/MS) techniques for the analysis of peptides and proteins.[1–5] These processes result in cleavage along the peptide backbone that is usually more uniform than collision induced dissociation (CID)[6,7] and infrared multi-photon dissociation (IRMPD),[8] which favor lower energy pathways. In addition, post-translational modifications such as phosphorylation are preserved during ETD and ECD.[9–14] In both processes, an electron is added to a multiply charged cation creating a charge reduced radical species that undergoes a non-ergodic process to form c- and z-ions by N-Cα bond cleavage along the peptide backbone.[1–5,15–17] A major drawback to both methods, however, is that they generally require multiply positively charged precursor ions.[1,4,18–21] Upon electron capture, an ion with a single positive charge would be reduced to a neutral molecule that cannot be analyzed by mass spectrometry.[22] Formation of multiply charged ions is advantageous because MS/MS fragmentation efficiency and sequence information often increase with increasing precursor ion charge state.[2,21,23–28] In addition higher charges on peptides and proteins generally require less energy to initiate fragmentation.[23–26] Multiple charging can shift the mass-to-charge ratios (m/z) of the ions to a range of the spectrum with optimal resolution[29] and can increase signal intensity in mass spectrometers that use image current detection.[30,31] However, certain peptides, such as acidic peptides, may not form doubly or even singly positively charged ions by electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI),[32–35] making analysis by ECD or ETD challenging. While deprotonated peptides (i.e., negative ions) can be analyzed by dissociation techniques such as electron detachment (EDD),[36] electron-induced (EID),[37] and negative electron transfer dissociation (NETD),[38] these processes generally suffer from low product ion intensities that are most notable when an electron interacts with a peptide anion (i.e., EDD and EID).
Peptides containing acidic amino acid residues are important in many biological processes. For example, acidic peptides are involved in bone growth,[39–42] stimulation of hormone releasing factor secretion,[43] blood coagulation[44,45] and the circulatory system,[46] development of the brain,[47] carbohydrate metabolism,[48–50] and differentiation of cells involved in immune response.[51,52] Potential vaccines for malaria[53] and HIV[54–56] have utilized acidic peptides.
One method of ionizing peptides that do not readily protonate is to form positively charged ions by addition of a metal ion. Sequence information from the resulting metallated peptide cations has been obtained in studies employing ECD[57–68] and ETD.[69–72] Flick and Williams[68] demonstrated the ability of trivalent lanthanide metal ions to produce multiply positively charged ions with peptides. During a recent ETD study[72] involving the addition of trivalent lanthanide metal cations to acidic peptides, we also noted that multiply charged metallated ions form by ESI even in the absence of a highly basic arginine amino acid residue (gas-phase basicity of arginine = 1006.6 kJ/mol[73]). In this prior study, metal-peptide complexes form with as much as a 4+ charge, [M + Pr + H]4+, simultaneously incorporating a trivalent metal cation and an extra hydrogen. Extensive peptide backbone cleavage and sequence information were obtained from ETD of these 4+ ions. However, our initial study focused primarily on one peptide and its analogs, so limited observations were made regarding effects of peptide size and sequence on ETD of these metallated peptide complexes.
Structural parameters including amino acid composition and chain length have been shown to affect electron-based dissociation of protonated peptides and metal-adducted peptide complexes.[62,69,74–76] In an ECD study involving model peptides containing two arginine residues, Chan and coworkers[74] found that [M + 2H]2+ produced less backbone cleavage for peptides with increased numbers of glutamic acid and asparagine. These researchers also noted that while a greater number of glutamic acid residues led to suppressed cleavage for [M + 2H]2+, [M + 3H]3+ underwent more dissociation and provided more sequence coverage. In two separate studies,[62,75] Chan and coworkers noted that increasing peptide chain length suppressed backbone cleavage for ECD of [M + 2H]2+ and [M + Cu]2+. In contrast, ECD of [M + 3H]3+ and [M + Cu + H]3+ produced abundant backbone cleavage for the same peptides. Vachet and Dong[69] investigated Cu(II)-peptide complexes and determined that an increase in the number of copper binding motifs such as histidine, aspartic acid, glutamic acid and methionine promoted cleavage by ETD. Wysocki, Coon, and coworkers[76] performed ETD on thousands of multiply protonated peptides; they observed that the intensities of the c-series changed as the lysine, arginine, and glutamic acid residues were positioned further away from the C-terminus. These researchers also noted that the effect of the position of the basic or acidic amino acid residue on ETD becomes less significant with a more highly charged precursor ion.
The current study explores the effects of chain length, as well as amino acid position and composition, on ETD of lanthanide metal-adducted acidic peptide ions without highly basic amino acid residues. Both biological and model peptides were employed. In our previous study,[72] the entire lanthanide series (except radioactive promethium) was investigated. Praseodymium, Pr(III), was found to give high quality and reproducible ETD spectra that are not complicated by the presence of multiple metal isotopes. Thus, Pr(III) was selected to represent the lanthanide series in the present work.
Experimental
All experiments were performed on a Bruker (Billerica, MA, USA) HCTultra PTM Discovery System high capacity quadrupole ion trap (QIT) mass spectrometer. Mixtures of metal salt and acidic peptide in a 10:1 Pr(III):peptide molar ratio were infused into the ESI source at a flow rate of 3 μL/min using a KD scientific (Holliston, MA, USA) syringe pump. Final peptide concentrations were 5 μM in acetonitrile:water at a 50:50 volume ratio. At the ESI source, the needle was held at ground. The capillary entrance, the stainless steel endplate, and the capillary entrance cap were set at a high voltage of −3.5 kV. Nitrogen drying gas was heated to 260°C and flowed at 5–10 L/min. Nitrogen was also the nebulizer gas and the pressure was optimized between 5 and 10 psi. All spectra shown are the result of signal averaging 200 scans.
The reagent anion for ETD experiments, fluoranthene, was generated in a negative chemical ionization source (nCI) with methane serving as the nCI reagent gas. The reagent anion was accumulated for 8–12 ms. The ion charge control (ICC) value was 300, 000–400, 000 to maximize electron transfer. The lower end m/z cutoff was set to 120 with anion/cation reaction times in the range of 180–250 ms. The “smart decomposition” function was enabled to facilitate further dissociation of any charge reduced (ETnoD) ions. Smart decomposition uses very low energy CID (resonant excitation) to overcome attractive forces (e.g., hydrogen or noncovalent bonding) that may hold peptide fragments together after the electron transfer process. Smart decomposition enhances observed ETD product ion formation. Previous work provides a more detailed explanation of smart decomposition.[72]
The acidic peptides were purchased from various vendors. Gastrin 1(1–14), hirudin(54–65) (desulfated), and adrenocorticotrophic hormone (ACTH(22–39)) were obtained from Bachem Americas (Torrance, CA, USA); human epidermal growth factor receptor peptide ((EGF)985–996) from Alfa Aesar (Ward Hill, MA, USA); casein kinase II peptide substrate from Sigma-Aldrich (St. Louis, MO, USA); and low molecular weight chromium binding (LMWCr) peptide from Gen Script (Piscataway, NJ, USA). Model peptides were synthesized in our laboratory with an Advanced ChemTech Model 90 peptide synthesizer (Louisville, KY, USA) using standard Fmoc protocol.[77] Praseodymium(III) nitrate salt was purchased from Alfa Aesar. A Barnstead (Dubuque, IA, USA) E-pure system was used to produce ultrapure Milli-Q 18 MΩ water.
Results and Discussion
The established peptide cleavage nomenclature of Roepstorff and Fohlman[78] as modified by Biemann[79] is used in labeling of spectra to help identify the number of hydrogen incorporated into product ions formed by ETD and CID. A prime symbol to the right indicates addition of hydrogen (i.e., zn/ = [zn + H]+) and to the left indicates loss of hydrogen (i.e., //zn = [zn – 2H]+). In the figures, the following colors are used to label the ions: non-metallated z-ions = red, non-metallated c-ions = blue, metal adducted z-ions = bright pink, metal adducted c-ions = green, and metal adducted y-ions = brown. Undissociated precursor ion is labeled in purple with a large purple diamond arrow head. Neutral loss products and electron transfer without dissociation (ETnoD) ions are labeled in black. ECD[1,2,5,15,17] and ETD[4,16] typically yield c- and z-ions that correspond to N-Cα peptide bond cleavage.
Biological peptides
Several biological acidic peptides were employed to investigate the effects of size and sequence on Pr(III) cationization by ESI and on ETD product ion formation. The peptides and their sequences are shown in Table 1. They contain a variety of amino acid residues with chain lengths ranging from seven to eighteen residues.
Table 1.
ESI ion formation and corresponding ETD product ions for acidic biological peptides cationized by Pr(III).
| Peptide name | Number of residues | Number of acidic residuesa | Sequenceb | Metallated ion formed by ESI | cn-ions | zn- ions |
|---|---|---|---|---|---|---|
| Low molecular weight chromium binding (LMWCr) peptide | 7 | 6 | E1E2E3E4GD6D7 | [M + Pr – H]2+ | [/cn + Pr]+, n = 4–6 | [/zn + Pr]+, n = 5–6 |
| Casein kinase II peptide substrate | 12 | 7 | RRRAD5D6SD8D9D10D11D12 | [M + Pr]3+ | cn//+, n = 2 [cn + Pr]+, n = 10, 11 [cn + Pr]2+, n = 10, 11 |
–c |
| [M + Pr + H]4+ | cn//+, n = 2–9 [cn + Pr]+, n = 9–11 [cn + Pr]2+, n = 9, 10 |
– | ||||
| Human epidermal growth factor receptor peptide(985–996), EGF(985–996) | 12 | 5 | D1VVD4AD6E7YLIPQ12 | [M + Pr – H]2+ | [/cn + Pr]+, n = 7–9, 11 | [/zn + Pr]+, n = 7–11 |
| [M + Pr]3+ | cn//+, n = 2–7 [/cn + Pr]+, n = 8–9, 11 |
zn/+, n = 3–5 [/zn + Pr]+, n = 11 [/zn + Pr]2+, n = 6–11 |
||||
| [M + Pr + H]4+ | cn//+, n = 2–6 [/cn + Pr]+, n = 6–9, 11 [/cn + Pr]2+, n = 9 |
zn/+, n = 1, 3–5 [/zn + Pr]+, n = 6–11 [/zn + Pr]2+, n = 11 |
||||
| Hirudin(54–65) | 12 | 6 | GD2FE4E5IPE8E9YLQ12 | [M + Pr – H]2+ | [/cn + Pr]+, n = 7–11 | [/zn + Pr]+, n = 7–11 |
| [M + Pr]3+ | cn//+, n = 3,4 [/cn + Pr]+, n = 7–11 [/cn + Pr]2+, n = 11 |
zn/+, n = 2–5 [/zn + Pr]+, n = 7–11 |
||||
| [M + Pr + H]4+ | cn//+, n = 2–5 [/cn + Pr]+, n = 7–11 |
zn/+, n = 2–5 [/zn + Pr]+, n = 7–11 |
||||
| Human gastrin I (1–14) | 14 | 6 | pEGPWLE6E7E8E9E10AYGW14 | [M + Pr – H]2+ | [/cn + Pr]+, n = 8–13 | [/zn + Pr]+, n = 8–11, 13 |
| [M + Pr]3+ | cn//+, n = 4–6 [/cn + Pr]+, n = 8–13 [/cn + Pr]2+, n = 11–13 |
zn/+, n = 1–6 [/zn + Pr]+, n = 7–11, 13 |
||||
| Adrenocorticotropic hormone, ACTH (22–39) | 18 | 6 | VYPNGAE7D8E9SAE12AFPLE17F18 | [M + Pr]3+ | cn//+, n = 3–11 [/cn + Pr]+, n = 9, 12, 13, 15–17 [/cn + Pr]2+, n = 15–17 |
[/zn + Pr]+, n = 9–15, 17 |
| [M + Pr + H]4+ | cn//+, n = 3–11 [/cn + Pr]+, n = 9–13, 15–17 [/cn + Pr]2+, n = 9, 15, 16 |
zn/+, n = 2, 3, 5–8 [/zn + Pr]+, n = 9–15, 17 |
Acidic residues refers to the number of aspartic acid (D), glutamic acid (E), and C-terminal residues in the peptide sequence. If an aspartic acid or glutamic acid residue is located on the C-terminus, then it is counted as only one acidic residue.
Bold font is used to indicate acidic residues. A number immediately following an acidic residue indicates its position in the peptide sequence.
- means that no ions were observed.
As the data in Table 1 indicates, two of the biological peptides, hirudin(54–65) and EGF(985–996) produced all three metallated ions by ESI: [M + Pr − H]2+, [M + Pr]3+, and [M + Pr + H]4+. These ions were also observed in our prior work with fibrinopeptide B.[72] The most acidic species studied, LMWCr peptide, generates no 3+ and 4+ ions but only [M + Pr – H]2+. The facile deprotonation is consistent with the fact that six of the residues of this heptapeptide are acidic. In contrast, casein kinase II substrate and ACTH(22–39) do not produce the deprotonated ion, but only the 3+ and 4+ ions, with [M + Pr + H]4+ occurring in greatest abundance for both peptides. Although casein kinase II substrate contains seven aspartic acid residues and an acidic C-terminus, there are three highly basic arginine residues at its N-terminus that can accept a proton. Hirudin(54–65), EGF(985–996), and ACTH(22–39) are still able to form [M + Pr + H]4+ in the absence of a highly basic arginine, lysine, or histidine residue. This may be attributed to the fact that these peptides contain proline (gas-phase basicity of proline = 886.0 kJ/mol[73,80]) that is the fifth most basic amino acid[81] and can sometimes accept a proton.[82]
The most extensive sequence coverage is obtained for ETD of [M + Pr + H]4+. Figure 1 shows ETD spectra of [M + Pr + H]4+ for EGF(985–996), hirudin(54–65), and casein kinase II substrate, while Supplemental Figure S1 gives the spectrum from ACTH(22–39). Efficient fragmentation with a high signal-to-noise ratio allows for easy identification of product ions. A mixture of complementary non-metallated and metallated c- and z-ions form, except for casein kinase II substrate that yields only c-ions, as seen in Figure 1(c). Most ions are singly charged although there are a few doubly charged ions. The c-series are cn//+ and [/cn + Pr]+, while the z-series are zn/+ and [/zn + Pr]+. All non-metallated c-ions have two extra hydrogens, while non-metallated z-ions contain one. Metallated c- and z-ions lose one hydrogen relative to the direct backbone cleavage product ion. For some peptides, both metallated and non-metallated ions can form at the same cleavage site. For example, in Figure 1(a), product ions with and without Pr(III) form between D6 and E7 for EGF(985-996). Cleavage occurs at almost every residue for the 4+ ions, except between R1 and R2 for casein kinase II substrate and except N-terminal to proline residues for hirudin(54–65), EGF(985–996), and ACTH(22–39). The side chain of proline forms a ring that incorporates the backbone, which prevents dissociation of the peptide into two pieces following cleavage of the N-Cα bond. Lack of cleavage N-terminal to proline has previously been observed in ECD and ETD studies of protonated peptides.[76,83–85]
Figure 1.
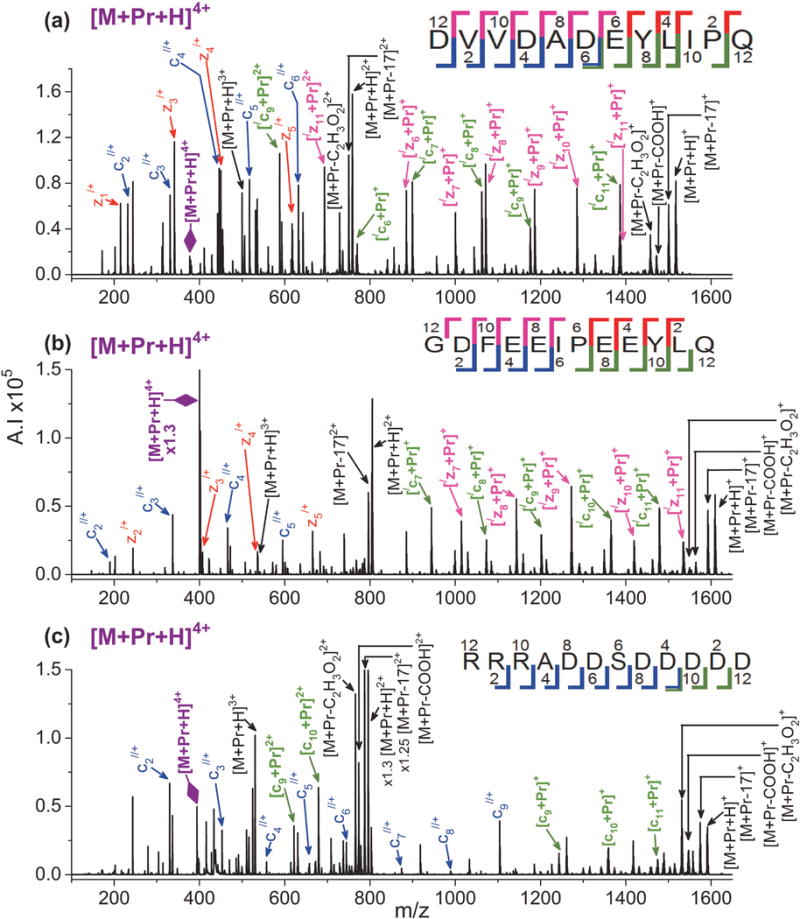
ETD mass spectra of [M + Pr + H]4+ for (a) EGF(985–996), (b) hirudin(54–65), and (c) casein kinase II substrate. In the figures, the following colors are used to label the ions: non-metallated z-ions = red, non-metallated c-ions = blue, metal adducted z-ions = bright pink, metal adducted c-ions = green, and metal adducted y-ions = brown. Undissociated precursor ion is labeled in purple with a large purple diamond arrow head. Neutral loss products and electron transfer without dissociation (ETnoD) ions are labeled in black.
For all the biological peptides, ETD on [M + Pr]3+ yields a mix of metallated and non-metallated c- and z-ions. Most are singly charged, but some doubly charged ETD product ions form as well. The c- and z-series are the same as those observed for [M + Pr + H]4+. Figure 2 shows the ETD spectra of [M + Pr]3+ for EGF(985–996), hirudin(54–65), and gastrin I(1–14), while the spectra for ACTH(22–39) and casein kinase II substrate appear in Supplemental Figures S2 and S3, respectively. For 3+ ions from five of the biological peptides, cleavage occurs at every residue (except N-terminal to proline) and the peptides can be sequenced. Interestingly, ETD of [M + Pr]3+ for casein kinase produces cleavages at only three sites (see Supplemental Figure S3), although [M + Pr + H]4+ (Figure 1(c)) cleaves at almost every residue. This may indicate that the three N-terminal arginine residues are sequestering the metal ion.
Figure 2.
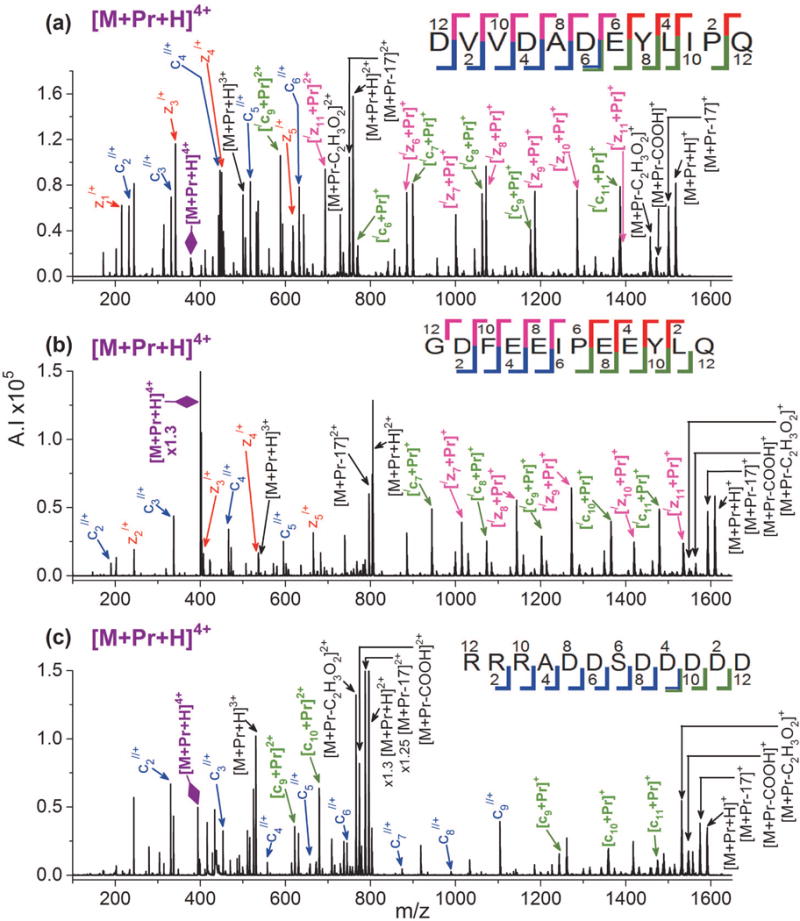
ETD mass spectra of [M + Pr]3+ for (a) EGF(985–996), (b) hirudin(54–65), and (c) gastrin I(1–14). Refer to the Figure 1 caption for an explanation of the color codes.
The least amount of sequence informative fragmentation is obtained from [M + Pr – H]2+. As shown in Figure 3, only metal adducted c- and z-ions form. All product ions are singly charged and lack a hydrogen, which can be explained by the fact that the precursor ion is already deprotonated. Our previous study with fibrinopeptide B found similar decreased product ion formation for ETD of [M + Pr – H]2+ compared to [M + Pr]3+ and [M + Pr + H]4+.[72] Cleavage between E3 and E4 is missing for LMWCr peptide and between E7 and E8 for human gastrin I(1–14). A similar lack of cleavage between adjacent glutamic acid residues was observed in the lower charged state ion (2+) in an ECD study involving trivalent lanthanide ions[68] and in our fibrinopeptide B study.[72] This effect appears to be most pronounced for central residues in a series of acidic residues and may relate to the metal ion bridging between acidic sites.
Figure 3.
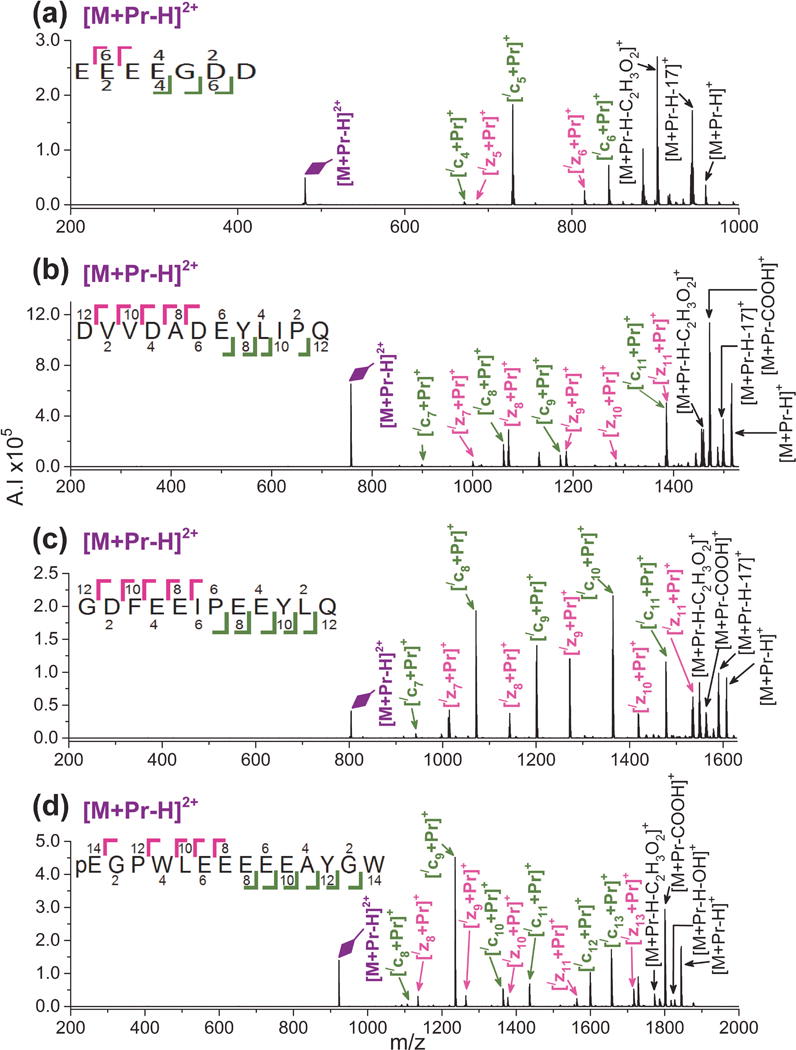
ETD mass spectra of [M + Pr – H]2+ for (a) LMWCr peptide, (b) EGF(985–996), (c) hirudin(54–65), and (d) human gastrinI(1–14). Refer to the Figure 1 caption for an explanation of the color codes.
The types of amino acid residues present in a peptide sequence can affect the ions formed by ETD. While casein kinase II substrate has a twelve residue sequence like EGF(985–996) and hirudin(54–65), the presence of the three highly basic arginine residues at its N-terminus leads to the formation of only c-ions by ETD for [M + Pr]3+ and [M + Pr + H]4+. This is interesting because the one highly basic arginine residue located at the C-terminus in fibrinopeptide B did not lead to its formation of only C-terminal product ions.[72] Instead, for fibrinopeptide B, members of both the c- and the z-series form for ETD of [M + Pr – H]2+, [M + Pr]3+, and [M + Pr + H]4+.
The ETD spectra of [M + Pr + H]4+, [M + Pr]3+, and [M + Pr – H]2+ are much more sequence informative than CID spectra of these ions. Example CID spectra are shown in Supplemental Figures S4 and S5. Typically, b- and y- ions form by CID, but spectra contain more neutral losses that complicate interpretation. The CID spectra also generally have lower product ion intensities and signal-to-noise ratios.
Model peptides
Several model peptides were employed to investigate the effects of peptide structural features on ETD of acidic peptides adducted with Pr(III). Factors under consideration include chain length, type and number of acidic amino acid residues, and the location of acidic residues. The heptapeptides investigated were XAAAAAA, AAAXAAA, AAAAAAX, XAAXAAX, and XXXXXXX (X7), where A is alanine and X is aspartic acid (D) or glutamic acid (E) residues. In addition, EEEEEE (E6), EEEEEEEE (E8), and DDDDDDDDDD (D10) were studied. For the model peptides, metallated precursor ion production by ESI and ETD product ion information is given in Table 2.
Table 2.
ESI ion and corresponding ETD product ions for acidic model peptides cationized by Pr(III).
| Peptide sequencea | Number of residues | Number of acidic residuesb | Metallated ion formed by ESI | cn-ions | zn-ions | yn-ions |
|---|---|---|---|---|---|---|
| D1AAAAAA7 | 7 | 2 | [M + Pr]3+ | [/cn + Pr]+, n = 4–6 [/cn + Pr]2+, n = 6 |
zn/+, n = 2 [/zn + Pr]+, n = 5, 6 [/zn + Pr]+, n = 6 |
[yn + Pr]+, n = 4–6 [yn + Pr]2+, n = 6 |
| E1AAAAAA7 | 7 | 2 | [M + Pr]3+ | [/cn + Pr]+, n = 4–6 [/cn + Pr]2+, n = 5, 6 |
[/zn + Pr]+, n = 5, 6 [/zn + Pr]2+, n = 6 |
[yn + Pr]+, n = 4–6 [yn + Pr]2+, n = 6 |
| AAAD4AAA7 | 7 | 2 | [M + Pr]3+ | [/cn + Pr]+, n = 4–6 [/cn + Pr]2+, n = 6 |
zn/+, n = 2 [/zn + Pr]+, n = 4–6 |
– |
| AAAE4AAA7 | 7 | 2 | [M + Pr]3+ | [/cn + Pr]+, n = 4–6 [/cn + Pr]2+, n = 5, 6 |
zn/+, n = 2 [/zn + Pr]+, n = 4–6 [/zn + Pr]2+, n = 6 |
– |
| AAAAAAD7 | 7 | 1 | –c | |||
| AAAAAAE7 | 7 | 1 | [M + Pr – H]2+ | – | [/zn + Pr]+, n = 5–6 | – |
| D1AAD4AAD7 | 7 | 3 | [M + Pr]3+ | cn//+, n = 1–3 [/cn + Pr]+, n = 4–6 [/cn + Pr]2+, n = 6 |
[/zn + Pr]+, n = 5, 6 [/zn + Pr – CO]+, n = 4 |
– |
| E1AAE4AAE7 | 7 | 3 | [M + Pr]3+ | cn//+, n = 1–3 [/cn + Pr]+, n = 4–6 [/cn + Pr]2+, n = 6 |
[/zn + Pr]+, n = 4–6 | – |
| E1E2E3E4E5E6 (E6) | 6 | 6 | [M +Pr – H]2+ | [/cn + Pr]+, n = 4, 5 | [/zn + Pr]+, n = 5 |
– |
| D1D2D3D4D5D6D7 (D7) | 7 | 7 | [M + Pr – H]2+ | cn//+, n = 6 [/cn + Pr]+, n = 5, 6 |
[/zn +Pr]+, n = 6 [/zn + Pr – C2H3O2]+, n = 6 |
– |
| E1E2E3E4E5E6E7 (E7) | 7 | 7 | [M + Pr – H]2+ | [/cn + Pr]+, n = 5, 6 | [/zn + Pr]+, n = 5, 6 | – |
| E1E2E3E4E5E6E7E8 (E8) | 8 | 8 | [M + Pr – H]2+ | [/cn + Pr]+, n = 5–7 | [/zn + Pr]+, n = 5–7 | – |
| D1D2D3D4D5D6D7D8D9D10 (D10) | 10 | 10 | [M + Pr – H]2+ | [/cn + Pr]+, n = 6–9 | [/zn + Pr]+, n = 6–9 | – |
Bold font is used to indicate acidic residues. A number immediately following an acidic residue indicates its position in the peptide sequence.
Acidic residues refers to the number of aspartic acid (D), glutamic acid (E), and C-terminal residues in the peptide sequence. If an aspartic acid or glutamic acid residue is located on the C-terminus, then it is counted as only one acidic residue.
- means that no ETD ions were observed.
Six of the heptapeptides (DAAAAAA, EAAAAAA, AAADAAA, AAAEAAA, DAADAAD, and EAAEAAE) produce predominantly [M + Pr]3+ by ESI. In addition, [M + Pr – H]2+ forms in low abundances but could not be isolated for further analysis by ETD. The other peptides yield predominantly [M + Pr – H]2+. The formation of only [M + Pr – H]2+ for peptides E6, D7, E7, E8, and D10 can be attributed to the fact that the sequence consists of only acidic side chains that preferentially deprotonate. Likewise, LMWCr peptide is also highly acidic and only forms [M + Pr – H]2+. A peptide sequence containing mostly aspartic or glutamic acid residues would undergo abundant deprotonation and generate [M + Pr – H]2+ despite the addition of a trivalent metal cation. None of the model peptides produce [M + Pr + H]4+, suggesting that the N-terminus as the only basic site is not sufficient to sequester a proton.
The heptapeptide AAAAAAD did not adduct Pr(III) despite multiple attempts involving several lots of peptide synthesized in our laboratory. (The peptide did protonate and deprotonate by ESI indicating that it had been correctly synthesized.) In contrast, AAAAAAE metallated readily by ESI to form [M + Pr − H]2+. Previous studies suggest the possibility of the lanthanide metal ion simultaneously coordinating to more than one acidic site.[68,72] Since the aspartic or glutamic acid residues are at the C-terminus of these two peptides, their side chain and C-terminal carboxylic acid sites are in close proximity but have different orientations.[86] The shorter side chain of the aspartic acid may limit its ability to achieve a conformation that will allow Pr(III) to coordinate to both carboxylic acid groups. Glutamic acid, however, has an extra methylene group on the side chain that may allow for greater flexibility in achieving a di-coordinated Pr(III)-AAAAAAE complex.
Several peptides with less than seven amino acid residues were investigated: DDD, EEE, AAAAEE, AAAADD, AAAEAA, and DAAADA. The tripeptides EEE and DDD did not adduct Pr(III). The hexapeptides formed predominantly [M + Pr – H]2+, but ETD only generates one or two cleavage products, which is not useful for sequencing the peptides. Supplemental Table 1 lists the ions formed by ESI and ETD product ions for these tri- and hexapeptides.
The peptides E6, E7, D7, E8, and D10 were chosen both to investigate ETD fragmentation of highly acidic metallated peptides and to explore the effect of chain length on ETD. Figure 4 shows the ETD spectra of [M + Pr – H]2+ for these peptides. Members of the c- and z-series dominate and all product ions are metal adducted. For E6, only [/cn + Pr]+, n = 4 and 5, and [/z5 + Pr]+ form, as shown in Figure 4(a). Cleavage at only three sites on the peptide backbone limits sequence information. In Figure 4(c), D7 produces the same number of ETD product ions as E6. However, in Figure 4(b), E7 forms one additional ion than E6, [/z5 + Pr]+, thus giving slightly more sequence information. Increasing the peptide chain length from seven to eight residues leads to much more peptide backbone cleavage with only cleavage between the fourth and fifth residues being absent in E8, as seen in Figure 4(d). For the polyglutamic acid peptides, the trend is that sequence informative fragmentation increases in the order E6 < E7 < E8. In Figure 4(e), ETD of [M + Pr – H]2+ for D10 yields cleavage at every residue except between the central acidic residues D5 and D6.
Figure 4.
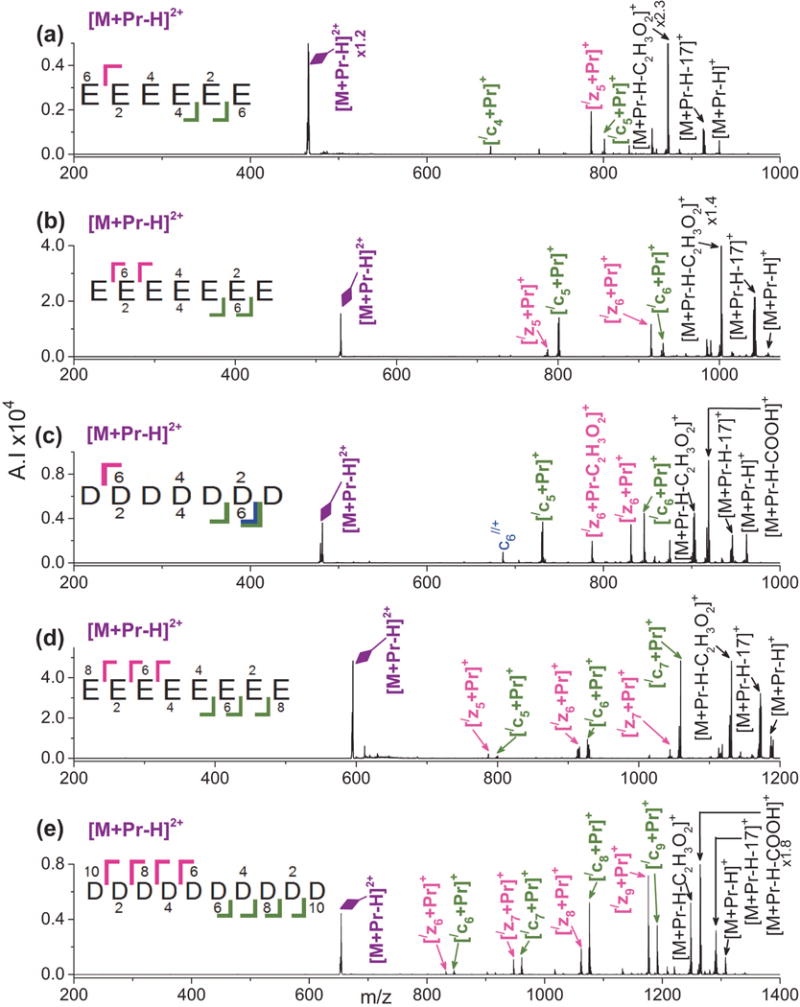
ETD mass spectra of [M + Pr – H]2+ for (a) E6, (b) E7, (c) D7, (d) E8, and (e) D10. Refer to the Figure 1 caption for an explanation of the color codes.
Our results indicate that increasing the peptide chain length increases ETD fragmentation for Pr(III)-peptide ions lacking residues with highly basic side chains. However, peptide sequence must also play a role because Chan and coworkers[75] reported an opposite effect. In an ECD study of protonated ions from peptides containing glutamic acid residues and two highly basic arginine residues, they observed that peptides with longer sequences (15 to 23 residues) underwent less backbone cleavage than peptides with shorter chain lengths of eight residues. This group postulated that an extensive network of hydrogen bonding involving the arginine residues helped to suppress the normal ECD mechanism for N-Cα bond cleavage. In another study involving Cu(II)-peptide complexes of basic peptides by Chan and coworkers,[62] the researchers also observed decreased backbone cleavage for larger peptides.
The heptapeptides DAAAAAA, EAAAAAA, AAADAAA, and AAAEAAA undergo efficient and informative ETD fragmentation. As show in Figure 5(a) and 5(b) for [M + Pr]3+, when the acidic residue is at the N-terminus, a mixture of metallated c-, z-, and y-ions form. Backbone cleavage occurs at every residue. When the acidic residue is in the middle of the sequence, only metal adducted c- and z-ions form with cleavage at every residue, as seen in Figure 5(c) and 5(d). For the heptapeptide with the acidic residue at the C-terminus, ETD of [AAAAAAE + Pr – H]2+ generates only metal-adducted C-terminal ions; i.e., [/zn + Pr]+, n = 5–6. Only two sites on the peptide backbone cleave and neither is adjacent to the acidic residue.
Figure 5.
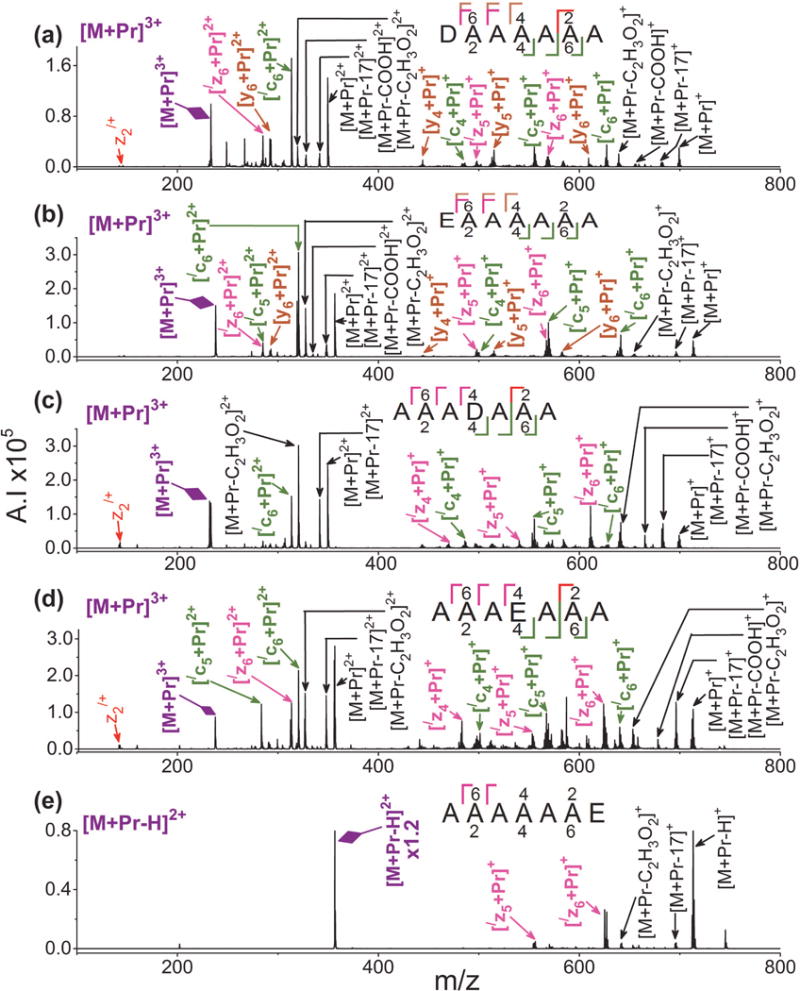
ETD mass spectra of [M + Pr]3+ for (a) DAAAAAA, (b) EAAAAAA, (c) AAADAAA, (d) AAAEAAA, and (e) [M + Pr – H]2+ for AAAAAAE. Refer to the Figure 1 caption for an explanation of the color codes.
As was the case with the biological peptides, [M + Pr]3+ generates more extensive ETD cleavage than [M + Pr – H]2+. For example, ETD of [M + Pr – H]2+ cleaves at a maximum of four sites for the heptapeptide E7, three for D7, and two for AAAAAAE, as shown in Figures 4 and 5. However, ETD of [M + Pr]3+ for the other heptapeptides, DAAAAAA, EAAAAAA, AAADAAA, AAAEAAA, DAADAAD, and EAAEAAE yields cleavage at every residue, as shown in Figures 5 and 6. Significantly less sequence coverage occurs for CID of [M + Pr]3+ for the model peptides. As shown in Supplemental Figure S6 for CID of [M +Pr]3+ for EAAAAAA, cleavage occurs at only three sites along the peptide backbone.
Figure 6.
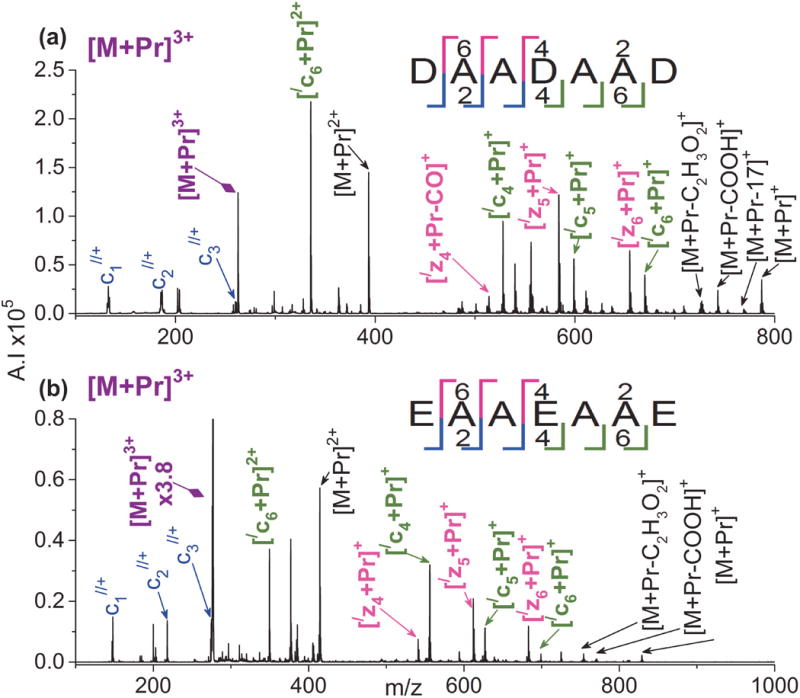
ETD mass spectra of [M + Pr]3+ for (a) DAADAAD and (b) EAAEAAE. Refer to the Figure 1 caption for an explanation of the color codes.
Heptapeptides DAADAAD and EAAEAAE were chosen to investigate the effect of multiple scattered acidic residues on ETD. As shown in Figure 6, ETD on [M + Pr]3+ produces a mixture of metal adducted c- and z-ions as well as non-metallated c-ions for both peptides. Full sequence coverage occurs, with the c-series being complete for both peptides. Thus, the presence of scattered acidic residues appears particularly effective at promoting fragmentation.
For the model peptides, the presence of aspartic acid versus glutamic acid had no noticeable effect on the ETD spectra of [M + Pr]3+. Our group has also found that the identity and position of acidic residues has negligible impact on ETD of doubly protonated acidic model peptides.[87] In addition, there is no evidence of increased cleavage abundance adjacent to the acidic residues. Chan and coworkers[88] also reported no enhanced cleavage C-terminal to aspartic or glutamic acid residues in their ECD work on protonated nonapeptides. However, their spectra suggest that there may be moderately enhanced cleavage on the N-terminal side of both residues. In addition, in an ETD study on thousands of protonated peptides, Wysocki, Coon, and coworkers[76] found that the probability of cleavage N-terminal to glutamic acid increases as the peptide chain length increases; peptide ions with an average length of 17 residues and an average charge of 3.5 showed enhanced cleavage N-terminal to glutamic acid when compared to peptides with an average of 15 residues and a charge of 3.8.
Metal adducted ions only appear after half of the sequence has been incorporated into the ETD product ion irregardless of the size of the peptide chain. For all peptides in this study, no metallated product ion contains less than half of the number of residues in its peptide. For example, for the heptapeptides, [/c4 + Pr]+, [/z4 + Pr]+, or [y4 + Pr]+ are the smallest metallated ETD product ions. This trend can also be observed for all the model heptapeptide spectra of Figure 5. This effect even occurs for longer peptides, including the biological ones. As an example, in Figure 1(a) showing ETD of [M + Pr + H]4+ for the dodecapeptide EGF(985–996), the smallest metal adducted product ions are [/c6 + Pr]+ and [/z6 + Pr]+. Spectra from our previous collision-induced dissociation (CID) study involving Cr(III) adduction to acidic peptides also exhibit this trend.[89] Trivalent metal adduction may involve simultaneous coordination of more than one amino acid residue with the metal cation. Peptide chains may need to contain at least four amino acid residues to properly interact with the metal ions. This is supported by the fact that neither of the tripeptides (D3 and E3) were able to form metal adducted ions by ESI.
The neutrals eliminated from ETD product ions include NH3 (17 Da), OH (17 Da), COOH (45 Da), and C2H3O2 (59 Da). Our QIT mass analyzer does not have sufficient resolution to distinguish between NH3 and OH elimination. For ECD and ETD on protonated peptides, 17 Da typically corresponds to the loss of NH3 from the amino group of the N-terminus.[22,90,91] However, Coon and coworkers[91] reported 17 Da loss as OH elimination from a serine side chain in an N-acetylated peptide; modification of the N-terminus suppressed NH3 loss. The absence or minimal elimination of 17 Da could be indicative of a peptide with a modified N-terminus. Gastrin I(1–14) from the current study and fibrinopeptide B from our gi study[72] both have a cyclized N-terminus with no amino group; these two peptides showed less 17 Da loss in comparison to the other acidic peptides studied. However, these peptides did exhibit some 17 Da elimination, which suggests that loss of OH can occur at aspartic acid and glutamic acid side chains. In addition, for ECD and ETD on protonated peptides, aspartic acid normally yields a side chain loss of COOH (45 Da) and glutamic acid has a loss of C2H3O2 (59 Da).[90–92] These neutral losses were also found in our ETD spectra of Pr(III)-adducted peptides. In general, the presence of the metal ion has negligible impact on the appearance of product ions involving neutral loss.
Conclusions
The current study illustrates the effects of peptide chain length and amino acid composition and position on ETD of acidic biological and model peptides without highly basic amino acid residues. Peptides with greater than six amino acid residues form multiply charged ions by Pr(III). ETD of [M + Pr – H]2+ generally provides more sequence informative fragmentation for longer peptides such as model peptide D10 or human gastrin I(1–14) than shorter peptides such as D7. ETD of [M + Pr]3+ can provide full sequence coverage for peptides containing as little as seven amino acid residues. For the acidic peptides in the current study as well as our previous work involving acidic fibrinopeptide B and analogs,[72] ETD of [M + Pr + H]4+ provides the most sequence informative fragmentation and leads to cleavage at every residue. However, production of this protonated and metallated 4+ precursor ion generally requires that the peptide contain at least one residue with a basic side chain. While only Pr(III) was employed in the current study, our prior work indicates that any of the trivalent lanthanide metal cations except promethium and europium would be suitable for sequencing acidic peptides. Cationization by trivalent lanthanide cations is a useful tool for analysis of acidic biological peptides containing seven or more residues by ETD and ECD.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge support from the National Institute of Health (1R15GM109401A). Financial support for the purchase of the Bruker HCTultra was provided by a National Science Foundation Chemistry Research Instrumentation Facility grant (CHE-0639003). The LMWCr peptide was provided by Dr. John B. Vincent.
Footnotes
Supporting Information
Supporting information may be found in the online version of this article.
References
- 1.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 2.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 3.Ge Y, Lawhorn B, ElNaggar M, Strauss E, Park J, Begley T, McLafferty F. Top down characterization of larger proteins (45 kDa) by electron capture dissociation mass spectrometry RID G-1783-2010. J Am Chem Soc. 2002;124:672–678. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- 4.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zubarev RA, Haselmann KF, Budnik B, Kjeldsen F, Jensen F. Towards an understanding of the mechanism of electron-capture dissociation: A historical perspective and modern ideas. Eur J Mass Spectrom. 2002;8:337–349. [Google Scholar]
- 6.Biemann K. Sequencing of peptides by tandem mass-spectrometry and high-energy collision-induced dissociation. Meth Enzymol. 1990;193:455–479. doi: 10.1016/0076-6879(90)93433-l. [DOI] [PubMed] [Google Scholar]
- 7.Senko MW, Speir JP, McLafferty FW. Collisional activation of large multiply charged ions using Fourier transform mass spectrometer. Anal Chem. 1994;66:2801–2808. doi: 10.1021/ac00090a003. [DOI] [PubMed] [Google Scholar]
- 8.Little DP, Speir JP, Senko MW, O’Connor PB, McLafferty FW. Infrared multiphoton dissociation of large multiply charged ions for biomolecule sequencing. Anal Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 9.Mirgorodskaya E, Roepstorff P, Zubarev RA. Localization of O-glycosylation sites in peptides by electron capture dissociation in a Fourier transform mass spectrometer. Anal Chem. 1999;71:4431–4436. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]
- 10.Shi SDH, Hemling ME, Carr SA, Horn DM, Lindh I, McLafferty FW. Phosphopeptide/phosphoprotein mapping by electron capture dissociation mass spectrometry. Anal Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 11.Kelleher NL, Zubarev RA, Bush K, Furie B, Furie BC, McLafferty FW, Walsh WT. Localization of labile posttranslational modifications by electron capture dissociation: The case of γ-carboxyglutamic acid. Anal Chem. 1999;71:4250–4253. doi: 10.1021/ac990684x. [DOI] [PubMed] [Google Scholar]
- 12.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 13.Stensballe A, Jensen ON, Olsen JV, Haselmann KF, Zubarev RA. Electron capture dissociation of singly and multiply phosphorylated peptides. Rapid Commun Mass Spectrom. 2000;14:1793–2000. doi: 10.1002/1097-0231(20001015)14:19<1793::AID-RCM95>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 14.Zhao C, Sethuraman M, Clavreul N, Kaur P, Cohen RA, O’Connor PB. Detailed map of oxidative post-translational modifications of human P21Ras using Fourier transform mass spectrometry. Anal Chem. 2006;78:5134–5142. doi: 10.1021/ac060525v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zubarev RA, Kruger NA, Fridriksson EK, Lewis MA, Horn DM, Carpenter BK, McLafferty FW. Electron capture dissociation of gaseous multiply-charged proteins is favored at disulfide bonds and other sites of high hydrogen atom affinity. J Am Chem Soc. 1999;121:2857–2862. [Google Scholar]
- 16.Chen X, Tureček F. The arginine anomaly: Arginine radicals are poor hydrogen atom donors in electron transfer induced dissociations. J Am Chem Soc. 2006;128:12520–12530. doi: 10.1021/ja063676o. [DOI] [PubMed] [Google Scholar]
- 17.Syrstad EA, Tureček F. Toward a general mechanism of electron capture dissociation. J Am Soc Mass Spectrom. 2005;16:208–224. doi: 10.1016/j.jasms.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 18.McLafferty FW, Horn DM, Breuker K, Ge Y, Lewis MA, Cerda BA, Zubarev RA, Carpenter BK. Electron capture dissociation of gaseous multiply charged ions by Fourier transform ion cyclotron resonance. J Am Soc Mass Spectrom. 2001;12:245–249. doi: 10.1016/S1044-0305(00)00223-3. [DOI] [PubMed] [Google Scholar]
- 19.Zubarev RA. Electron capture dissociation LC/MS/MS for bottom-up proteomics. Methods Mol Biol. 2009;492:413–416. doi: 10.1007/978-1-59745-493-3_25. [DOI] [PubMed] [Google Scholar]
- 20.Coon JJ. Collisions or electrons? Protein sequence analysis in the 21st century. Anal Chem. 2009;81:3208–3215. doi: 10.1021/ac802330b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2007;6:1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Tureček F, Julian RR. Peptide radicals and cation radicals in the gas phase. Chem Rev. 2013;113:6691–6733. doi: 10.1021/cr400043s. [DOI] [PubMed] [Google Scholar]
- 23.Dongre AR, Jones JL, Somogyi A, Wysocki VH. Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: Evidence for the mobile proton model. J Am Chem Soc. 1996;118:8365–8374. [Google Scholar]
- 24.Kjeldsen F, Giessing AMB, Ingrell CR, Jensen ON. Peptide sequencing and characterization of post-translational modifications by enhanced ion-charging and liquid chromatography electron-transfer dissociation tandem mass spectrometry. Anal Chem. 2007;79:9243–9252. doi: 10.1021/ac701700g. [DOI] [PubMed] [Google Scholar]
- 25.Wells JM, Stephenson JL, Jr, McLuckey SA. Charge dependence of protonated insulin decompositions. Int J Mass Spectrom. 2000;203:A1–A9. [Google Scholar]
- 26.Madsen JA, Brodbelt JS. Comparison of infrared multiphoton dissociation and collision–induced dissociation of supercharged peptides in ion traps. J Am Soc Mass Spectrom. 2009;20:349–358. doi: 10.1016/j.jasms.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Chalkley RJ, Medzihradszky KF, Lynn AJ, Baker PR, Burlingame AL. Statistical analysis of peptide electron transfer dissociation fragmentation mass spectrometry. Anal Chem. 2010;82:579–584. doi: 10.1021/ac9018582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma V, Eng JK, Feldman S, von Haller PD, MacCoss MJ, Noble WS. Precursor charge state prediction for electron transfer dissociation tandem mass spectra. J Proteome Res. 2010;9:5438–5444. doi: 10.1021/pr1006685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall AG, Hendrickson CL. Charge reduction lowers mass resolving power for isotopically resolved electrospray ionization Fourier transform ion cyclotron resonance mass spectra. Rapid Commun Mass Spectrom. 2001;15:232–235. [Google Scholar]
- 30.Marshall AG, Hendrickson CL. High-resolution mass spectrometers. Ann Rev Anal Chem. 2008;1:579–599. doi: 10.1146/annurev.anchem.1.031207.112945. [DOI] [PubMed] [Google Scholar]
- 31.Perry RH, Cooks RG, Noll RJ. Orbitrap mass spectrometry: Instrumentation, ion motion and applications. Mass Spectrom Rev. 2008;27:661–699. doi: 10.1002/mas.20186. [DOI] [PubMed] [Google Scholar]
- 32.Lapko VN, Jiang XY, Smith DL, Song PS. Posttranslational modification of oat phytochrome A: Phosphorylation of a specific serine in a multiple serine cluster. Biochemistry. 1997;36:10595–10599. doi: 10.1021/bi970708z. [DOI] [PubMed] [Google Scholar]
- 33.Yagami T, Kitagawa K, Futaki S. Liquid secondary-ion mass spectrometry of peptides containing multiple tyrosine-O-sulfates. Rapid Commun Mass Spectrom. 1995;9:1335–1341. doi: 10.1002/rcm.1290091403. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Watson HM, Gao J, Sinha SH, Cassady CJ, Vincent JB. Characterization of the organic component of low-molecular-weight chromium-binding substance and its binding of chromium. J Nutr. 2011;141:1225–1232. doi: 10.3945/jn.111.139147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jai-nhuknan J, Cassady CJ. Negative ion postsource decay time-of-flight mass spectrometry of peptides containing acidic amino acid residues. Anal Chem. 1998;70:5122–5128. doi: 10.1021/ac980577n. [DOI] [PubMed] [Google Scholar]
- 36.Ganisl B, Valovka T, Hartl M, Taucher M, Bister K, Breuker K. Electron detachment dissociation for top-down mass spectrometry of acidic proteins. Chem Eur J. 2011;17:4460–4469. doi: 10.1002/chem.201003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalli A, Grigorean G, Håkansson K. Electron induced dissociation of singly deprotonated peptides. J Am Soc Mass Spectrom. 2011;22:2209–2221. doi: 10.1007/s13361-011-0233-6. [DOI] [PubMed] [Google Scholar]
- 38.Huzarska M, Ugalde I, Kaplan DA, Hartmer R, Easterling ML, Polfer NC. Negative electron transfer dissociation of deprotonated phosphopeptide anions: Choice of radical cation reagent and competition between electron and proton transfer. Anal Chem. 2010;82:2873–8. doi: 10.1021/ac9028592. [DOI] [PubMed] [Google Scholar]
- 39.Albeck S, Aizenberg J, Addadi L, Weiner S. Interactions of various skeletal intracrystalline components with calcite crystals. J Am Chem Soc. 1993;115:11691–11697. [Google Scholar]
- 40.Hunter GK, Goldberg HA. Nucleation of hydroxyapatite by bone sialoprotein. Proc Natl Acad Sci U S A. 1993;90:8562–8565. doi: 10.1073/pnas.90.18.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunter GK, Goldberg HA. Modulation of crystal formation by bone phosphoproteins: Role of glutamic acid-rich sequences in the nucleation of hydroxyapatite by bone sialoprotein. Biochem J. 1994;302(Pt 1):175–179. doi: 10.1042/bj3020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walters D, Smith B, Belcher A, Paloczi G, Stucky G, Morse D, Hansma P. Modification of calcite crystal growth by abalone shell proteins: An atomic force microscope study. Biophys J. 1997;72:1425–1433. doi: 10.1016/S0006-3495(97)78789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rebar RW, Miyake A, Low TL, Goldstein AL. Thymosin stimulates secretion of luteinizing hormone-releasing factor. Science. 1981;214:669–671. doi: 10.1126/science.7027442. [DOI] [PubMed] [Google Scholar]
- 44.Ebert RF, Bell WR. Assay of human fibrinopeptides by high-performance liquid chromatography. Anal Biochem. 1985;148:70–78. doi: 10.1016/0003-2697(85)90629-3. [DOI] [PubMed] [Google Scholar]
- 45.Voet D, Voet JG. Biochemistry. John Wiley & Sons; New York: 1995. [DOI] [PubMed] [Google Scholar]
- 46.Malinda K, Goldstein A, Kleinman H. Thymosin beta(4) stimulates directional migration of human umbilical vein endothelial cells. Faseb J. 1997;11:474–481. doi: 10.1096/fasebj.11.6.9194528. [DOI] [PubMed] [Google Scholar]
- 47.Gómez-Márquez J, Anadón R. The beta-thymosins, small actin-binding peptides widely expressed in the developing and adult cerebellum. The Cerebellum. 2002;1:95–102. doi: 10.1007/BF02941895. [DOI] [PubMed] [Google Scholar]
- 48.Vincent JB. Elucidating a biological role for chromium at a molecular level. Acc Chem Res. 2000;33:503–510. doi: 10.1021/ar990073r. [DOI] [PubMed] [Google Scholar]
- 49.Speetjens JK, Parand A, Crowder MW, Vincent JB, Woski SA. Low-molecular weight chromium-binding substance and biomimetic [Cr3O(O2CH2CH3)6(H2O)3]+ do not cleave DNA under physiologically-relevant conditions. Polyhedron. 1999;18:2617–2624. [Google Scholar]
- 50.Davis CM, Vincent JB. Isolation and characterization of a biologically active chromium oligopeptide from bovine liver. Arch Biochem Biophys. 1997;339:335–343. doi: 10.1006/abbi.1997.9878. [DOI] [PubMed] [Google Scholar]
- 51.Low TL, Hu SK, Goldstein AL. Complete amino acid sequence of bovine thymosin beta 4: A thymic hormone that induces terminal deoxynucleotidyl transferase activity in thymocyte populations. Proc Natl Acad Sci USA. 1981;78:1162–1166. doi: 10.1073/pnas.78.2.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weller FE, Mutchnick MG, Goldstein AL, Naylor PH. Enzyme immunoassay measurement of thymosin [beta] 4 in human serum. J Immunother. 1988;7:91–96. [PubMed] [Google Scholar]
- 53.Chougnet C, Troye-Blomberg M, Deloron P, Kabilan L, Lepers JP, Savel J, Perlmann P. Human immune response in plasmodium falciparum malaria. synthetic peptides corresponding to known epitopes of the Pf155/RESA antigen induce production of parasite-specific antibodies in vitro. J Immunol. 1991;147:2295–2301. [PubMed] [Google Scholar]
- 54.Nara PL, Hwang KM, Rausch DM, Lifso JD, Eiden LE. CD4 antigen-based antireceptor peptides inhibit infectivity of human immunodeficiency virus in vitro at multiple stages of the viral life cycle. Proc Natl Acad Sci USA. 1989;86:7139–7143. doi: 10.1073/pnas.86.18.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lifson JD, Hwang KM, Nara PL, Fraser B, Padgett M, Dunlop NM, Eiden LE. Synthetic CD4 peptide derivatives that inhibit HIV infection and cytopathicity. Science. 1988;241:712–716. doi: 10.1126/science.2969619. [DOI] [PubMed] [Google Scholar]
- 56.Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 57.Fung YME, Liu H, Chan TWD. Electron capture dissociation of peptides metalated with alkaline-earth metal ions. J Am Soc Mass Spectrom. 2006;17:757–771. doi: 10.1016/j.jasms.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 58.Chen X, Chan WYK, Wong PS, Yeung HS, Chan TWD. Formation of peptide radical cations (M+·) in electron capture dissociation of peptides adducted with group IIB metal ions. J Am Soc Mass Spectrom. 2011;22:233–244. doi: 10.1007/s13361-010-0035-2. [DOI] [PubMed] [Google Scholar]
- 59.Liu H, Håkansson K. Electron capture dissociation of tyrosine O-sulfated peptides complexed with divalent metal cations. Anal Chem. 2006;78:7570–7576. doi: 10.1021/ac061352c. [DOI] [PubMed] [Google Scholar]
- 60.Chen X, Fung YME, Chan WYK, Wong PS, Yeung HS, Chan TWD. Transition metal ions: Charge carriers that mediate the electron capture dissociation pathways of peptides. J Am Soc Mass Spectrom. 2011;22:2232–2245. doi: 10.1007/s13361-011-0246-1. [DOI] [PubMed] [Google Scholar]
- 61.Liu H, Håkansson K. Divalent metal ion-peptide interactions probed by electron capture dissociation of trications. J Am Soc Mass Spectrom. 2006;17:1731–1741. doi: 10.1016/j.jasms.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 62.Chen X, Wang Z, Li W, Wong YLE, Chan TWD. Effect of structural parameters on the electron capture dissociation and collision-induced dissociation pathways of copper(II)-peptide complexes. Eur J Mass Spectrom. 2015;21:649–657. doi: 10.1255/ejms.1382. [DOI] [PubMed] [Google Scholar]
- 63.Chen X, Liu G, Wong EYL, Deng L, Wang Z, Li W, Chan TWD. Dissociation of trivalent metal ion (Al3+, Ga3+, In3+ and Rh3+)–peptide complexes under electron capture dissociation conditions. Rapid Commun Mass Spectrom. 2016;30:705–710. doi: 10.1002/rcm.7502. [DOI] [PubMed] [Google Scholar]
- 64.Voinov VG, Hoffman PD, Bennett SE, Beckman JS, Barofsky DF. Electron capture dissociation of sodium-adducted peptides on a modified quadrupole/time-of-flight mass spectrometer. J Am Soc Mass Spectrom. 2015;26:2096–2104. doi: 10.1007/s13361-015-1230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kleinnijenhuis AJ, Mihalca R, Heeren RMA, Heck AJR. Atypical behavior in the electron capture induced dissociation of biologically relevant transition metal ion complexes of the peptide hormone oxytocin. Int J Mass Spectrom. 2006;253:217–224. [Google Scholar]
- 66.van der Burgt YEM, Palmblad M, Dalebout H, Heeren RMA, Deelder AM. Electron capture dissociation of peptide hormone changes upon opening of the tocin ring and complexation with transition metal cations. Rapid Commun Mass Spectrom. 2009;23:31–38. doi: 10.1002/rcm.3849. [DOI] [PubMed] [Google Scholar]
- 67.Iavarone AT, Paech K, Williams ER. Effects of charge state and cationizing agent on the electron capture dissociation of a peptide. Anal Chem. 2004;76:2231–2238. doi: 10.1021/ac035431p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flick TG, Donald WA, Williams ER. Electron capture dissociation of trivalent metal ion-peptide complexes. J Am Soc Mass Spectrom. 2013;24:193–201. doi: 10.1007/s13361-012-0507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong J, Vachet RW. Coordination sphere tuning of the electron transfer dissociation behavior of Cu(II)-peptide complexes. J Am Soc Mass Spectrom. 2012;23:321–329. doi: 10.1007/s13361-011-0299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Asakawa D, Takeuchi T, Yamashita A, Wada Y. Influence of metal–peptide complexation on fragmentation and inter-fragment hydrogen migration in electron transfer dissociation. J Am Soc Mass Spectrom. 2014;25:1029. doi: 10.1007/s13361-014-0855-6. [DOI] [PubMed] [Google Scholar]
- 71.Asakawa D, Wada Y. Electron transfer dissociation mass spectrometry of peptides containing free cysteine using group XII metals as a charge carrier. J Phys Chem B. 2014;118:12318–12325. doi: 10.1021/jp502818u. [DOI] [PubMed] [Google Scholar]
- 72.Commodore JJ, Cassady CJ. The effects of trivalent lanthanide cationization on the electron transfer dissociation of acidic fibrinopeptide B and its analogs. J Am Soc Mass Spectrom. 2016;27:1499–1509. doi: 10.1007/s13361-016-1428-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hunter EP, Lias SG. In: NIST Chemistry WebBook, NIST Standard Reference Database Number 69. Linstrom PJ, Mallard WG, editors. NIST, National Institute of Standards and Technology; Gaithersburg MD, 20899: Feb 5, 2016. retrieved. [Google Scholar]
- 74.Chan WYK, Chan TWD. Natural structural motifs that suppress peptide ion fragmentation after electron capture. J Am Soc Mass Spectrom. 2010;21:1235–1244. doi: 10.1016/j.jasms.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 75.Wong PSJ, Chen X, Deng L, Wang Z, Li W, Wong YLE, Chan TWD. Suppression of peptide ion dissociation under electron capture: Role of backbone amide hydrogen. Rapid Commun Mass Spectrom. 2015;29:1757–1764. doi: 10.1002/rcm.7275. [DOI] [PubMed] [Google Scholar]
- 76.Li W, Song C, Bailey DJ, Tseng GC, Coon JJ, Wysocki VH. Statistical analysis of electron transfer dissociation pairwise fragmentation patterns. Anal Chem. 2011;83:9540–9545. doi: 10.1021/ac202327r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chan WC, White PD. Fmoc Solid Phase Peptide Synthesis A Practical Approach. Oxford University Press Inc.; New York: 2000. [Google Scholar]
- 78.Roepstorff P, Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 79.Biemann K. Contributions of mass-spectrometry to peptide and protein-structure. Biomed Environ Mass Spectrom. 1988;16:99–111. doi: 10.1002/bms.1200160119. [DOI] [PubMed] [Google Scholar]
- 80.Ewing NP, Zhang X, Cassady CJ. Determination of the gas-phase basicities of proline and its di- and tripeptides with glycine: The enhanced basicity of prolylproline. J Mass Spectrom. 1996;31:1345–1350. doi: 10.1002/(SICI)1096-9888(199612)31:12<1345::AID-JMS430>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 81.Harrison AG. The gas-phase basicities and proton affinities of amino acids and peptides. Mass Spectrom Rev. 1997;16:201–217. [Google Scholar]
- 82.Martens JK, Grzetic J, Berden G, Oomens J. Gas-phase conformations of small polyprolines and their fragment ions by IRMPD spectroscopy. Int J Mass Spectrom. 2015;377:179–187. [Google Scholar]
- 83.Chalkley RJ. Improving peptide identification using empirical scoring systems. Methods Mol Biol. 2013:173–182. doi: 10.1007/978-1-62703-392-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Creese AJ, Cooper HJ. The effect of phosphorylation on the electron capture dissociation of peptide ions. J Am Soc Mass Spectrom. 2008;19:1263–1274. doi: 10.1016/j.jasms.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JEP, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li Z, Matus MH, Velazquez HA, Dixon DA, Cassady CJ. Gas-phase acidities of aspartic acid, glutamic acid, and their amino acid amides. Int J Mass Spectrom. 2007;265:213–223. [Google Scholar]
- 87.Feng C. PhD Dissertation. The University of Alabama; Tuscaloosa, AL, USA: 2014. Electron Transfer Dissociation Mass Spectrometry Studies of Peptides. [Google Scholar]
- 88.Fung YME, Duan L, Chan TWD. A comparative study of the collision-induced dissociation and the electron capture dissociation of model peptides using electrospray ionization Fourier-transform mass spectrometry. Eur J Mass Spectrom. 2004;10:449–457. doi: 10.1255/ejms.648. [DOI] [PubMed] [Google Scholar]
- 89.Pu D, Vincent JB, Cassady CJ. The effects of chromium(III) coordination on the dissociation of acidic peptides. J Mass Spectrom. 200;43:773–781. doi: 10.1002/jms.1374. [DOI] [PubMed] [Google Scholar]
- 90.Cooper HJ, Hudgkins RR, Håkansson K, Marshall AG. Characterization of amino acid side chain losses in electron capture dissociation. J Am Soc Mass Spectrom. 2002;13:241–249. doi: 10.1016/S1044-0305(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 91.Xia Q, Lee M, Rose C, Marsh A, Hubler S, Wenger C, Coon J. Characterization and diagnostic value of amino acid side chain neutral losses following electron-transfer dissociation. J Am Soc Mass Spectrom. 2011;22:255. doi: 10.1007/s13361-010-0029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Savitski MM, Nielsen ML, Zubarev RA. Side-chain losses in electron capture dissociation to improve peptide identification. Anal Chem. 2007;79:2296–2302. doi: 10.1021/ac0619332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


