Abstract
Let-7a miRNA is downregulated in various cancers. However, in hepatocellular carcinoma (HCC) patients infected with hepatitis B virus (HBV), the relationship between let-7a and HBV replication has not been fully elucidated. Liver specimens were collected from 23 HCC patients with chronically active HBV. The serum levels of the HBV antigens hepatitis B surface antigen (HBsAg) and hepatitis B virus e antigen (HBeAg), and the HBV antibodies, anti-HBs, anti-HBe and anti-hepatitis B core antigen (anti-HBc) were measured using the microparticle enzyme immunoassay. Let-7a levels and HBV DNA copy numbers were measured by quantitative real-time PCR (qRT-PCR) and analyzed statistically. A let-7a specific antisense oligonucleotide was introduced to the HBV-producing cell line HepG2.2.15 and a change in HBV DNA copy numbers was assessed by qRT-PCR. HCC patients with highly active HBV replication (>106 DNA copies/mL) showed higher levels of serum HBsAg and anti-HBc than patients with less active HBV replication (<103 DNA copies/mL). The level of let-7a was lower in malignant tissues than in adjacent normal tissues. However, patients with highly active HBV replication demonstrated a significantly higher level of let-7a in hepatocarcinoma tissue than patients with less active HBV replication (P < 0.05). A higher level of let-7a was observed in the HBV-producing cell line HepG2.2.15 than in HepG2 cells (P < 0.05), and let-7a down-regulation by antisense oligonucleotides led to a reduction in HBV DNA copy numbers (P < 0.05), indicating a correlation between the let-7a level and HBV replication. Down-regulation of let-7a reduces HBV replication and could prevent the development of HCC, suggesting it could be an effective therapeutic treatment for HBV infection.
Impact statement
Although interferon and nucleic acid analogues effectively suppress HBV replication in HBV patients, there is no treatment which eradicates the virus.
Moreover, the therapeutic effect can be reduced by virus mutations or drug resistance. Let-7a is a miRNA initially found in the nematode as a master regulator of developmental processes, but also exists in humans. It has been reported that the transcription of let-7a was much lower in HCC than in normal liver tissues and specific miRNA could directly promote virus replication. Therefore we hypothesized that transcription of let-7a promotes HBV replication, which might compromise the therapeutic effects of antivirus treatments. In our present study, we demonstrated a correlation between let-7a transcription and HBV replication in surgical specimens obtained from patients with HCC, as well as in HCC cell lines. Our finding might be the base for a new approach to improve HBV infection treatments in the future.
Keywords: Hepatitis B virus replication, hepatocellular carcinoma, let-7a, microRNA
Introduction
Hepatitis B virus (HBV) is one of the most prevalent pathogens in China with an incidence of 84.3 per 100,000 in China between 2005 and 2010,1 and a worldwide mortality of 600,000 people per year.2
HBV infections can lead to liver diseases such as acute and chronic hepatitis, cirrhosis, as well as hepatocellular carcinoma (HCC). It has been estimated that in China approximately 383,000 people die from HCC every year, accounting for 51% of deaths caused by liver cancer worldwide.3 At present, the consensus approach to prevent the progression to hepatocirrhosis and HCC is antiviral therapy.4 Although therapy with interferon (IFN) and nucleic acid analogues effectively suppresses HBV replication, there is no treatment to eradicate the virus.5,6 Moreover, the therapeutic effect could be compromised to a great extent by virus mutation or upon drug withdrawal. Thus, the development of new and more effective therapeutic strategies is an urgent requirement.
A miRNA is a noncoding single-strand RNA recently discovered in a wide range of animals and plants that regulates gene expression by interacting with target transcripts. The lethal-7 (let-7) miRNA family was initially found in nematodes to be the master regulator of developmental processes. The physiological function of let-7 has been described as a regulator of cell cycle exits after temporal upregulation in Caenorhabditis elegans. In let-7 deficient C. elegans mutants, cells divide without control, which suggests that let-7 is a tumor suppressor.7 Later research revealed that let-7 genes are located in the deleted regions of cancer cells.8 Let-7 expression was found to be reduced in lung cancer, but its over-expression led to cancer cell growth inhibition.9
An abnormal let-7a expression is frequently observed in inflammatory tissues, tumors or in cell samples taken from patients with chronic active hepatitis, cirrhosis or HCC. It has been shown that the expression of let-7a was much lower in HCC tissues than in normal liver tissue.10 In host cells, viruses utilize miRNA to enhance their own replication and also to evade host immune defense.11 Binding of miR146a in HBV infected HepG2.2.15 cells led to silencing of type I IFN-induced antiviral factors, thereby providing IFN resistance.12 Similarly, the IFN-γ receptor 1 has been shown to be downregulated by miR-548.13 Another study reported that over-expression of miR-1 in HBV-infected HepG 2.2.15 cells enhanced HBV replication by arresting the cell cycle at G1 and inhibition of cell proliferation by actions on E2F transcription factor 5 and histone deacetylase 4 in addition to host specific HBV promotor binding farnesoid X receptor a (FXRA) upregulation.14 It was hypothesized that higher expression of let-7a promotes HBV replication, which compromises the therapeutic effects of antivirus treatment. In the present study we demonstrated a correlation between let-7a expression and HBV replication in specimens obtained from patients with HCC who underwent surgery, as well as in cultured HCC cell lines, that might be the basis for developing HBV virus and HCC targeting therapies in the future.
Methods
Specimens and cell lines
Surgical specimens of liver tissue were collected from 23 HCC patients with chronically active HBV from January 2010 to October 2013 in our hospital. Simultaneously, normal adjacent tissues in 10 patients were also surgically removed and collected. Signed informed consent forms were provided by all patients included in the study. All specimens were stored at −80℃. The HBV-producing cell line HepG2.2.15 and the human hepatoma cell line HepG2 were purchased from ATCC (Manassas, VA). Cells were grown in Dulbecco's modified Eagle medium (DMEM) medium which contained 10% fetal bovine serum and 200 µg/mL G418 at 37℃ in an incubator supplied with 5% CO2 in a humidified atmosphere.
RNA extraction and cDNA synthesis
Tissues were lysed with TRIzol reagent sourced from Invitrogen, Carlsbad, USA and the resulting lysates were collected in RNAase-free tubes. Small RNAs including miRNA, siRNA, shRNA and snRNA were then extracted using a mirVana™ miRNA isolation kit (Ambion, Naugatuck, CT) and quantified by real-time PCR (qRT-PCR) using the Applied Biosystems 7300 HT Sequence Detection System (Applied Biosystems, USA). For cDNA synthesis, total RNA was extracted using the RNeasy Mini kit (Qiagen, Hilden, Germany). Any residual DNA in the RNA samples was digested with an RNase-free DNase Set (Qiagen). A high fidelity cDNA synthesis kit (Roche, Basel, Switzerland) was used to synthesize first-strand cDNA with anchored-oligo (dT) 18 primers.
Quantification of HBV mRNA and let-7a using qRT-PCR
The primers for the HBV mRNA quantification were designed using Premier v5.0 software (Primer Biosoft, Palo Alto, CA) and are listed below:
HBV mRNA F1 5′-CAA CTT GTC CTG GTT ATC GC-3′
HBV mRNA R1 5′-AAG CCC TAC GAA CCA CTG AA-3′
RT-PCR was carried out at 95℃ for 30 s, and then 40 cycles at 95℃ for 5 s and 30 s for 60℃ in a TaKaRa Thermal Cycler Dice® (TaKaRa, Japan) using SYBR® Premix Ex Taq™ II (Tli RNaseH Plus).
The level of a specific 250 bp sequence of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA served as the internal control using the primers:
GADPH-F1 5′-CAA CGG ATT TGG TCG TAT TGG G-3′
GADPH-R1 5′-CCT GGA AGA TGG TGA TGG GAT T-3′
The level of let-7a was quantified using the TaqMan Gene Expression Assay (assay ID 000377) (ABI, Foster City, CA), which included human let-7a and U6 snRNA (assay ID 001973) being used as an endogenous control; 20 pmol/µL of each primer was used. cDNA was pre-denatured for 2 min at 93℃. Ten and 30 cycles of RT-PCR reactions were performed as follows: primer annealing 45 s at 93℃ with extension for 60 s at 55℃ and primer annealing for 30 s at 93℃ with extension for 45 s at 55℃, respectively. The negative control was RT-PCR without a template, which was carried out as a control for each experiment.
Determination of HBV DNA copy numbers
Virus DNA was extracted from liver tissue samples and cultured cells with DNAzol (Invitrogen), and quantified using a HBV DNA FQ-PCR kit (Qiagen). The sequences of primers used were:
F1 5′-ATCCTGCTGCTATGCCTCATCTT-3′ (23 bp)
R1 5′-ACAGTGGGGGAAAGCCTACGAA-3′ (23 bp)
The extracted DNA was mixed with appropriate reagents and amplified in an ABI7700 fluorescence detection system (ABI). Ten and 30 cycles of RT-PCR reactions were performed as follows: pre-denatured at 93℃ for 2 min, primer annealing for 45 s at 93℃ with a 60 s extension at 55℃, and primer annealing for 30 s at 93℃, with an extension for 45 s at 55℃, respectively. The number of copies of HBV DNA was determined using a custom algorithm.
Transfection of inhibitors for let-7a miRNA
A let-7a specific antisense miRNA inhibitor and a non-specific antisense miRNA inhibitor (Ambion) were transfected into HepG2.2.15 and HepG2 cells, with the transfection agent siPORT (Ambion). Transfected cells were cultured in DMEM medium, which contained 200 µg/mL G418 and 10% fetal bovine serum, and harvested on days 1, 2, 5, and 7. After cells were lysed, the supernatant was collected and subjected to RT-PCR to determine the inhibition rate. The efficiency of inhibition was calculated as follows: (non-specific antisense miRNA inhibitor—let-7a specific antisense miRNA inhibitor)/non-specific antisense miRNA inhibitor × 100%.
Determination of the serum HBV antigens’ and antibodies’ concentrations using MEIA
Blood samples were collected from patients and the sera isolated by centrifugation at 1000g for 20 min at 4℃ after which they were stored at −20℃ or −80℃ for subsequent analyses. The levels of the HBV antibodies anti-HBc, anti-HBe and anti-HBs, and levels of the HBV antigens hepatitis B virus e antigen (HBeAg) and hepatitis B surface antigen (HbsAg) were measured by microparticle enzyme immunoassay (MEIA) using an automated biochemistry analyzer AxSYM (Abbott Diagnosis, Abbott Park, IL). HBsAg was considered to be positive in HBV patients when the fluorescent ratio of sample to negative control (S/N) was ≥2.00, and HBeAg was considered to be positive when the fluorescent ratio of sample to cut off value ratio (S/CO) was ≥ 1.00.
Analysis of let-7a target sequences in HBV
Let-7a target analysis was carried out using the ViTa Virus miRNA Target database (http://vita.mbc.nctu.edu.tw/).
Statistical analysis
Statistical analyses were performed with SPSS v18.0 (SPSS, Inc., Chicago, IL). Student t-tests were used to compare normally distributed data between various groups, which was expressed as the median (range) and mean ± standard deviation (x ± SD). One-way analysis of variance (ANOVA) was used to analyze the data collected at different time points. A P value ≤ 0.05 was taken to denote statistical significance.
Results
Serum HBV antigen and antibody concentrations correlated with the HBV DNA copy numbers
The correlation between the HBV antigen and antibody concentrations in HCC patient’s serum and the copy numbers of HBV DNA in liver tissues were examined. In patients found to have an HBV DNA level < 103 copies/mL, HBsAg and anti-HBc concentrations were lower compared to patients with a high DNA level (≥106 copies/mL). Both serum concentrations positively correlated with the copy numbers of HBV DNA (r = 0.2251, P = 0.040 and r = 0.2271, P = 0.039, respectively) (Table 1).
Table 1.
The HBV antigen and antibody concentrations in HCC patients’ serum and the copy numbers of HBV DNA in liver tissues
| HCC patients (n = 23) | HBV DNA (copies/mL) |
|
P | R |
|---|---|---|---|---|
| <103 (n = 10) | ≥106 (n = 13) | |||
| Age (mean ± SD) | 42.3 ± 5.5 | 44.2 ± 4.5 | 0.372 | |
| Gender | ||||
| Male (N) | 6 | 8 | 1.000 | |
| Female (N) | 4 | 5 | ||
| HbsAg (ng/mL) (median [range]) | 2.56 (0.26, 250) | 250 (12.48, 250) | 0.040 | 0.225 |
| Anti-HBs (mIU/mL) (median [range]) | 0.98 (0, 9.3) | 0 (0, 6.6) | 0.293 | 0.065 |
| HBeAg (PEIU/mL) (median [range]) | 0.45 (0.12, 3.26) | 10.31 (2.04, 28.34) | 0.461 | 0.032 |
| Anti-HBe (PEIU/mL) (median [range]) | 8.35 (0.75, 29.31) | 7.58 (0.04, 21.36) | 0.483 | 0.029 |
| Anti-HBc (PEIU/mL) (median [range]) | 0.87 (0.01, 9.39) | 9.73 (4.51, 11.24) | 0.039 | 0.227 |
P < 0.05 was considered to be statistically significant; r: correlation coefficient.
Higher expression levels of let-7a were correlated with an increased production of HBV mRNA
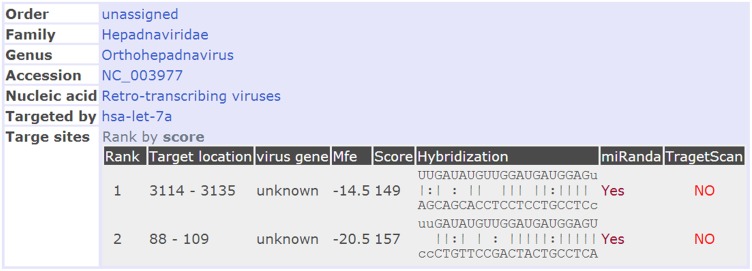
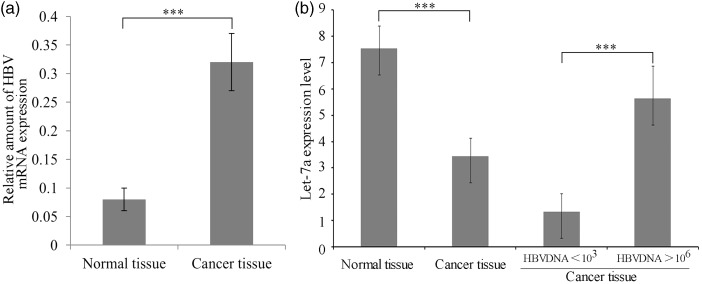
An in silico analysis revealed two regions of the HBV mRNA (nucleotides 88–109 and 3114–3125) to be the targets of let-7a miRNA (Figure 1). HBV mRNA is eventually reverse transcribed into viral DNA in the infected cells; therefore the let-7a level might correlate with the copy numbers of viral DNA in patients. We determined the expression levels of let-7a miRNA in liver tumor tissues taken from 12 patients with strong HBV replication (>106 DNA copies/mL) and in 11 patients with less active HBV replication (<103 DNA copies/mL) (Figure 2). The expression of let-7a was greater in adjacent normal tissue than in tumor tissue in each patient. However, a relatively higher expression level of let-7a was found in tumor tissue taken from patients with highly active HBV replication (P < 0.05), suggesting that let-7a miRNA might upregulate HBV mRNA production in patients with HCC.
Figure 1.
The let-7a target sequences of HBV mRNA. (A color version of this figure is available in the online journal.)
Figure 2.
Expression of HBV mRNA and let-7a miRNA in adjacent normal tissues and malignant tissues taken from HCC patients. (a) Comparison of HBV replication in normal and cancer tissues. (b) Left side: comparison of let-7a transcription in normal and cancer tissue, right side: difference of let-7a transcription in cancer tissues with low (<103) and high (>106) HBV DNA replication levels; ***indicates P < 0.001
Effect of let-7a inhibition on HBV replication in HepG2.2.15 cells
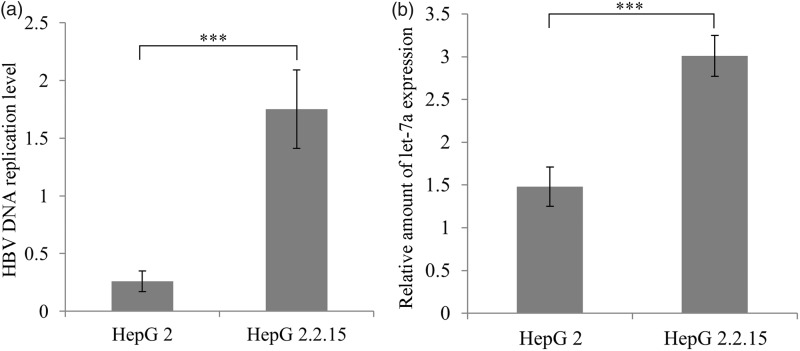
Quantification of replication of HBV revealed that HBV DNA was about seven times less detectable in HepG2 cells than in HepG2.2.15 cells (P < 0.001); this quantification was accompanied by a two-fold expression of let-7a expression in HepG2.2.15 compared to HepG2 cells (P < 0.001) (Figure 3).
Figure 3.
Levels of (a) HBV DNA replication and (b) let-7a transcription in HepG2 and HBV-producing HepG2.2.15 cells. ***indicates P < 0.001
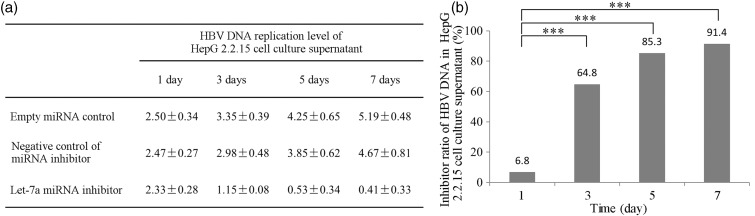
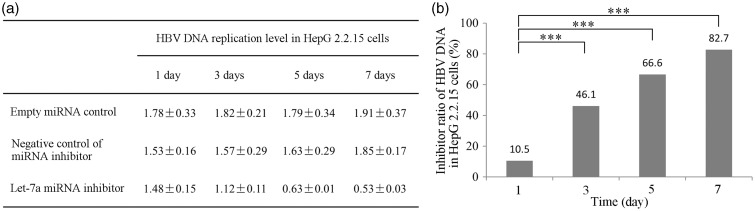
To correlate the level of let-7a expression and HBV DNA replication, let-7a was downregulated in HepG2.2.15 by transfecting them with a let-7a specific antisense inhibitor and a non-specific antisense miRNA inhibitor as the control. Since HepG2.2.15 cells produce and secrete HBV virus particles, we measured the copy numbers of HBV DNA both in the cells and the cell culture supernatant. When HepG2.2.15 cells were transfected with a let-7a specific antisense inhibitor, there was significantly reduced copy numbers of HBV DNA in a time-dependent manner, both in the cells and the culture supernatant compared to the controls.
The efficiency of inhibition was 6.8%, 64.8%, 85.3%, and 91.4% in cell culture supernatant, and 10.5%, 46.1%, 66.6%, and 82.7% in the cells at days 1, 3, 5, and 7, respectively (Figures 4 and 5), suggesting that a decreased let-7a miRNA expression is inhibitory for HBV replication.
Figure 4.
Effects of let-7a down-regulation on HBV DNA replication in the supernatant of HepG2.2.15 cell culture media. (a) HBV replication levels at 1, 3, 5, and 7 days after transfection with empty miRNA control, negative miRNA control and let-7a miRNA inhibitor vectors. (b) Inhibition of HBV replication at 1, 3, 5, and 7 days after transfection with let-7a miRNA containing vector. ***Indicates P < 0.001
Figure 5.
Effects of let-7a down-regulation on HBV DNA replication in HepG2.2.15 cells. (a) HBV replication levels at 1, 3, 5, and 7 days after transfection with empty miRNA control, negative miRNA control and let-7a miRNA inhibitor vectors. (b) Inhibition of HBV replication at 1, 3, 5 and 7 days after transfection with let-7a miRNA containing vector. ***indicates P < 0.001
Discussion
Let-7a miRNA has been widely detected in hepatic tissues affected by chronically active hepatitis B, cirrhosis, and cancer. In the present study, we showed that normal liver tissues adjacent to tumor obtained from HCC patients expressed higher levels of let-7a miRNA than malignant tissues, and that let-7a expression positively correlated with HBV replication in malignant tissues. Previous studies found multiple forms of HBsAg in adjacent normal tissues without fragmented small inclusion bodies that are observed in tumor cells, suggesting that these normal tissues were in the process of tumorigenesis.15 High concentrations of serum HBV DNA and HBsAg are important risk factors known to be associated with HCC.16,17 Considering the positive correlation of let-7a expression and HBV replication, let-7a miRNA could be used potentially as a new tool to inhibit HBV replication and to prevent the development of HCC.
In agreement with our finding that the levels of let-7a miRNA was reduced in malignant liver tissues (Figure 2), the expression of let-7a miRNA was reported to be reduced in colon18 and gastric cancer.19
Zhang et al. reported that the let-7a miRNA expression was lower in gastric tumors than in normal gastric tissue, while the protein concentration of the RAS oncogene was significantly higher in gastric tumors, implicating an association of let-7a with gastric cancer.19 Furthermore, Johnson et al. showed that all three human RAS genes contain let-7 complementary sides in the 3′-UTRs of their mRNAs, which explained the down-regulation of RAS proteins in lung tumors by mRNA destabilization via let-7 activity.20
Liu et al. reported that let-7a miRNA up-regulates the expression of ubiquitin specific protease 35 (USP35) in various cancer cell lines and tissues, over-expression of which inhibited tumor xenograft growth in vivo and cell proliferation in vitro by inhibiting TNFα-induced NF-κB activation by stabilizing A20 binding inhibitors (ABIN-2 protein).21
In previous research it has been noted that let-7a expression is downregulated in tissues of HCC patients with HBV infections.10,22,23 which is in agreement with our findings (Figure 2(b)). However, we found that the expression levels of let-7a miRNA were, though still reduced compared to normal tissue, higher in malignant liver tissues obtained from patients with particularly high HBV replication rates (HBV DNA > 106 copies/mL) than in tissues obtained from patients with less marked HBV replication (HBV DNA < 103 copies/mL). To confirm the correlation, we used HepG2.2.15 cells derived from human hepatoblastoma HepG2 cells that had been transfected with a plasmid containing HBV DNA.24 HepG2.2.15 cells stably secrete HBsAg, HBeAg, and HBV virus particles into the supernatant of culture medium.25 We found that HepG2.2.15 expressed a higher level of let-7a miRNA than HepG2 cells. In addition, down-regulation of let-7a miRNA by transfecting HepG2.2.15 with an antisense inhibitor demonstrated a positive correlation between the let-7a expression level and the copy numbers of HBV DNA. According to these findings, we suggest that although increased let-7a expression inhibits cell proliferation and tumor invasion, it also promotes HBV replication in liver cells.
Taken together, let-7a similarly to miR-1 has been shown to inhibit proliferation of hepatic cells via the USP35-ABIN-2 pathway,21 and a growing body of evidence suggests that restoration of let-7 expression may be a useful target option in cancers where its expression has been lost. However, according to our findings, though its down-regulation may favor cell proliferation and the cancer process, on the other hand, the let-7a inhibition in HepG2.2.15 cells was accompanied by reduced HBV DNA replication thereby reducing the risk of developing HCC over time.
Conclusions
There was a positive correlation between let-7a miRNA expression and HBV DNA copy numbers in liver tissues removed from HCC patients, and inhibition of let-7a downregulated HBV replication in HepG2.2.15 cells. Since a high level of HBV DNA might be associated with a higher HCC risk in patients who had been infected by HBV, let-7a miRNA inhibition might be applied as an antivirus therapy after HBV infections with high HBV DNA copy numbers to prevent the development of HCC over time.
Acknowledgments
This work was supported by the Shanghai municipal bureau of health youth research projects (2010Y057).
Authors’ note
All authors confirm that the content has not been published elsewhere and does not overlap with or duplicate their published work.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors' contributions
DNQ and JC were responsible for the conception and design of the study. DNQ, JC, JL, ZGL, WRJ, JPH, ZBQ, WJY, and LJW were responsible for acquisition of data. DNQ and WRJ performed the data analysis. DNQ drafted the manuscript. All authors participated in interpretation of the findings. DNQ revised and commented the draft, and all authors read and approved the final version of the manuscript.
References
- 1.Yan YP, Su HX, Ji ZH, Shao ZJ, Pu ZS. Epidemiology of hepatitis B virus infection in china: current status and challenges. J Clin Transl Hepatol 2014; 2: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt S, Bundschuh M, Scutaru C, Klingelhoefer D, Groneberg DA, Gerber A. Hepatitis B: global scientific development from a critical point of view. J Viral Hepat 2014; 21: 786–93. [DOI] [PubMed] [Google Scholar]
- 3.Wang FS, Fan JG, Zhang Z, Gao B, Wang HY. The global burden of liver disease: the major impact of China. Hepatology 2014; 60: 2099–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lun-Gen L. Antiviral therapy of liver cirrhosis related to hepatitis B virus infection. J Clin Transl Hepatol 2014; 2: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maepa MB, Roelofse I, Ely A, Arbuthnot P. Progress and prospects of anti-HBV gene therapy development. Int J Mol Sci 2015; 16: 17589–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H, Hou Z, Han Q, Zhang C, Zhang J. The anti-HBV effect mediated by a novel recombinant eukaryotic expression vector for IFN-alpha. Virol J 2013; 10: 270–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000; 403: 901–6. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004; 101: 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004; 64: 3753–6. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Shi W, Gao Y, Yang B, Jing X, Shan S, Wang Y, Du Z. Analysis of microRNA expression profiles in human hepatitis B virus-related hepatocellular carcinoma. Clin Lab 2013; 59: 1009–15. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan CS. New roles for large and small viral RNAs in evading host defences. Nat Rev Genet 2008; 9: 503–7. [DOI] [PubMed] [Google Scholar]
- 12.Hou ZH, Han QJ, Zhang C, Tian ZG, Zhang J. miR146a impairs the IFN-induced anti-HBV immune response by downregulating STAT1 in hepatocytes. Liver Int 2014; 34: 58–68. [DOI] [PubMed] [Google Scholar]
- 13.Xing TJ, Xu HT, Yu WQ, Wang B, Zhang J. MiRNA-548ah, a potential molecule associated with transition from immune tolerance to immune activation of chronic hepatitis B. Int J Mol Sci 2014; 15: 14411–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, Roggendorf M, Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology 2011; 53: 1476–85. [DOI] [PubMed] [Google Scholar]
- 15.Hsia CC, Axiotis CA, Di Bisceglie AM, Tabor E. Transforming growth factor-alpha in human hepatocellular carcinoma and coexpression with hepatitis B surface antigen in adjacent liver. Cancer 1992; 70: 1049–56. [DOI] [PubMed] [Google Scholar]
- 16.Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, Huang GT, Iloeje UH, Group R-HS. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 2006; 295: 65–73. [DOI] [PubMed] [Google Scholar]
- 17.Lin CL, Kao JH. Risk stratification for hepatitis B virus related hepatocellular carcinoma. J Gastroenterol Hepatol 2013; 28: 10–7. [DOI] [PubMed] [Google Scholar]
- 18.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull 2006; 29: 903–6. [DOI] [PubMed] [Google Scholar]
- 19.Zhang HH, Wang XJ, Li GX, Yang E, Yang NM. Detection of let-7a microRNA by real-time PCR in gastric carcinoma. World J Gastroenterol 2007; 13: 2883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell 2005; 120: 635–47. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Wang L, Chen W, Zhao S, Yin C, Lin Y, Jiang A, Zhang P. USP35 activated by miR let-7a inhibits cell proliferation and NF-kappaB activation through stabilization of ABIN-2. Oncotarget 2015; 6: 27891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng M, Hou J, Hu J, Wang S, Chen M, Chen L, Ju Y, Li C, Meng S. Hepatitis B virus mRNAs functionally sequester let-7a and enhance hepatocellular carcinoma. Cancer Lett 2016; 383: 62–72. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow P, Chung AY, Jooi LL, Lee CG. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol 2010; 53: 57–66. [DOI] [PubMed] [Google Scholar]
- 24.Sureau C, Romet-Lemonne JL, Mullins JI, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell 1986; 47: 37–47. [DOI] [PubMed] [Google Scholar]
- 25.Zhang HL, Dai LH, Wu YH, Yu XP, Zhang YY, Guan RF, Liu T, Zhao J. Evaluation of hepatocyteprotective and anti-hepatitis B virus properties of Cichoric acid from Cichorium intybus leaves in cell culture. Biol Pharm Bull 2014; 37: 1214–20. [DOI] [PubMed] [Google Scholar]