Abstract
Here, we provide the dataset associated with our research article ‘label-free quantitative proteomic analysis of harmless and pathogenic strains of infectious microalgae, Prototheca spp.’ (Murugaiyan et al., 2017) [1]. This dataset describes liquid chromatography–mass spectrometry (LC–MS)-based protein identification and quantification of a non-infectious strain, Prototheca zopfii genotype 1 and two strains associated with severe and mild infections, respectively, P. zopfii genotype 2 and Prototheca blaschkeae. Protein identification and label-free quantification was carried out by analysing MS raw data using the MaxQuant-Andromeda software suit. The expressional level differences of the identified proteins among the strains were computed using Perseus software and the results were presented in [1]. This DiB provides the MaxQuant output file and raw data deposited in the PRIDE repository with the dataset identifier PXD005305.
Specifications Table
| Subject area | Biology |
|---|---|
| More specific subject area | Label-free quantitative proteomics, Bovine mastitis-associated infectious microalgae, Prototheca. spp. |
| Type of data | Raw data, table and Excel output files |
| How data was acquired | LC-MS using an UltiMate 3000 HPLC system (Dionex) connected online to an LTQ-Orbitrap Velos (Thermo Scientific) |
| Data format | Raw, processed |
| Experimental factors |
|
| Experimental features |
Whole cell proteins were extracted from Prototheca cultured strains cultured until mid-logarithmic phase of growth. For each sample protein concentrations were determined using the Bradford assay (Bio-Rad). Proteins were reduced, alkylated and digested with trypsin in solution. Following LC–MS analysis, protein identification and quantification was performed with MaxQuant software, the label-free quantitation was carried out using Perseus software. |
| Data source location | Berlin, Germany |
| Data accessibility | Data available at PRIDE:PXD005305. |
Value of the data
-
•
The data further validate the protein identification presented in Murugaiyan et al. [1].
-
•
Data from the LC–MS analysis will provide researchers with detailed information on proteins associated with non-infectious, mildly and severely infectious strains of Prototheca spp.
-
•
Prototheca spp. represents an “orphan species” whose genome sequence has not yet been sequenced, therefore, this raw data is useful for quick analysis once the genome sequence has become available.
1. Data
This mass spectrometry data-in-brief is associated with the research article aimed towards identification of differentially expressed proteins among three different strains of Prototheca spp., Prototheca zopfii genotype 1 (GT1), genotype 2 (GT2) and Prototheca blaschkeae [1]. The dataset comprises raw data, results of protein identification using MaxQuant-Andromeda software suit and a list of proteins identified as differentially expressed between non-infectious, infectious and mildly infectious strains of Prototheca spp. The raw data can be downloaded from the PRIDE repository (identifier PXD005305), a compilation of the identified proteins is presented in Supplementary table 1 and the differentially expressed proteins are listed in Table 1.
Table 1.
List of proteins identified as differentially expressed.
| S.No | UniProt Acc. No. | Protein name | −Log2(fold change) |
||
|---|---|---|---|---|---|
| P. zopfii GT2 vs P. zopfii GT1 | P. blaschkeae vs P. zopfii GT1 | P. zopfii GT2 vs P. blaschkeae | |||
| 1 | E1ZQV2 | Heat shock protein 70 | −1.0⁎ | −0.4⁎ | −0.6⁎ |
| 2 | E1ZLA8 | Acetyl-coenzyme A synthetase | −6.8⁎ | −6.8⁎ | 0.0 |
| 3 | A0A087SCT6 | Citrate synthase | −3.6⁎ | −3.6⁎ | 0.0 |
| 4 | E1ZL24 | Putative uncharacterized protein | −4.6⁎ | −4.6⁎ | 0.0 |
| 5 | A0A087SSM0 | Actin | −0.6⁎ | +0.1 | −0.7⁎ |
| 6 | A0A087SFG0 | Cysteine synthase, chloroplastic/chromoplastic | −3.9⁎ | +1.7 | −5.6⁎ |
| 7 | A0A087SP16 | FK506-binding protein 1 | −1.4⁎ | −0.1 | −1.3⁎ |
| 8 | E1ZK88 | Ubiquitin | −1.1⁎ | +0.3 | −1.4⁎ |
| 9 | A0A087SJV3 | Aldehyde dehydrogenase family 2 member B4 | +0.5⁎ | −0.5⁎ | +1.0⁎ |
| 10 | E1ZG37 | Putative uncharacterized protein | +0.6⁎ | −3.8⁎ | +4.4⁎ |
| 11 | A0A087SS91 | Aconitate hydratase, mitochondrial (Aconitase) | +0.6⁎ | −7.3⁎ | +8.0 |
| 12 | E1ZTB0 | Fructose-bisphosphate aldolase | +8.3⁎ | +8.8⁎ | −0.6⁎ |
| 13 | E1ZCI5 | Putative uncharacterized protein | +0.5⁎ | +0.7⁎ | −0.3 |
| 14 | E1ZT42 | V-type H+ ATPase subunit A | +0.5⁎ | +0.4⁎ | +0.1 |
| 15 | A0A087SJM7 | 40S ribosomal protein S10 | +6.9⁎ | 0.0 | +6.9⁎ |
| 16 | E1ZQY4 | 40S ribosomal protein S5 | +3.3⁎ | 0.0 | +3.3⁎ |
| 17 | A0A087SBU8 | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | +6.4⁎ | 0.0 | +6.4⁎ |
| 18 | A0A087SNV1 | 60S ribosomal protein L12-1 | +6.7⁎ | 0.0 | +6.7⁎ |
| 19 | A0A087SKG6 | 60S ribosomal protein L6 | +4.4⁎ | 0.0 | +4.4⁎ |
| 20 | A0A087SN43 | 6-phosphogluconate dehydrogenase, decarboxylating (EC 1.1.1.44) | +4.5⁎ | +0.7 | +3.8⁎ |
| 21 | A0A087SJX6 | Argininosuccinate synthase | +3.6⁎ | 0.0 | +3.6⁎ |
| 22 | A0A087SPA9 | Carbamoyl-phosphate synthase large chain | +4.6⁎ | +1.1 | +3.4⁎ |
| 23 | A0A087SHS8 | Eukaryotic initiation factor 4A-10 | +0.4⁎ | −0.2 | +0.6⁎ |
| 24 | E1ZFZ5 | Glutamate dehydrogenase | +3.1⁎ | 0.0 | +3.1⁎ |
| 25 | A0A087SQ68 | Phosphate carrier protein, mitochondrial | +3.1⁎ | 0.0 | +3.1⁎ |
| 26 | E1ZGA3 | 40S ribosomal protein S27 | +3.3⁎ | +1.2 | +2.1 |
| 27 | E1Z7R4 | Heat shock protein 70 | +5.3⁎ | +2.2 | +3.1 |
| 28 | E1ZSM6 | Putative uncharacterized protein | +3.3⁎ | +1.2 | +2.1 |
| 29 | A0A087SF19 | Adenosylhomocysteinase | +1.7 | −2.4⁎ | +4.2⁎ |
| 30 | A0A087SK74 | Elongation factor 1-alpha | +0.2 | −0.6⁎ | +0.8⁎ |
| 31 | E1Z5R3 | Putative uncharacterized protein | −1.6 | −5.3⁎ | +3.8⁎ |
| 32 | E1ZJM1 | Tubulin beta chain | 0.0 | −0.6⁎ | +0.6⁎ |
| 33 | A0A087SE71 | Elongation factor Tu | −1.5 | −4.3⁎ | +2.8 |
| 34 | A0A087SG29 | Glucose-6-phosphate isomerase | −3.2 | −5.3⁎ | +2.1 |
| 35 | A0A087SSF2 | Nucleoside diphosphate kinase 1 | −2.0 | −4.5⁎ | +2.5 |
| 36 | A0A087SL21 | Ubiquitin-60S ribosomal protein L40-2 | −3.7 | −8.2⁎ | +4.5 |
| 37 | A0A087SI38 | Acetyl-coenzyme A synthetase | 0.0 | +4.6⁎ | −4.6⁎ |
| 38 | A0A087SBN0 | ATP synthase subunit beta (Delta-aminolevulinic acid dehydratase) | 0.0 | +0.5⁎ | −0.5⁎ |
| 39 | A0A087SQR3 | Chaperonin CPN60, mitochondrial | +0.2 | +0.9⁎ | −0.7⁎ |
| 40 | A0A087SBQ6 | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic | 0.0 | +6.8⁎ | −6.8⁎ |
| 41 | A0A087SND2 | Heat shock 70 kDa protein, mitochondrial | −0.1 | +0.6⁎ | −0.7⁎ |
| 42 | A0A087ST26 | Phosphoglycerate kinase | 0.0 | +5.5⁎ | −5.5⁎ |
| 43 | A0A087SNN6 | Stress-induced-phosphoprotein 1 | 0.0 | +3.7⁎ | −3.7⁎ |
| 44 | A0A087SIY9 | Succinyl-CoA ligase [ADP-forming] subunit alpha-1, mitochondrial | 0.0 | +4.7⁎ | −4.7⁎ |
| 45 | A0A087S9W3 | Histone H4 | 0.0 | +2.9⁎ | −2.9 |
| 46 | E1ZRV3 | Putative uncharacterized protein | +0.7 | +4.3⁎ | −3.6 |
| 47 | E1ZMD2 | Putative uncharacterized protein | 0.0 | +2.4⁎ | −2.4 |
| 48 | A0A087SAK4 | Chaperone protein ClpB1 | −0.8 | +2.0 | −2.8⁎ |
| 49 | A0A087S9L8 | Enolase | −3.7 | +1.7 | −5.4⁎ |
| 50 | A0A087SI84 | GTP-binding nuclear protein | −0.6 | +0.4 | −1.0⁎ |
| 51 | E1ZD41 | Putative uncharacterized protein | +3.3 | −0.7 | +4.0⁎ |
(+) indicates upregulated and (−) indicates downregulated.
Statistical significance was calculated using two-way Student-t test and error correction (p value <0.05) using the method of Benjamini–Hochberg [2].
2. Experimental design
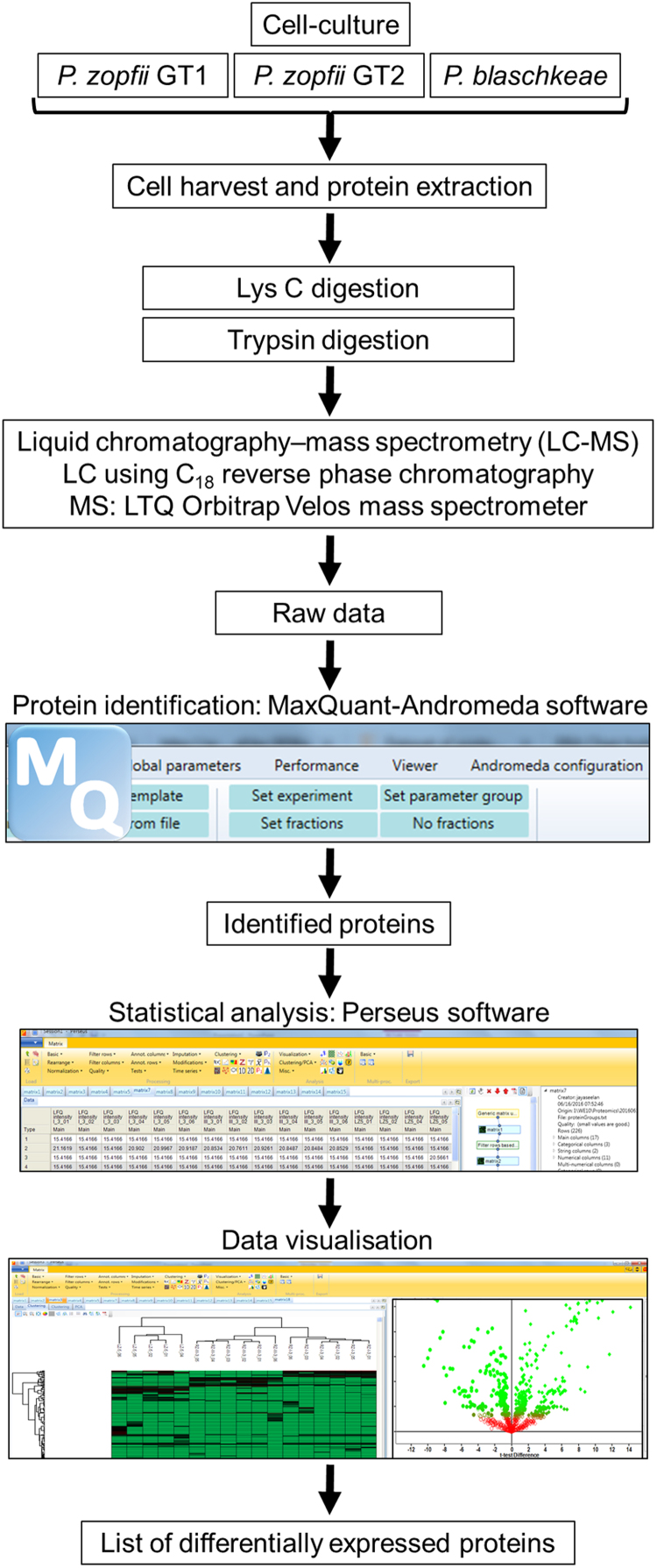
The dataset presented here was obtained from using the label-free proteomic analysis of three different strains of Prototheca species, P. zopfii genotype 1, genotype 2 and P. blaschkeae representing non-infectious, infectious and moderately infectious strains, respectively. In total 17 samples representing six independent cultures for each (only five in P. zopfii genotype 2) were used to generate the dataset (experimental design is shown in Fig. 1). A Student-t test, p-value <0.05% and 1% false discovery rate (FDR) was applied for identification of differentially expressed proteins between (a) P. zopfii genotype 2 and P. zopfii genotype 1; (b) P. blaschkeae and P. zopfii genotype 1; and (c) P. zopfii genotype 2 and P. blaschkeae.
Fig. 1.
Schematic overview of the overall analysis workflow.
3. Materials and methods
3.1. Prototheca strains
The following three strains from the culture collection of the Institute of Animal Hygiene and Environmental Health, Freie Universität Berlin, Germany were utilized for this study [3].
-
a.
P. zopfii genotype 1 (SAG 2063T), non-infectious environmental strain.
-
b.
P. zopfii genotype 2 (SAG 2021T), clinical strain.
-
c.
P. blaschkeae (SAG 2064T), clinical strain.
3.2. Cell culture and protein extraction
Following the retrieval from the culture collection, the strains were first streaked in Sabouraud dextrose solid media, incubated at 37 °C until the appearance of visible colonies. The species and genotypes were reconfirmed using MALDI profiling as described [4]. The cell culture and protein extraction was carried out as described [1].
3.3. Mass spectrometry analysis
The proteins were subjected to in-solution trypsin digested as described [1]. The resultant peptides were purified using solid phase extraction procedure [5], separated by nanoscale C18 reverse-phase liquid chromatography using the Dionex Ultimate 3000 nanoLC (Dionex/Thermo Fisher Scientific, Idstein, Germany) and directly ionised by electrospray ionization and measured after transfer into an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). MS survey scan (m/z 300–1700, resolution 60,000) was acquired in the Orbitrap and the 20 most intensive precursor ions were fragmented.
3.4. Data analysis
Data from MS/MS spectra was searched using MaxQuant-Andromeda software suit [6], [7], [8] against the Uniprot FASTA dataset of Chlorella variabilis and Auxenochlorella protothecoides proteome with the parameters settings as described in [1]. Table 2 shows the experimental design and sample file naming format and the dataset associated to the MaxQuant analysis is shown in Supplementary table 2.
Table 2.
Experimental design and raw data file naming format.
| S. No | Sample name | Strain designation | Replicates | raw data file designation |
|---|---|---|---|---|
| 1 | P. zopfii genotype 1 | SAG 2063T | 1 | I_3_01 |
| 2 | 2 | I_3_02 | ||
| 3 | 3 | I_3_03 | ||
| 4 | 4 | I_3_04 | ||
| 5 | 5 | I_3_05 | ||
| 6 | 6 | I_3_06 | ||
| 7 | P. blaschkeae | SAG 2064T | 1 | III_3_01 |
| 8 | 2 | III_3_02 | ||
| 9 | 3 | III_3_03 | ||
| 10 | 4 | III_3_04 | ||
| 11 | 5 | III_3_05 | ||
| 12 | 6 | III_3_06 | ||
| 13 | P. zopfii genotype 2 | SAG 2021T | 1 | LZ5_01 |
| 14 | 2 | LZ5_02 | ||
| 15 | 3 | sample lost during transit | ||
| 16 | 4 | LZ5_04 | ||
| 17 | 5 | LZ5_05 | ||
| 18 | 6 | LZ5_06 |
The statistical analysis was carried out using Perseus 1.4.1.3 (Available online: http://141.61.102.17/ perseus_doku/doku.php?id=start) as described [1]. The differences in protein expression computed in three different ways i) mildly infectious vs environmental strain, ii) severe infection-associated vs environmental strain and iii) severely infectious vs mildly infectious strain were presented in Murugaiyan et al. [1].
3.5. Mass Spectrometry dataset deposit
The mass spectrometry data was deposited at the ProteomeXchange (PX) Consortium [9], [10], [11] via the PRIDE (PRoteomics IDEntifications) partner repository at the European Bioinformatics Institute (http://www.ebi.ac.uk/pride/) and is now accessible with the dataset identifier PXD005305.
Acknowledgements
We would like to thank Michael Kühl for excellent technical assistance. We acknowledge the assistance of the Bio-MS unit of the Core Facility BioSupraMol supported by the Deutsche Forschungsgemeinschaft (DFG). The author Murat Eravci was supported by the Deutsche Forschungsgemeinschaft (DFG, SFB958). We thank the PRIDE team for their assistance in the MS data deposition.
Footnotes
Transparency data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.dib.2017.04.006.
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.dib.2017.04.006.
Transparency document. Supplementary material
Supplementary material
Appendix A. Supplementary material
Supplementary material
References
- 1.Murugaiyan J., Eravci M., Weise C., Roesler U. Label-free quantitative proteomic analysis of harmless and pathogenic strains of infectious microalgae, Prototheca spp. Int. J. Mol. Sci. 2017;18:59. doi: 10.3390/ijms18010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamini Y., Hochberg Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 3.Roesler U., Moller A., Hensel A., Baumann D., Truyen U. Diversity within the current algal species Prototheca zopfii: a proposal for two Prototheca zopfii genotypes and description of a novel species, Prototheca blaschkeae sp. nov. Int. J. Syst. Evol. Microbiol. 2006;56:1419–1425. doi: 10.1099/ijs.0.63892-0. [DOI] [PubMed] [Google Scholar]
- 4.Murugaiyan J., Ahrholdt J., Kowbel V., Roesler U. Establishment of a matrix-assisted laser desorption ionization time-of-flight mass spectrometry database for rapid identification of infectious achlorophyllous green micro-algae of the genus Prototheca. Clin. Microbiol. Infect. 2012;18:461–467. doi: 10.1111/j.1469-0691.2011.03593.x. [DOI] [PubMed] [Google Scholar]
- 5.Rappsilber J., Ishihama Y., Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 6.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 7.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 9.Vizcaíno J.A., Cote R.G., Csordas A., Dianes J.A., Fabregat A., Foster J.M., Griss J., Alpi E., Birim M., Contell J., O׳Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.W., Wang R., Hermjakob H. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vizcaíno J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J.:A., Sun Z., Farrah T., Bandeira N., Binz P.A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R.J., Kraus H.J., Albar J.P., Martinez-Bartolomé S., Apweiler R., Omenn G.S., Martens L., Jones A.R., Hermjakob H. ProteomeXchange provides globally co-ordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;30(3):223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material