Abstract
North American amphibians have recently been impacted by two major emerging pathogens, the fungus Batrachochytrium dendrobatidis (Bd) and iridoviruses in the genus Ranavirus (Rv). Environmental factors and host genetics may play important roles in disease dynamics, but few studies incorporate both of these components into their analyses.
Here, we investigated the role of environmental and genetic factors in driving Bd and Rv infection prevalence and severity in a biodiversity hot spot, the southeastern United States.
We used quantitative PCR to characterize Bd and Rv dynamics in natural populations of three amphibian species: Notophthalmus perstriatus, Hyla squirella and Pseudacris ornata. We combined pathogen data, genetic diversity metrics generated from neutral markers, and environmental variables into general linear models to evaluate how these factors impact infectious disease dynamics. Occurrence, prevalence and intensity of Bd and Rv varied across species and populations, but only one species, Pseudacris ornata, harbored high Bd intensities in the majority of sampled populations. Genetic diversity and climate variables both predicted Bd prevalence, whereas climatic variables alone predicted infection intensity.
We conclude that Bd is more abundant in the southeastern United States than previously thought and that genetic and environmental factors are both important for predicting amphibian pathogen dynamics. Incorporating both genetic and environmental information into conservation plans for amphibians is necessary for the development of more effective management strategies to mitigate the impact of emerging infectious diseases.
Introduction
Infectious disease is a well-known driver of animal declines worldwide (e.g. [1–2]). Ectothermic vertebrates, particularly reptiles and amphibians, have been exceptionally impacted by emerging infectious diseases (EIDs) [1]. Observations of massive, worldwide, pathogen-associated amphibian die-offs date back to the 1970s and 80s [3]. In North America, amphibians have been impacted by two major emerging pathogens, the fungus Batrachochytrium dendrobatidis (Bd) and a class of iridoviruses collectively known as Ranavirus (Rv) [4–7]. Bd was identified as a disease agent in 1998 [8] and Rv was classified as an emerging pathogen after 1993 [9–10]. In North America, Bd has mainly impacted anurans in the families Ranidae and Bufonidae, causing mass mortality events for species in these families. Most infections have occurred in the western United States and have impacted already threatened and endangered species (e.g. [1,4, 11–12]). Rv has caused die-offs in at least 20 different reptile and amphibian species throughout the United States, including species under state and federal protection [4, 13–14].
In the southeastern United States (hereafter, Southeast), emerging pathogens impacting ectothermic vertebrates have been poorly characterized but have the potential to significantly impact the area. The Southeast is exceptionally rich in amphibian and reptile diversity, hosting more than half of species occurring in the United States [15]. EIDs therefore have potential to severely impact this region’s ecosystems and overall biodiversity. Presently, EIDs have been documented in multiple amphibian and reptile groups occurring in the Southeast [4, 13, 16], yet disease monitoring has been limited. One species, the Gopher frog (Lithobates capito), is susceptible to Rv infections in the lab [14] and infection and die-offs have been documented for wild populations [16–17]. However, impacts of pathogens on other species in this area are more enigmatic [13, 17–19]. Because disease monitoring has been limited, museum specimens and other archived biological samples are critical for retrospective pathogen detection and can aid in uncovering when pathogens were first introduced and where they were found in the past [20].
Climatic variables are significant drivers of pathogen prevalence in wildlife populations, and amphibians and their pathogens are no exception (e.g. [7, 21–22]). Temperature and precipitation are the two major environmental factors that appear to be drivers of Bd dynamics. Studies have shown a negative relationship between temperature and Bd occurrence, prevalence and intensity both in the lab [23–24] and in natural populations (e.g. [7, 25–27]). Additionally, variation in precipitation and humidity have been implicated in the occurrence and prevalence of Bd, with increased precipitation and humidity driving patterns (e.g. [23, 28]). Most work to date on Rv has focused on documenting infections. To our knowledge, there is a paucity of work investigating how environmental factors serve as drivers of Rv. Only two studies look at this pattern; one in which salamanders were experimentally infected under different temperature regimes and a negative relationship between Rv infection intensity and temperature was found [29] and one testing ranid frogs under differing temperature regimes [30].
There is increasing empirical evidence for a genetic basis to disease resistance in wild vertebrate populations [31–32], although more studies are needed to test this hypothesis in amphibian taxa [33]. To date, reduced genetic diversity has been found to increase susceptibility to one pathogen (Rv) in a single amphibian species based on neutral microsatellite loci [34]. Additionally, a handful of studies have explored the underlying genetic basis for disease resistance to Bd. Tobler and Schmidt [35] looked at among-population susceptibility to Bd in a European frog and inferred that differences in susceptibility among populations had a genetic basis due to differential population responses in a common garden experiment. Similarly, Savage et al. [36] found that neutral genetic diversity was negatively correlated with Bd infection prevalence in a North American frog. Immunogenetic analyses have also found significant associations between specific major histocompatibility complex (MHC) class II alleles and Bd tolerance in the lab and in natural systems for multiple species of anurans [37–39]. These studies suggest that host genetic diversity underlies differential amphibian population responses to EIDs, but are based on a limited number of taxa.
Despite the importance of environmental and genetic factors in explaining amphibian disease dynamics when investigated separately, few studies incorporate both into a single analysis. Ribas et al. [24] demonstrated in the lab that both temperature and expression of skin peptides determined how anuran hosts responded to Bd infection. Savage et al. [39] found that environmental factors were responsible for predicting Bd infection intensity, while both genetic and environmental factors influenced Bd prevalence. This was the first analysis to combine both genetic and environmental factors in a predictive model for EIDs in a natural amphibian system. More studies are needed to confirm the importance of both genetic and environmental factors for explaining amphibian EIDs.
Here, we assess whether EIDs are impacting amphibian populations in the southeastern United States by characterizing Bd and Rv dynamics using archived samples from three amphibian species: a salamander, Notophthalmus perstriatus and two tree frogs, Hyla squirella and Pseudacris ornata. Each species exhibits unique life history traits and all three occur throughout the Southeast, offering varying perspectives into pathogen dynamics among diverse taxa. Notopthalmus perstriatus has a complex life cycle, spending two out of its three life stages in the water [40]. Previous studies have observed that N. perstriatus and other Notophthalmus are susceptible to both Bd [19] and Rv [41]. Hyla squirella is a common, highly arboreal species that breeds in large aggregates during summer months (Elliot et al. 2009), and little is known about the presence or impact of Bd and Rv in this species. In contrast, P. ornata is an increasingly uncommon species (B. Means pers. comm.), breeds in winter at low densities [42], and is highly susceptible to Rv in experimental infection trials [43]. Field studies of P. ornata tadpoles in northern Florida have also confirmed Rv infections in the wild [17]. Previous population genetic analyses found that N. perstriatus form distinct East-West groups that do not share haplotypes [40], H. squirella genetic structure is heavily determined by habitat structure [44], and P. ornata genetic structure varies among populations and the species may have been widespread across the Southeast in the past [45]. By combining these genetic data with environmental and disease variables for each sampled population, we simultaneously assessed the importance of host genetics and environmental variables on predicting disease impact and spread in amphibian populations of the Southeast.
Materials and methods
Sample collection
All appropriate permits were acquired for the desired field and lab work. Vertebrate animal use was approved by University of Central Florida's IACUC, #06-01W, 09-13W, 09-21W. Samples were collected over various months throughout the Southeast Atlantic Coastal Plain from 1997 to 2010 (Table A in S1 File). Notophthalmus perstriatus samples were collected in Florida and Georgia from 1997 to 2000, with additional samples collected in 2008–2010. Samples were collected during various months over the entire collection period [40]. Hyla squirella samples were collected in Florida and Georgia in 2010 during summer months [44]. Pseudacris ornata samples were collected in Florida, Georgia, Alabama, South Carolina and North Carolina between 2006 and 2009 during winter months [45]. Toe clips were taken from anurans and tail clips were taken from salamanders. Each animal was then released where it was found. For N. perstriatus, tissue was either stored in saturated salt buffer (NaCl; 25 mM EDTA, pH 7.5; 20% DMSO), or in DrieRite Desiccant [40]. For H. squirella, tissue was stored in in anhydrous calcium sulfate [44]. P. ornata tissue samples were also stored in anhydrous calcium sulfate [45]. While multiple storage methods were used, long-term storage occurred in a -20C freezer, we ran a random assemblage of samples (10 samples from each species group) and tested elutions via a Microdrop assay in a BioTek plate reader to check for a comparison of the DNA extractions. No significant difference was found between species in amount of DNA present in elutions (P = .06).
Pathogen detection
DNA was extracted from whole tissue samples using DNeasy Blood and Tissue kits (Qiagen) or through the phenol—chloroform method [40, 44–45] and DNA elutions were stored at -20 C or cooler. Taqman quantitative (q)PCR was performed on extracted DNA using the Bio-Rad CFX96 Real-Time System and analyzed with Bio-Rad CFX Manager 3.1 software. Reaction volumes were 25 μL for all standards, samples and controls, consisting of: 8 μL of Bio-Rad Super Mix, 2 μL of 10 μM Forward primer (0.8 μM/ μL), 2 μL of 10 μM Reverse primer (0.8 μM/ μL), 3 μL of Molecular Grade water, 5 μL of 1 μM probe (Bd or Rv; 0.2 μM/ μL) and 5 μL of standard DNA template or sample DNA template. Cycling conditions were as follows: 95°C for 5 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 min. Bd reactions used primers and probes developed by Boyle et al. [46] and Rv reactions used primers and probes developed by Allender, Bunick and Mitchell [47]. Bd and Rv reactions were run separately on individual 96-well plates. For absolute pathogen quantification, standard curves were generated from serial dilutions of synthetic pathogen DNA (gBlock Gene Fragments) run in duplicate [48–49]. Two negative controls (molecular grade water) were included with each run, as well as a positive control. Samples were first run in pools, which consisted of 5 μL of DNA template from each individual within a population combined, to test for the presence of positives within a population. A result was considered positive if the DNA from 5 μL of the pooled sample amplified before cycle 39 for at least two runs. If a population tested positive, each individual was then tested. All positive samples were run twice and the average of the two values was used in downstream analysis. In rare cases when two runs were inconsistent (one positive and one negative, or more than an order of magnitude difference in infection intensity), a third run was performed and the two most consistent results were retained.
Pathogen data analyses
We used two metrics to catalog Bd and Rv infection across the species ranges for these three amphibians. Prevalence was calculated by dividing the number of infected individuals by the total population sample size, and 95% Clopper—Pearson binomial confidence intervals were calculated using the package binom in R (see S1 File for Data A (R code)). To compare prevalence across population and species, we analyzed two-way contingency tables using Fisher's Exact Tests with simulated P-values based on 2000 replicates. The second metric, infection intensity, was calculated as the mean number of Genome Equivalents (GE) among duplicate runs. Mean infection intensity per population was measured as the mean infection intensity among infected individuals only. To compare average infection intensity across populations and species, a two-way ANOVA was performed in R v. 3.1.3 [50]. Prevalence and intensity maps were created in ArcGIS v. 10.2.2 to visualize the spatial distribution of infections.
Genetic and environmental disease modeling
For species that tested positive for Bd or Rv in multiple populations, we used general linear models (GLMs) to predict pathogen prevalence (with binomial error; [51]) and the natural log of intensity based on genetic and environmental variables, as well as location. For genetic predictor variables, we used average expected heterozygosity (HE) and allelic richness (AR) as these were found to be important predictors of infection by Savage et al. [36]. Genetic diversity estimates were calculated by investigating diversity at 7 nuclear microsatellites for P. ornata [45], as this species was the only one used in modeling due to its infection prevalence recovered (Table B in S1 File). For environmental predictor variables, we used average precipitation, average temperature, and maximum and minimum temperature per population site for each month of the year. Location factors included latitude and longitude for each population site. Environmental data (current data interpolated from 1960–2000 data) was acquired through Worldclim/Bioclim layers in ArcGIS [52]. Environmental and location data were assessed with Principle Component Analysis (PCA). Additive and interactive models were created and assessed via variance inflation factor (vif) using the car package in R, and only models with values ≤4 were included [53]. GLMs meeting this criterion were ranked using Akaike information criterion (AICc), and the most informative model was chosen using the lowest AICc value [53].
Results
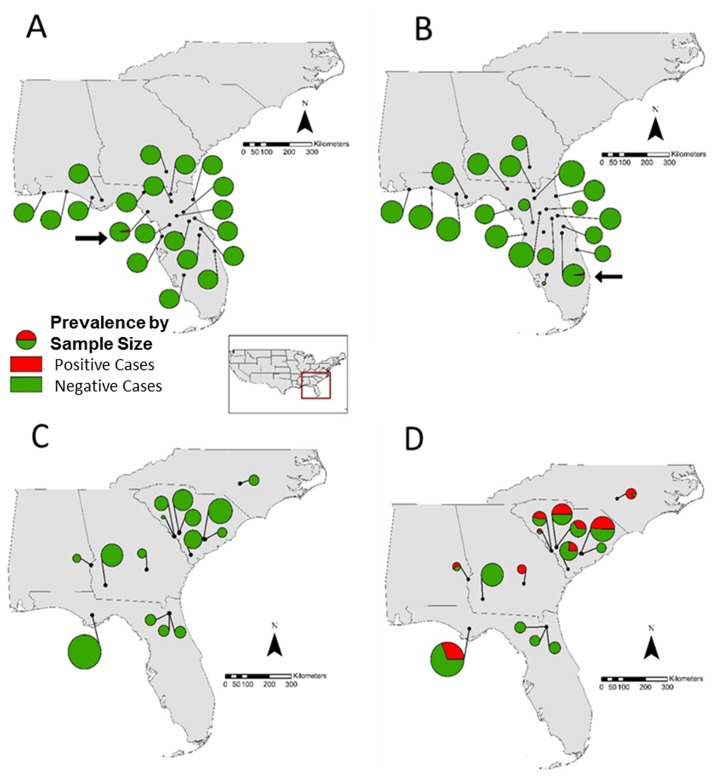
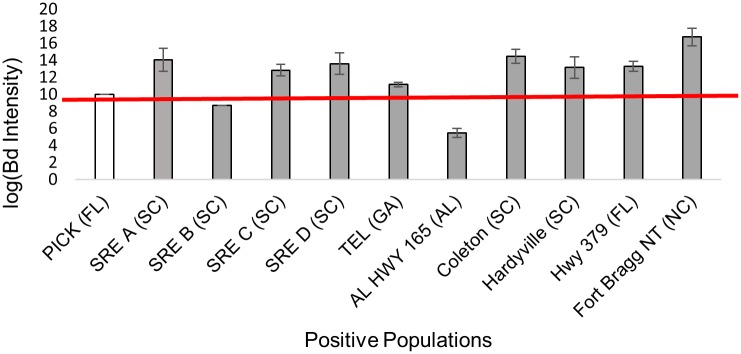
We tested 401 N. perstriatus tissue samples from 11 populations, none of which were infected with Bd or Rv. Among 580 H. squirella tissue samples from 20 populations, one individual was infected with Bd (Table C in S1 File; Fig 1a) and one individual from a different population tested positive for Rv (Table C in S1 File; Fig 1b). Neither Bd nor Rv prevalence were significantly different among H. squirella populations (Fisher Exact test P = 0.64). Finally, among 327 P. ornata tissue samples from 15 populations, 103 individuals from 10 populations were Bd positive (Table A in S1 File; Fig 1c), whereas none tested positive for Rv (Table C in S1 File; Fig 1d). Bd prevalence among infected P. ornata populations ranged from 0.24 to 1.0 (Table C in S1 File). Bd prevalence varied significantly among P. ornata populations (Fisher Exact test, P = 0.0005). Additionally, pooled Bd prevalence was significantly different in P. ornata compared to H. squirella (Fisher Exact test, P < 0.00001). Average Bd infection intensity among infected populations ranged from 226 to almost eighteen million genome equivalents (GE; Fig 2). Of the 11 infected populations, nine harbored average infections of over 10,000 GE (Table C in S1 File; Fig 2). All but one infected populations had 40% of individuals harboring loads above 10,000 GE and two populations had 100% of individuals with infections above 10,000 GE. Bd infection intensity was significantly different among infected P. ornata populations (two-way ANOVA, P < 0.00001).
Fig 1. Map of sample localities and (A) Bd prevalence in Hyla squirella, (B) Rv prevalence in H. squirella, (C) Bd prevalence in Pseudacris ornata and (D) Rv prevalence in P. ornate.
Circle size is relative to population size. Arrows point to infected H. squirella populations. Green represents proportion of negative cases of the indicated pathogen while red represents proportion of positive cases.
Fig 2. Log-transformed Bd average intensities for sampled populations of H. squirella and P. ornata with standard error of the mean (SEM).
White bars indicate average intensity among infected individuals from H. squirella populations and black bars denote average intensity among infected individuals from P. ornata populations. The red line marks an infection intensity of 10,000 GE.
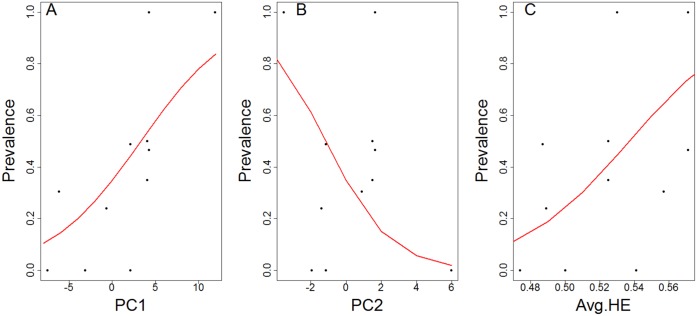
Because P. ornata was the only species harboring pathogens in more than one population, and Bd was the only pathogen detected, we limited GLM analyses to Bd dynamics within P. ornata. PCA of environmental variables across sampled P. ornata populations revealed that the first two components explained 89% of variation in our data (Fig A in S1 File). PC1 is associated with decreasing temperature and some increasing precipitation variables and positively associated with latitude. PC2 is positively associated with winter precipitation. We assessed only additive models with AICs for prevalence and intensity as interactive models showed high variance inflation. We chose the most informative models based on the lowest AIC score in the set (Table 1). PC1, PC2 and average heterozygosity significantly influenced Bd prevalence (Fig 3; Table D in S1 File). Based on our general linear models, there is a significant relationship between decreasing temperature and an increase in Bd prevalence (P = 0.0000002; Fig 3A). Moreover, there is a significant negative relationship between winter precipitation and Bd prevalence (P = 0.000591; Fig 3B). Surprisingly, our model identified a significant relationship between increased average heterozygosity and increased Bd prevalence (P = 0.0000698; Fig 3C). Environmental factors (PC1) significantly influenced Bd intensity (Fig B and Table E in S1 File). Linear regression displayed a significant relationship between a decrease in temperature and an increase of Bd intensity (P = 0.011; Fig B in S1 File).
Table 1. Five most informative general linear models and the null models for Bd prevalence and Bd intensity.
| Bd Prevalence models | AICc | dAICc | Df | Weight |
| PC1+PC2+AvgHE | 62.4 | 0 | 4 | 0.750 |
| PC1+PC2+AvgHE+AR | 64.6 | 2.2 | 5 | 0.249 |
| PC1+PC2+AR | 75.6 | 13.2 | 3 | 0.001 |
| PC1+PC2 | 80.4 | 18.0 | 2 | <0.001 |
| PC1+AvgHE | 82.6 | 20.3 | 3 | <0.001 |
| NULL | 112.6 | 50.3 | 1 | <0.001 |
| Bd Intensity models | AICc | dAICc | Df | Weight |
| PC1 | 87.0 | 0 | 3 | 0.5192 |
| PC1+AR | 89.6 | 2.6 | 4 | 0.1425 |
| PC1+AvgHE | 90.7 | 3.7 | 4 | 0.0823 |
| PC1+PC2 | 91.3 | 4.3 | 4 | 0.0601 |
| AvgHE | 91.8 | 4.8 | 3 | 0.0478 |
| NULL | 91.5 | 4.5 | 2 | 0.0559 |
AvgHE = average heterozygosity, AR = allelic richness, dAICc = delta Akaike information criterion. The difference in AICc between the current model and the most informative model in the set. df = degrees of freedom.
Fig 3. Logistic relationships between Bd prevalence and PC1 (A), PC2 (B), and average heterozygosity (C) in populations of P. ornata.
Increasing values of PC1 correspond most strongly to decreasing values of temperature. Increasing values of PC2 correspond most strongly to increasing winter precipitation. In all panels, each dot corresponds to the observed prevalence in a population and each line corresponds to the best fit logistic model of the relationship between the two variables shown.
Discussion
Our study used both environmental and genetic variables to create a predictive model for chytridiomycosis disease dynamics in the southeastern United States. We identified both concordant and discordant patterns of pathogen prevalence and infection intensity in N. perstriatus, H. squirella, and P. ornata compared to previous studies [18–19, 54]. First, our data showed limited Rv occurrence and high variation in Bd infection prevalence within and among our three focal species. Second, we found surprisingly high Bd infection intensity in P. ornata and H. squirella, a strikingly different result compared to the extremely low intensities previously detected [54]. Finally, our overall model found that both genetic and environmental variables predict Bd prevalence but only environmental variables predict infection intensity. These results are important for understanding the enigmatic story of Bd infection in this region, and provide a framework for future management of declining amphibians in the Southeast. Overall, this study offers insight into pathogen infection history and amphibian disease dynamics in the southeastern United States, a hotspot of amphibian diversity.
Our three focal amphibian species showed largely unique patterns of Bd and Rv infection prevalence compared to previous studies on the same or similar species. Interestingly, Rv was previously found within ponds where N. perstriatus populations occur [17], and a closely related Notophtahlmus species in the Southeast, N. viridescens, was found to have high Bd infection prevalence within sampled populations [19]. In contrast, we found no evidence of Bd or Rv in N. perstriatus, suggesting the presence of genetic, temporal or seasonal differences for our sampled populations. Hyla squirella infection dynamics were also surprising as this species breeds in large aggregates in habitats frequented by known Bd and Rv vector species, particularly Lithobates catesbianus [42, 55]. We predicted high pathogen prevalence in H. squirella, but instead only found two infected individuals (one Bd infected and one Rv infected) among all sampled populations. Interestingly, these are the first documented Bd and Rv infections in H. squirella despite previous sampling efforts [19]. These results indicate low overall infection prevalence in H. squirella, but the high Bd infection intensity we detected in one individual suggests the potential for negative impacts on populations where infection does occur. The only focal species with high pathogen prevalence was P. ornata, which exhibited strikingly different infection patterns for Rv and Bd compared to H. squirella. While Rv infection was undetectable in P. ornata, high Bd infection prevalence and intensity occurred in the majority of sampled populations. These results mirror previous studies testing P. ornata and other members of Pseudacris for pathogens in the Southeast [19, 54].
Life history variation among amphibian species often contributes to variation in pathogen infection prevalence [56]. The distinct life histories among our three study species may be an underlying factor contributing to the significant differences in Bd prevalence and intensity that we observed. In particular, high susceptibility to Bd infection in P. ornata may be due to the trait of breeding exclusively during rainy periods in the winter months [42], unlike the other two species that are summer breeders. Our data, along with data from several other studies, correlate Bd infection with cooler temperatures and higher precipitation in winter months [7, 26–27, 56–57]. Thus, high contact rates among P. ornata individuals during winter breeding aggregations when Bd experiences preferred temperatures may be driving the observed high infection prevalence and intensity. Our results, combined with previous monitoring efforts, serve as a valuable baseline should infection outbreaks occur for other southeastern amphibian species and may help elucidate reasons for any enigmatic declines.
Our Bd infection intensity data seem to contradict previous studies suggesting that values greater than 10,000 GE lead to mortality, regardless of the amphibian species [58–59]. We found high Bd intensities that surpassed this threshold for the single infected H. squirella individual and within most infected P. ornata individuals (Fig 2). Although high Bd infection prevalence in Pseudacris populations is well documented in the literature [19–20, 54], only one other study quantified Bd intensity for P. ornata; they found low values (<102 GE) for all individuals sampled throughout the United States [54]. We uncovered a pattern that is much more extreme; average Bd intensities were millions of GE for all but three infected populations (Fig 2) despite the absence of any observed disease signs or mortality events (T. Hether pers. comm.). Our data therefore demonstrate that the 10,000 GE proposed mortality threshold is not a standard applicable to every species. Indeed, our findings reinforce recent Bd studies in Brazilian amphibian communities [60] and New York State amphibian communities [61] which showed varying infection intensities across a range of species and seasons without observing any mortality or disease signs in the individuals sampled.
To uncover the driving forces behind the observed patterns of Bd infection among our sampled P. ornata populations, we incorporated climatic variables and genetic diversity factors into a comprehensive model. Bd intensity increased at lower air temperatures, consistent with similar analyses in other species and regions (e.g. [27, 36, 56, 62]. Further, the same variables that influenced Bd prevalence for P. ornata in our study (temperature, precipitation and average heterozygosity) also explained Bd prevalence in L. yavapaiensis in Arizona [36]. However, in contrast to the negative correlation between average heterozygosity and Bd prevalence found for L. yavapaiensis, we found average heterozygosity was positively correlated with Bd prevalence for P. ornata (Fig 3C). This pattern is in direct contrast with expectations, as numerous studies across wildlife disease systems have found higher genetic diversity within populations leads to decreased infection prevalence and increased disease resistance (e.g. [34, 63–64]. Our models show that for P. ornata, the opposite is true: increased genetic diversity correlates positively with Bd prevalence. Two possible explanations exist for this pattern. First, because average heterozygosity increases with larger effective population size, there could be better facilitation of pathogen spread due to density-dependent disease outbreaks [65–66]. This explanation is unlikely for P. ornata, however, as this species is generally uncommon and has historically small population sizes [42]. Another possible explanation is that Bd swept through P. ornata populations before our sampling occurred, and selection favoring Bd tolerant individuals was strong enough to push tolerant genotypes towards fixation, resulting in decreased heterozygosity. Bd has been present in the United States long enough to make this “genetic purging” [67] scenario plausible; Ouelett et al. [20] and Talley et al. [68] found evidence of Bd infections existing in North America as far back as the late 1800s. Our results could thus represent indirect evidence of genetic tolerance to Bd evolving in natural amphibian populations [39], although further genetic sampling, molecular tests of selection and experimental evidence of Bd tolerance are necessary to resolve this hypothesis.
Our data strongly suggest that both genetic and environmental factors should be incorporated, when possible, into models when trying to predict dynamics of infectious pathogens in natural populations. Management plans often only consider genetic or environmental factors when planning for long-term species persistence, but it is becoming increasingly clear that both are important for predicting pathogen impacts. While our study only focuses on amphibians, this modeling framework is applicable and important for other wildlife disease systems. Our results also suggest that Bd may be more of a concern for the Southeast than previously thought, at least for some species. There are no documented instances of disease-driven morbidity or mortality in P. ornata, yet a majority of sampled individuals were heavily infected with Bd and there is evidence of population declines in recent decades (B. Means pers.comm.). Cryptic chytridiomycosis may therefore be an unobserved but causal factor behind population declines and patterns of genetic diversity. Alternately, the Bd strain(s) present in the Southeast may currently exist as commensals or sub-lethal pathogens in P. ornata. Even under the latter scenario, monitoring Bd and other pathogen dynamics is important for future P. ornata conservation efforts. If novel biotic or abiotic stressors appear, the additional toll of harboring massive Bd intensities may be a tipping point towards extirpation. This may be especially true for populations with low genetic diversity, even if that loss of diversity is due to selection for pathogen tolerance. Means and Means (unpublished data) highlight that habitat destruction and degradation are threatening P. ornata population persistence, and more recently Means et al. [17] found wild P. ornata tadpoles to be heavily infected with Rv. Our results suggest Bd is a threat for adult frogs, particularly if the same populations are impacted by Rv prior to metamorphosis.
Overall, our study highlights how species are differentially impacted by EIDs in the Southeast and how models can be used to infer which environmental and genetic factors are drivers of infection. Infectious disease is often implicated in amphibian population declines only after morbidity and mortality is observed, making the trigger for a disease outbreak difficult to determine retrospectively. It has therefore become increasingly important to characterize and monitor species that have yet to display signs of disease in order to generate a baseline of pathogen dynamics should any future disease outbreak occur. Museum collections and specimens collected for non-disease studies are invaluable for assessing conditions faced by amphibians in the past [20], and here we utilized these resources to document the presence of two infectious pathogens in two frog species without any prior evidence of disease. Whether ubiquitous Bd infections in P. ornata reflect post-epidemic adaptation, non-pathogenic Bd strains, or virulent, ongoing chytridiomycosis that has gone undetected will require additional analyses. Regardless, our modeling results highlight the combined importance of host genetic variation and climate for determining Bd prevalence. Other climatic factors, such as seasonality, may also play a big part in disease dynamics and should be considered in future studies. Our results begin the journey to uncovering amphibian pathogen dynamics and can be used to develop more robust predictive models to assess where pathogens will likely spread and to inform species managers, as well as target suitable future re-introduction sites for amphibians that have been hit the hardest by disease-related declines.
Supporting information
(DOCX)
Acknowledgments
We thank Zachary Rivas and Sanabel Mahmoud for providing assistance with lab work and Jacob Degner and Sarah May for providing tissue samples. We also thank Taina Torres for assistance with the prevalence figures. This research was supported by grants from the US Fish and Wildlife Service and Florida Fish and Wildlife Conservation Commission’s Nongame Wildlife Program to E.A.H., Sigma Xi Grants-in-Aid research #G200810150636 to T.D.H., and the University of Central Florida. All applicable institutional and/or national guidelines for the care and use of animals were followed.
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
This work was funded by the US Fish and Wildlife Service: EAH, Florida Fish and Wildlife Conservation Commission’s Nongame Wildlife Program: EAH, and Sigma Xi Grants-in-Aid: TDH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Daszak P, Berger L, Cunningham AA, Hyatt AD, Green DE, Speare R. Emerging infectious diseases and amphibian population declines. Emerg Infect Diseas. 1999; 5: 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hochachka W M, Dhondt AA. Density-dependent decline of host abundance resulting from a new infectious disease. Proc Natl Acad Sci. 2000; 97 5303–5306. 10.1073/pnas.080551197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barinaga N. Where have all the froggies gone. Science 1990. 247: 1033–1034 10.1126/science.247.4946.1033 [DOI] [PubMed] [Google Scholar]
- 4.Green DE, Converse KA, Schrader AK. Epizootiology of sixty-four amphibian morbidity and mortality events in the USA, 1996–2001. Ann N. Y. Acad Sci. 2002; 969: 323–339 [DOI] [PubMed] [Google Scholar]
- 5.Briggs CJ, Knapp RA,Vredenburg VT. Enzootic and epizootic dynamics of the chytrid fungal pathogen of amphibians. Proc Natl Acad Sci. 2010; 107: 9695–9700. 10.1073/pnas.0912886107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller D, Gray M, Storfer A. Ecopathology of Ranaviruses infecting amphibians. Viruses 2011;3: 2351–2373. 10.3390/v3112351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savage AE, Sredl MJ, Zamudio KR. Disease dynamics vary spatially and temporally in a North American amphibian. Biol Conserv. 2011;144: 1910–1915. [Google Scholar]
- 8.Berger L, Speare R, Daszak P, Green DE, Cunningham AA, Goggin CL, et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci. 1998; 95: 9031–9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher MC, Garner TWJ, Walker SF. Global Emergence of Batrachochytrium dendrobatidis and Amphibian Chytridiomycosis in Space, Time, and Host. Ann Rev Microbiol. 2009; 63: 291–310. [DOI] [PubMed] [Google Scholar]
- 10.Gray M, Miller D; Hoverman J. (2009) Ecology and pathology of amphibian ranaviruses. Diseas Aquat Organ. 2009; 87: 243–266. [DOI] [PubMed] [Google Scholar]
- 11.Pearl CA, Bull EL, Green DE, Bowerman J, Adams MJ, Hyatt A, et al. Occurrence of the Amphibian Pathogen Batrachochytrium Dendrobatidis in the Pacific Northwest. J Herpetol. 2007;41: 145–149. [Google Scholar]
- 12.Muths E, Pilliod DS, Livo LJ. Distribution and environmental limitations of an amphibian pathogen in the Rocky Mountains, USA. Biol Conserv. 2008;141: 1484–1492. [Google Scholar]
- 13.Johnson AJ, Pessier AP, Wellehan JFX, Childress A, Norton TM, Stedman NL, et al. Ranavirus infection of free-ranging and captive box turtles and tortoises in the United States. J Wildl Diseas. 2008; 44: 851–863 [DOI] [PubMed] [Google Scholar]
- 14.Hoverman JT, Gray MJ, Haislip NA, Miller DL. Phylogeny, life history, and ecology contribute to differences in amphibian susceptibility to ranaviruses. EcoHealth. 2011; 8: 301–319. 10.1007/s10393-011-0717-7 [DOI] [PubMed] [Google Scholar]
- 15.Hanson C. (2010) Southern Forests for the Future. World Resources Institute, Washington, DC. [Google Scholar]
- 16.Landsberg J, Kiryu Y, Tabuchi M, Waltzek T, Enge K, Reintjes-Tolen S, et al. Co-infection by alveolate parasites and frog virus 3-like ranavirus during an amphibian larval mortality event in Florida, USA. Diseas Aquat Organ. 2013;105: 89–99. [DOI] [PubMed] [Google Scholar]
- 17.Means R, Means RPM, Miller DL, Gray MJ, Johnson SA, Means DB, et al. A conservation strategy for the imperiled Striped Newt (Notophthalmus perstriatus) in the Apalachicola National Forest, Florida. Third Annual Report to the US Forest Service. 2013
- 18.Chatfield MWH, Moler P, Richards-Zawacki CL. The Amphibian Chytrid Fungus, Batrachochytrium dendrobatidis, in Fully Aquatic Salamanders from Southeastern North America (ed J Sturtevant). PLoS One. 2012; 7: e44821 10.1371/journal.pone.0044821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothermel B, Walls S, Mitchell J, Dodd C, Irwin L, Green D, et al. Widespread occurrence of the amphibian chytrid fungus Batrachochytrium dendrobatidis in the southeastern USA. Diseas Aquat Organ. 2008;82: 3–18. [DOI] [PubMed] [Google Scholar]
- 20.Ouellet M, Mikaelian I, Pauli BD, Rodrigue J, Green DM. Historical Evidence of Widespread Chytrid Infection in North American Amphibian Populations. Conserv Biol. 2005;19: 1431–1440. [Google Scholar]
- 21.Gremillion-Smith C, Woolf A. Epizootiology of skunk rabies in North America. Jour. Of Wild. Diseas. 1988; 24(4):620–6. [DOI] [PubMed] [Google Scholar]
- 22.Altizer S, Hochachka WM, Dhondt AA. Seasonal dynamics of mycoplasmal conjunctivitis in eastern North American house finches. J Anim Ecol. 2004; 73: 309–322. [Google Scholar]
- 23.Woodhams DC, Alford RA, Marantelli G. Emerging disease cured by elevated body temperature. Diseas Aquat Organ. 2003;55: 65–66 [DOI] [PubMed] [Google Scholar]
- 24.Ribas L, Li MS, Doddington BJ, Robert J, Seidel JA, Kroll JS, et al. Expression profiling the temperature-dependent amphibian response to infection by Batrachochytrium dendrobatidis. PLoS One. 2009;4: e8408 10.1371/journal.pone.0008408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger L, Speare R, Daszak P, Green DE, Cunningham AA, Goggin CL, et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci. 1998; 95: 9031–9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodhams DC, Hyatt AD, Boyle DG, Rollins-Smith LA. The northern leopard frog Rana pipiens is a widespread reservoir species harboring Batrachochytrium dendrobatidis in North America. Herpetol Rev. 2008;39: 66 [Google Scholar]
- 27.Sapsford SJ, Alford RA, Schwarzkopf L. Elevation, temperature, and aquatic connectivity all influence the infection dynamics of the amphibian chytrid fungus in adult frogs. PloS One. 2013;8: e82425 10.1371/journal.pone.0082425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson ML, Berger L, Phillips L, Speare R. Fungicidal effects of chemical disinfectants, UV light, desiccation and heat on the amphibian chytrid, Batrachochytrium dendrobatidis. Diseas. Aquat. Organ. 2003;57:255–60. [DOI] [PubMed] [Google Scholar]
- 29.Rojas S, Richards K, Jancovich JK, Davidson EW. Influence of temperature on Ranavirus infection in larval salamanders Ambystoma tigrinum. Diseas Aquat Organ. 2005;63: 95–100. [DOI] [PubMed] [Google Scholar]
- 30.Echaubard P, Leduc J, Pauli B, Chinchar V G, Robert J, Lesbarreres D. Environmental dependency of amphibian—Ranavirus genotypic interactions: evolutionary perspectives on infectious diseases. Evol Appl. 2014; 7: 723–733. 10.1111/eva.12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiegertjes GF, Stet RM, Parmentier H K, van Muiswinkel WB. Immunogenetics of disease resistance in fish: a comparative approach. Devel Comput Immunol. 1996;20: 365–381. [DOI] [PubMed] [Google Scholar]
- 32.Meagher S. Genetic diversity and Capillaria hepatica (Nematoda) prevalence in Michigan deer mouse populations. Evolution 1999;53: 1318–1324. [DOI] [PubMed] [Google Scholar]
- 33.Richmond JQ, Savage AE, Zamudio KR, Rosenblum EB. Toward immunogenetic studies of amphibian chytridiomycosis: linking innate and acquired immunity. Bioscience. 2009;59: 311–320. [Google Scholar]
- 34.Pearman PB, Garner TW. Susceptibility of Italian agile frog populations to an emerging strain of Ranavirus parallels population genetic diversity. Ecol Letter. 2005;8: 401–408. [Google Scholar]
- 35.Tobler U, Schmidt BR. Within-and among-population variation in chytridiomycosis-induced mortality in the toad Alytes obstetricans. PLoS One. 2010;5: e10927 10.1371/journal.pone.0010927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savage AE, Becker CG, Zamudio KR. Linking genetic and environmental factors in amphibian disease risk. Evol Appl. 2015;8: 560–572. 10.1111/eva.12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savage AE, Zamudio KR. MHC genotypes associate with resistance to a frog-killing fungus. Proc Natl Acad Sci. 2011;108: 16705–16710. 10.1073/pnas.1106893108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bataille A, Cashins SD, Grogan L, Skerratt LF, Hunter D, McFadden M, et al. Susceptibility of amphibians to chytridiomycosis is associated with MHC class II conformation. Proc Royal Soc London B: Biol Sci. 2015; 282: 20143127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savage AE, Zamudio KR. Adaptive tolerance to a pathogenic fungus drives major histocompatibility complex evolution in natural amphibian populations. Proc Natl Acad Sci. 2016;283: 20153115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.May SE, Medley KA, Johnson SA, Hoffman EA. Combining genetic structure and ecological niche modeling to establish units of conservation: A case study of an imperiled salamander. Biol Conserv. 2011;144: 1441–1450. [Google Scholar]
- 41.Means R, Means RPM, Miller DL, Gray MJ, Johnson SA, Means DB, et al. conservation strategy for the imperiled Striped Newt (Notophthalmus perstriatus) in the Apalachicola National Forest, Florida. First Annual Report to the US Forest Service. 2011.
- 42.Elliot L, Gerhardt C, Davidson C. Frogs and Toads of North America. New York, New York: 2009. [Google Scholar]
- 43.Brenes RM. Mechanisms Contributing to the Emergence of Ranavirus in Ectothermic Vertebrate Communities. PhD Thesis, University of Tennessee, Knoxville. 2013.
- 44.Hether TD, Hoffman EA. Machine learning identifies specific habitats associated with genetic connectivity in Hyla squirella: Frogs in space. J Evol Biol. 2012; 25: 1039–1052. 10.1111/j.1420-9101.2012.02497.x [DOI] [PubMed] [Google Scholar]
- 45.Degner JF, Silva DM, Hether TD, Daza JM, Hoffman EA. Fat frogs, mobile genes: unexpected phylogeographic patterns for the ornate chorus frog (Pseudacris ornata): Genetic structure in the Ornate Chorus Frog. Molec Ecol. 2010; 2501–2515. [DOI] [PubMed] [Google Scholar]
- 46.Boyle DG, Boyle DB, Olsen V, Morgan JAT, Hyatt AD. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Diseas Aquat Organ. 2004; 60: 141–148. [DOI] [PubMed] [Google Scholar]
- 47.Allender MC, Bunick D, Mitchell MA. Development and validation of TaqMan quantitative PCR for detection of frog virus 3-like virus in eastern box turtles (Terrapene carolina carolina). Virol Methods. 2013; 188: 121–125. [DOI] [PubMed] [Google Scholar]
- 48.Gunawardana M, Chang S, Jimenez A, Holland-Moritz D, Holland-Moritz H, La Val TP, Lund C, et al. Isolation of PCR quality microbial community DNA from heavily contaminated environments. J Microbiol Methods. 2014; 102: 1–7. 10.1016/j.mimet.2014.04.005 [DOI] [PubMed] [Google Scholar]
- 49.Sandkam B, Young CM, Breden F. Beauty in the eyes of the beholders: colour vision is tuned to mate preference in the Trinidadian guppy (Poecilia reticulata). Molec Ecol. 2015;24: 596–609. [DOI] [PubMed] [Google Scholar]
- 50.R Core Team (2016) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL http://www.R-project.org/ [Google Scholar]
- 51.Warton DI, Hui FK. The arcsine is asinine: the analysis of proportions in ecology. Ecology. 2011;92:3–10. [DOI] [PubMed] [Google Scholar]
- 52.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International journal of climatology. 2005. December 1;25(15):1965–78. [Google Scholar]
- 53.Burnham KP, Anderson DR. Model Selection and Inference: A Practical Information-Theoretic Approach. Springer, New York: 2002. [Google Scholar]
- 54.Peterson CE, Lovich RE, Phillips CA, Dreslik MJ, Lannoo MJ. Prevalence and seasonality of the smphibian chytrid fungus Batrachochytrium dendrobatidis along widely separated longitudes across the United States. EcoHealth. 2016;1–15. [DOI] [PubMed] [Google Scholar]
- 55.Garner TW, Perkins MW, Govindarajulu P, Seglie D, Walker S, Cunningham AA, et al. The emerging amphibian pathogen Batrachochytrium dendrobatidis globally infects introduced populations of the North American bullfrog, Rana catesbeiana. Biol Letters. 2006; 2: 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kriger KM, Hero JM. The chytrid fungus Batrachochytrium dendrobatidis is non-randomly distributed across amphibian breeding habitats. Divers Distrib. 2007a; 13: 781–788. [Google Scholar]
- 57.Woodhams DC, Alford RA. Ecology of chytridiomycosis in rainforest stream frog assemblages of tropical Queensland. Conserv Biol. 2005;19: 1449–1459. [Google Scholar]
- 58.Vredenburg VT, Knapp RA, Tunstall TS, Briggs CJ. Dynamics of an emerging disease drive large-scale amphibian population extinctions. Proc Natl Acad Sci. 2010;107: 9689–9694. 10.1073/pnas.0914111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kinney VC, Heemeyer JL, Pessier AP, Lannoo MJ. Seasonal pattern of Batrachochytrium dendrobatidis infection and mortality in Lithobates areolatus: Affirmation of Vredenburg's “10,000 Zoospore Rule”. PLoS One. 2011; 6: e16708 10.1371/journal.pone.0016708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Preuss JF, Lambertini C, Leite DDS, Toledo LF, Lucas EM. Crossing the threshold: an amphibian assemblage highly infected with Batrachochytrium dendrobatidis in the southern Brazilian Atlantic forest. Stud Neotrop Fauna Environ. 2016;51: 68–77. [Google Scholar]
- 61.Lenker MA, Savage AE, Becker CG, Rodriguez D, Zamudio KR. Batrachochytrium dendrobatidis infection dynamics vary seasonally in upstate New York, USA. Diseas Aquat Organ. 2014;111: 51–60. [DOI] [PubMed] [Google Scholar]
- 62.Kriger KM, Hero JM. Large-scale seasonal variation in the prevalence and severity of chytridiomycosis. J Zool. 2007b; 271: 352–359. [Google Scholar]
- 63.Lande R. Genetics and demography in biological conservation. Science. 1988; 241: 1455–1460. [DOI] [PubMed] [Google Scholar]
- 64.Spielman D, Brook BW, Briscoe DA, Frankham R. Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv Genet. 2004;5: 439–448. [Google Scholar]
- 65.Hochachka W M, Dhondt AA. Density-dependent decline of host abundance resulting from a new infectious disease. Proc Natl Acad Sci. 2000; 97 5303–5306. 10.1073/pnas.080551197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langwig KE, Frick WF, Reynolds R, Parise KL, Drees KP, Hoyt JR, et al. Host and pathogen ecology drive the seasonal dynamics of a fungal disease, white-nose syndrome. Proc R Soc Lond B Biol Sci. 2015;282: 20142335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crnokrak P, Barrett SC. Perspective: purging the genetic load: a review of the experimental evidence. Evolution. 2002; 56: 2347–2358. [DOI] [PubMed] [Google Scholar]
- 68.Talley BL, Muletz CR, Vredenburg VT, Fleischer RC, Lips KR. A century of Batrachochytrium dendrobatidis in Illinois amphibians (1888–1989). Biol Conserv. 2015;182: 254–261. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.