Abstract
The indigenous people of the Tibetan Plateau have been the subject of much recent interest because of their unique genetic adaptations to high altitude. Recent studies have demonstrated that the Tibetan EPAS1 haplotype is involved in high altitude-adaptation and originated in an archaic Denisovan-related population. We sequenced the whole-genomes of 27 Tibetans and conducted analyses to infer a detailed history of demography and natural selection of this population. We detected evidence of population structure between the ancestral Han and Tibetan subpopulations as early as 44 to 58 thousand years ago, but with high rates of gene flow until approximately 9 thousand years ago. The CMS test ranked EPAS1 and EGLN1 as the top two positive selection candidates, and in addition identified PTGIS, VDR, and KCTD12 as new candidate genes. The advantageous Tibetan EPAS1 haplotype shared many variants with the Denisovan genome, with an ancient gene tree divergence between the Tibetan and Denisovan haplotypes of about 1 million years ago. With the exception of EPAS1, we observed no evidence of positive selection on Denisovan-like haplotypes.
Author summary
The Tibetan population has been residing on high plateau for thousands of years and developed unique adaptation to the local environment. To investigate the demographic history of Tibetans and search for possible adaptive genetic variants, we performed whole-genome sequencing of 27 Tibetan individuals. We found evidence of genetic separation between Han and Tibetans around since 44 and 58 thousand years ago; however, these two populations maintained a high rate of gene flow until 9 thousand years ago. In addition to replicating two previously discovered candidate genes (EGLN1 and EPAS1) for high altitude adaptation, we also found three new candidate genes, including PTGIS, VDR and KCTD12. We confirmed the high similarity of EPAS1 gene region between Tibetans and Denisovans, but did not detect any evidence of high altitude adaptation from Denisovan gene alleles otherwise.
Introduction
Adaptation to high altitude is among the most notable examples of natural selection in our species. Tibetans exhibit an exceptional capacity to compensate for decreased oxygen availability (hypoxia), UV exposure, cold, and limited food sources as exemplified by continuous habitation at greater than 4000 meters above sea level on the Qinghai-Tibetan Plateau over several millennia[1–4].
Many candidate genes have been highlighted as likely contributors to Tibetans’ trans-generational success at high altitude[5–9]. These findings are largely based upon the overlap of a priori genes, specifically those involved in hypoxia tolerance, and genomic footprints of adaptation identified in patterns within single nucleotide polymorphism (SNP) microarrays[10–12] or exome sequencing data[13]. In a few cases, putatively adaptive tagging variants in hypoxia inducible factor (HIF) pathway genes (EPAS1[10,13], EGLN1 and PPARA[12]) are associated with relatively lower hemoglobin concentration exhibited by many Tibetans at altitude. With the exception of the EGLN1 locus[14,15], however, precise functional variants within these genomic regions are unknown.
Recent advances in whole-genome sequencing (WGS) technology provide progress toward identifying the functional variants that underlie high-altitude adaptation. Rather than focusing on a relatively small portion of the genome using candidate gene approaches, WGS selection analyses often have sufficient power to comprehensively interrogate the genome in an unbiased manner. Moreover, WGS offers the opportunity to perform exhaustive searches for adaptive variation, yielding complete surveys of non-protein coding regulatory, conserved, and structural variants possibly tagged or missed in microarray analyses. WGS data are also important from an evolutionary standpoint, providing unbiased insights into long-standing questions regarding adaptive processes, including the role of adaptive introgression, as recently revealed through the targeted sequence comparisons between Tibetans and the Denisovan genome at the EPAS1 gene locus[16].
To identify regions contributing to high-altitude adaptation and regions with archaic introgressions with high resolution, we performed a comprehensive genomic analysis of WGS data in 27 Tibetans[17]. We estimated the demographic history of Tibetans using MSMC and ∂a∂i, and performed the Composite of Multiple Signals test for fine-mapping loci and variants with recent selective sweep. We also estimated genome-wide levels of archaic admixture, and generated a fine-scale map of Denisovan-like introgression in Tibetans.
Results
Global genomic analysis and demographic history estimate
We conducted principal components analysis (PCA) on all SNVs with observed non-reference allele frequency greater than 5% among the combined samples of Tibetans and five 1000 Genomes Project[18] populations (Peruvian, Punjabi, Chinese, Yoruba and European) (Supplementary Methods). Tibetans and Han Chinese were closely clustered in the first 4 principal components (S1A and S1B Fig) similar to our previous observation[12], but were clearly differentiated along 5th principal component (S1C Fig). Similarly, ADMIXTURE analysis[19] (Supplementary Methods) on the same six populations shows that when the number of ancestral populations (K) was set to less than 6, Tibetans and Han Chinese were genetically similar. However, when K was set to 6 or greater, these two populations exhibited distinct genetic profiles (S2 Fig). The estimated proportion of Tibetan genetic ancestry was greater than 99% for 19 out of the 27 Tibetan individuals. For the remaining Tibetan individuals, the average estimated proportion of Tibetan genetic ancestry was 83.1%. For the demographic history analysis, we excluded Tibetan individuals with less than 99% Tibetan genetic ancestry. FST between Tibetans and Chinese based on our SNV data was 0.0148 using the Hudson estimator[20] and 0.0149 using the Weir and Cockerham estimator [21], similar to our previous analyses based on SNP microarray data[22], which is slightly higher than the FST between Chinese and Japanese populations but lower than the FST between pairs of HapMap populations from different continents [23].
We next annotated the functional impact of SNVs that are frequent in Tibetans but uncommon in other populations. We selected all germline SNVs with an alternative allele frequency above 10% in Tibetans but below 1% among Yoruba, Han Chinese, and Europeans from the 1000 Genomes Project. In total, we identified 10,702 such SNVs, among which there were 65 nonsynonymous variants that occurred within 58 genes (S1 Table). We predicted the impact of these protein-altering SNVs on genome functions using PolyPhen-2 and the Conservation-controlled Amino Acid Substitution Matrix (CASM) in VAAST 2.0[24]. Of the 65 nonsynonymous variants, Polyphen-2 predicted 12 to be functional and CASM predicted a distinct set of 3 variants to be functional. One of the variants, Chr2:233244223 A->C (p.T104P; all genomic coordinates in this manuscript are relative to GRCh37 reference sequence), is in the gene ALPP with an observed frequency of 0.14% in dbSNP and 14.81% in Tibetans. ALPP encodes an alkaline phosphatase, which is mainly expressed in placental and endometrial tissues. Rare nonsynonymous variants in ALPP have previously been associated with decreased risk of spontaneous abortion and in vitro fertilization failure[25].
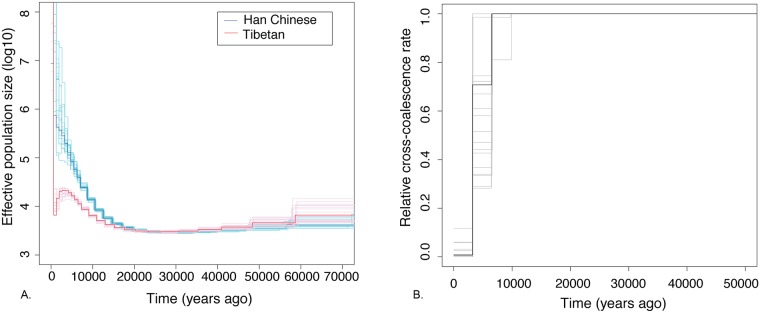
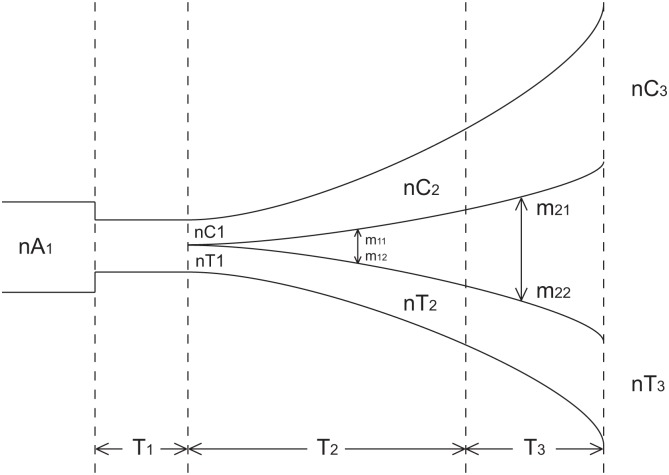
We inferred the demographic history of Tibetans using the multiple sequential Markovian Coalescent (MSMC)[26] (Fig 1) and ∂a∂i[27] (Fig 2; Table 1) (Supplementary Methods). MSMC estimated that the relative cross coalescence rate (a measurement of the amount of gene flow between two populations) between Han and Tibetans fell below 80% around 7 kya (bootstrap 95% CI: 3kya to 10kya). We then derived a more detailed demographic model using ∂a∂i, which predicted that the initial divergence between Han and Tibetan was much earlier, at 54kya (bootstrap 95% C.I 44 kya to 58 kya). However, for the first 45ky, the two populations maintained substantial gene flow (6.8x10-4 and 9.0x10-4 per generation per chromosome). After 9.4 kya (bootstrap 95% C.I 8.6 kya to 11.2 kya), the gene flow rate dramatically dropped (1.3x10-11 and 4x10-7 per generation per chromosome), which is consistent with the estimate from MSMC. FST predicted by the best-fitting demographic model in ∂a∂i produced estimates consistent with observed FST (0.0147 using Hudson’s FST estimator and 0.0148 using the Weir and Cockerham’s estimator).
Fig 1. MSMC estimate of population histories for Tibetans and Han Chinese.
(A) The effective population size as the function of time in the past for Tibetans (red) and Han (blue), estimated using 4 Han and 4 Tibetan genomes with greater than 99% corresponding genetic ancestries. (B) The relative coalescence rate between Tibetans and Han as the function of time in the past, estimated using 1 Han and 1 Tibetan genomes with greater than 99% corresponding genetic ancestries. Solid color indicates the estimates from the actual data, and the corresponding lighter colors indicate the estimates from 20 bootstrapped datasets.
Fig 2. Illustration of the best-fitting ∂a∂i model.
Table 1. Point estimate and 95% confidence interval of the parameters of the best fitting ∂a∂i model (see Fig 2 for the explanation of the parameters).
| point estimate | lower 95% CI | upper 95% CI | |
|---|---|---|---|
| nA1 | 12804 | 12619 | 13445 |
| nC1 | 500 | 412 | 607 |
| nC2 | 75203 | 70047 | 86703 |
| nC3 | 1326988 | 490163 | 5940902 |
| nT1 | 2445 | 2098 | 5251 |
| nT2 | 13292 | 10292 | 14033 |
| nT3 | 77743 | 63131 | 148702 |
| T1 (years) | 6355 | 4365 | 38991 |
| T2 (years) | 44588 | 35784 | 47330 |
| T3 (years) | 9419 | 8573 | 11166 |
| m11 (/gen/chrom) | 6.8E-04 | 5.8E-04 | 9.0E-04 |
| m12 (/gen/chrom) | 9.0E-04 | 8.0E-04 | 1.0E-03 |
| m21 (/gen/chrom) | 1.3E-11 | 8.6E-12 | 1.9E-11 |
| m22 (/gen/chrom) | 4.0E-07 | 2.3E-18 | 6.5E-06 |
Whole-genome positive selection scans in the Tibetan genome
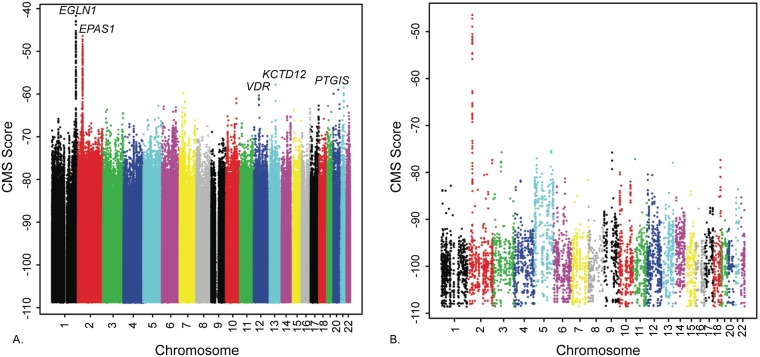
We used the Composite of Multiple Signals (CMS) test[28] to map regions under positive selection in the Tibetan genome (Fig 3). At each genomic variant, CMS evaluates the likelihood of five test statistics (iHS, XP-EHH, ΔiHH, FST and ΔDAF) under two models: a null model that assumes neutral evolution and an alternative model that incorporates a variety of scenarios involving recent strong positive selection (see S1 Supplementary Methods). CMS then combines information from the five test statistics in a Bayesian framework to create an aggregated CMS score. CMS and its components (iHS, FST and XP-EHH) have been successfully applied to identify adaptive sweeps originating both from the same population[12,28,29] and from archaic introgression[30–32].
Fig 3. Manhattan plot of the CMS score across the autosome.
The x-axis represents the chromosome number and each dot represents one SNV. A) All autosomal SNVs; B) SNVs that are present in the high-coverage reference Denisovan genome and Tibetans but uncommon (MAF<5%) in Yoruba, Europeans, Native Americans and Asians. CMS scores all have negative values, with higher scores corresponding to stronger positive selection signals.
We first calculated the CMS score at each SNV using the 27 Tibetan genomes, with 62 Han Chinese genomes from the 1000 Genomes (1KG) Project[18,33] included as a comparison population (Supplementary Methods). In order to evaluate the statistical significance of our top candidates, we performed coalescence simulations with cosi[34] under a neutral model, using the demographic model that we estimated with ∂a∂i[27] (Supplementary methods). Comparing the CMS scores from our observed data and the null distribution from coalescence simulations, we identified 377 candidate SNVs with FDR < 0.3; 100 of these SNVs are within 350kb of EGLN1 and 199 are within 350kb of EPAS1. We listed all the SNVs with FDR<0.3 in S2 Table.
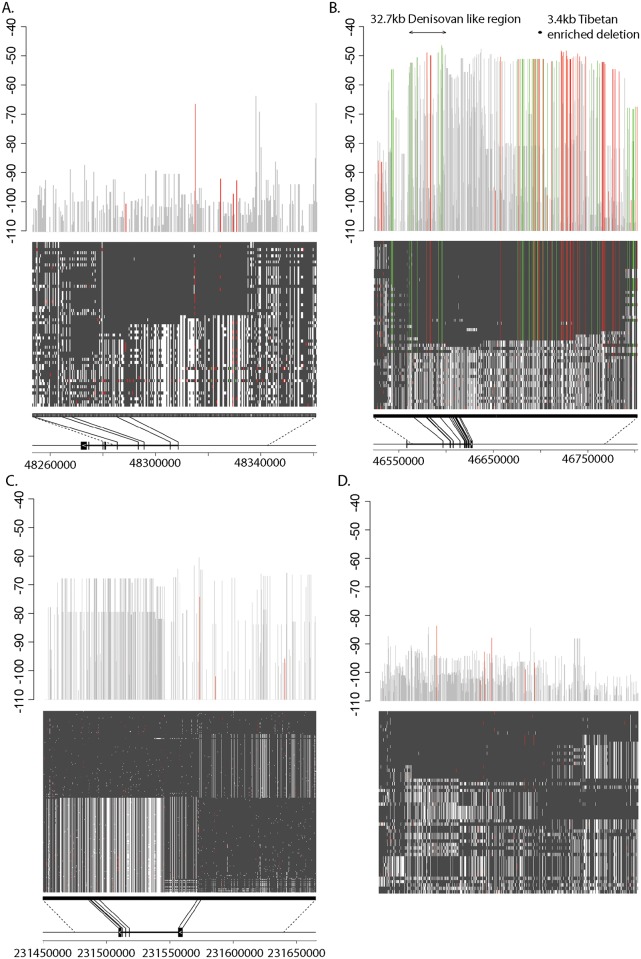
Because CMS scores between nearby SNVs are often correlated due to linkage disequilibrium, we also performed a region-based CMS test by dividing the genome into 200kb consecutive regions[12], with the test statistic equal to the highest CMS score within each region. Table 2 lists the top 10 regions with the highest CMS scores. The top regions are in the proximity of EPAS1 and EGLN1 genes, which were previously reported to be responsible for high-altitude adaptation among Tibetans[12,13]. One of the regions overlaps the gene VDR, which encodes the vitamin D3 receptor. Mutations in this gene can cause an abnormality in vitamin D metabolism, which may in turn lead to rickets[35] and preterm birth[36]. We examined the evidence of positive selection on the three SNVs with the best-CMS scores in this 200kb region. Based on the Complete Genomics (CG) 1000 Genome panel data[18,33], the alternative alleles of top 2 SNVs (Chr12:48363253 G->C and Chr12:48359527) are infrequent in Han Chinese (MAFs: 1.6% and 3.2%), but the MAFs were comparable between Tibetans (31.5% and 31.5%) and Europeans (32.1% and 31.4%). The third ranking SNV (Chr12: 48337328 C->T) has a minor allele frequency of 2.4% in Han Chinese, 0.0% in Europeans, 17.0% in Yoruba and 25.9% in Tibetans. The iHS value at this location is 4.22 standard deviations higher than the genome-wide mean, which is about 13,000 times more likely under the alternative model than the null. Fig 4a displays the haplotype map in Tibetans surrounding this region. The focal SNV is 394 bp upstream of the first exon of the transcript AK309587, overlapping the binding site of three transcription factors: TCF7L2, MAFK and MAFF[37]. Both a DNaseI hypersensitivity assay and H3K27me3 histone marker signature in the K562 cell line indicates this is a potential noncoding regulatory region.
Table 2. Top 10 regions with the highest CMS scores (hg19 coordinates), with connecting 200-kb windows merged.
| Chr | Start | End | Highest CMS score | Genes in the region |
|---|---|---|---|---|
| 1 | 231200001 | 232000000 | -41.7 | EGLN1, SPRTN, TSNAX, SNRPD2P2, EXOC8, DISC1, DISC2, TSNAX-DISC1, GNPAT, C1orf131, TRIM67 |
| 2 | 46400001 | 46800000 | -46.4 | EPAS1, RHOQ, PRKCE, ATP6V1E2 |
| 13 | 77200001 | 77600000 | -57.8 | KCTD12,BTF3P11,CLN5,FBXL3 |
| 21 | 37800001 | 38000000 | -58.5 | CLDN14 |
| 20 | 48000001 | 48200000 | -59.0 | KCNB1,PTGIS |
| 7 | 35400001 | 35600000 | -59.8 | None |
| 20 | 1800001 | 2000000 | -59.9 | PDYN,SIRPA |
| 12 | 48200001 | 48400000 | -60.3 | VDR,COL2A1,HDAC7,TMEM106C |
| 10 | 101600001 | 101800000 | -61.0 | ABCC2,DNMBP-AS1,DNMBP |
| 7 | 52400001 | 52800000 | -61.7 | None |
Fig 4. Haplotype map and CMS score in four genomic regions.
The upper bar plot demonstrates the CMS values at each SNV. The middle plot shows the haplotype structure of 27 Tibetan genomes in the region; each row represents one genome and each column is one SNV aligned with its CMS score in the upper figure. Both red and green color indicates uncommon variants (MAF<5%) in Yoruba, Europeans, Native Americans and Asians; variants in green in addition must be in the high-coverage reference Denisovan genome. When applicable, the lower plot represents the gene model for the protein-coding gene in the region, with each vertical line representing one exon. a) VDR gene region (Chr12:48257328–48357328); b) EPAS1 gene region (Chr2: 46533376–46792633). The arrowed block above the bar plot indicates a previously identified 32.7kb region enriched for Denisovan variants; the dot above the bar plot indicates a previously identified deletion common in Tibetans (chr2: 46694276–46697683); c) EGLN1 gene region (Chr1: 231457651–231657496); d) a 200-kb genomic region that presumably underwent no positive selection (showing Chr21: 32800690–33000356, the 200-kb region with the median CMS score).
Another top 10 ranking region overlapped the KCTD12 gene (potassium channel tetramerization domain containing 12), a component of the G-protein-coupled receptors for γ-aminobutyric acid (GABA) receptor[38]. This region contains 3 SNVs with FDR < 0.05 from our single-variant CMS test. The strongest signal (Chr13: 77399462) among these three SNVs has an observed minor allele frequency of 18.5% in Tibetans, 0.06% in dbSNP (build 142) and 0% among Yoruba, Europeans, and Han Chinese in the CG 1000 Genomes panel. The normalized integrated haplotype score (iHS) value at this location was 4.90 standard deviations higher than the genome-wide mean, which is ~48,000 times more likely to occur under the alternative model than under the null model. This indicates that the haplotype homozygosity around the derived allele is much higher than around the ancestral allele, a scenario consistent with strong positive selection. The three SNVs are located in genomic regions with potential regulatory function. Chr13: 77399462 T->C and Chr13:77398842 G->A overlap with histone mark H3K27Ac annotation and are within enhancer regions, while Chr13:77405193 G->A is at a DNase I hypersensitive site and has a strong signature of H3K27me3 in the K562 (myelogenous leukemia) cell line[37,39].
We next examined Gene Ontology (GO)[40] annotations associated with genes in the top 0.2% of 200kb windows. We evaluated the statistical significance for over-representation of any GO term by repeated sampling without replacement from all 200kb regions in our dataset (Table 3). The following GO terms related to hypoxia responses were over-represented: GO:0071456 (cellular response to hypoxia), GO:0036294 (cellular response to decreased oxygen levels), and GO:0071453(cellular response to oxygen levels). In addition to two genomic regions surrounding EPAS1 and EGLN1, we identified one additional gene related to hypoxia response in GO:0071456: prostaglandin I2 synthase (PTGIS). PTGIS converts prostaglandin H2 to prostacyclin, an effective vasodilator and inhibitor of platelet activation[41]. The SNV with the highest CMS score in this region (Chr20: 48175598 C->T) has a normalized XP-EHH value of 3.67, which was 51 times more likely under the alternative model than the null. The derived allele frequency was 85.2% in Tibetan, 42.7% in Han Chinese, 32.1% in Europeans, and 8.9% in Yoruba. The SNV is in the first intron of PTGIS, overlapping DNase I hypersensitivity regions, potential ZNF217 transcription factor binding sites and H3K36me3 histone mark signatures in the NT2-D1 (pluripotent embryonic carcinoma) cell line[37].
Table 3. Over-represented GO terms within the top 0.2% CMS windows with q-value <0.05.
| GO term | description | q-value | Genes |
|---|---|---|---|
| GO:0050892 | intestinal absorption | 0.0295875 | VDR, ABCG8, ABCG5 |
| GO:0071456 | cellular response to hypoxia | 0.0295875 | PTGIS, EPAS1, EGLN1, PRKCE |
| GO:0036294 | cellular response to decreased oxygen levels | 0.0295875 | PTGIS, EPAS1, EGLN1, PRKCE |
| GO:0071453 | cellular response to oxygen levels | 0.0295875 | PTGIS, EPAS1, EGLN1, PRKCE |
Because of the lower variant-calling qualities at genomic insertions and deletions (indels), we could not obtain reliable results from haplotype-based tests (XP-EHH, iHS and ΔiHH) on indels. Instead, we used the Population Branch Statistic (PBS)[13] to search for signatures of positive selection. To calculate the significance levels of the PBS scores, we estimated the null distribution on the same simulation dataset as in the CMS test. We found that indels with the best PBS signals tended to fall into regions having the highest CMS scores. Specifically, 6 out of the top 10 indels are also within the top 10 200kb regions with the highest CMS scores, all in the EPAS1 and EGLN1 regions (S3 Table; p = 1.1x10-15).
Mapping of variants under positive selection in the EPAS1 and EGLN1 regions
We fine-mapped SNVs under positive selection in the EPAS1 region by combining the CMS scores with the population allele frequencies in major continental population groups (Europeans, Han, Native Americans and Yoruba) (Fig 4b). We discovered 199 SNVs with FDR < 0.3 from the CMS test (CMSq<0.3) in the EPAS1 region (S4 Table), which spans 321 kb from the first intron of EPAS1 to 257 kb downstream of the last exon of EPAS1. Of these 199 SNVs, 64 were in the intronic region and the remaining 135 were in the 3’ downstream region. These SNVs are enriched for alleles present in the Denisovan genome and rare in non-Tibetan populations (MAF<0.05 among Yoruba, Europeans, Native Americans and Asians). Specifically, 16.6% of EPAS1 CMSq<0.3 SNVs meet this criterion, as compared to 0.3% genome-wide (p<2x10-16).
A previous study identified a 32.7kb region (Chr2: 46567916–46600661) in EPAS1 that was highly differentiated between Han and Tibetans and also has a high proportion of Denisovan-like variants[16]. In our CMS test result, 2 out of the top 3 SNVs with the highest CMS score in the EPAS1 region (46597756 and 46598025 on Chromosome 2) were in this 32.7kb region. After excluding rare variants (SNVs with MAF <0.05 in both Han and Tibetans), the average FST between Han and Tibetans in this 32.7kb region was 0.28. However, the haplotype extends far beyond the previously reported 32.7 kb region and contains genomic regions at least 150 kb downstream of EPAS1. Fig 4b illustrates the Tibetan haplotypes in EPAS1 and its 3’ region. The linkage disequilibrium between SNVs (both Denisovan-like and not) was high among Tibetans (S5 Table), suggesting a slow decay of haplotype homozygosity. The small indel with the highest PBS score was a 4bp deletion located in the 2nd intron of EPAS1 (Chr2:46577800–46577803). Three out of the four nucleotides affected by this deletion are highly conserved in mammals. This indel overlaps an activating H3K27Ac mark in seven cell lines and the binding sites of three transcription factors: the polymerase subunit POLR2A, the HIF co-activator EP300[42], and GATA2, which is key in the control of erythroid differentiation[43]. The observed frequency of the derived allele (deletion) was 62.9% in Tibetans, 0.8% in Han Chinese and 0% in Europeans, corresponding to a PBS score of 0.600, which was ranked 3rd among all the genomic variant (indels and SNVs) in the EPAS1 gene.
In addition to the 32.7 kb block, we also identified a second 39.0kb candidate block downstream of EPAS1 coding region on the haplotype (Chr2: 46675505–46714553); this block was highly differentiated between Han and Tibetans and enriched for Denisovan variants (S3 Fig). Within this block, 88.2% (60 out of 68) of common Tibetan SNVs (derived allele frequency >50%) are present in the Denisovan genome, as compared to 81.4% (48 out of 59) in the previously reported 32.7kb region. The proportion of SNVs shared with the Neanderthal genome is 26.5% (18 out of 68) for the new 39.0kb block as compared to 25.4% (15 out of 59) for the previous 32.7kb block. In this block, the average FST between Han and Tibetans for SNVs common in at least one of the two populations is 0.304 (as compared to 0.279 for the previously reported region). Intriguingly, this region also fully contains a 3.4kb Tibetan-enriched deletion region that showed footprints of positive selection in a previous study[44] (Fig 4b). Among our Tibetan samples, this deletion has an allele frequency of 59.2% and is in high LD (r2 = 0.86) with the SNV with the highest CMS in the EPAS1 region (Chr2: 46597756; S6 Table).
We estimated the origin age of the selective sweep on the adaptive EPAS1 haplotype to be 12 kya (95% CI: 7 kya to 28 kya) using a maximum likelihood approach based on coalescence simulations of the demographic model described in Table 1 (see S1 Supplementary Methods for details). Our estimate is consistent with a previous estimate of 12 kya to 15 kya[44]. Using this point estimate of selection start time and assuming that the adaptive haplotype indeed originated from Denisovans or a closely related population, we further inferred that the introgression of this haplotype most likely occurred between 32 and 12 kya. We also estimated the gene-tree divergence time between the adaptive haplotype in Tibetans and the available Denisovan DNA sequence to be 997 kya (95% CI: [868, 1,139]). From the gene-tree divergence and calibrated models of Denisovan demographic history[45], we estimated a population divergence time of 868 kya (95%CI: 952 kya to 238 kya) between the Densivan population and the archaic population that contributed the adaptive EPAS1 haplotype.
Previously, we identified two non-synonymous variants in the EGLN1 gene (c.12C>G, p.D4E; c.380G>C, p.C127S) which together act as a co-adapted gene complex contributing to high-altitude adaptation in Tibetans[14]. In the current dataset, the c.12C>G variant was absent from any other populations in the 1KG dataset and had high FST (0.70) and ΔDAF (0.70) values, indicative of strong positive selection. The c.380G>C variant has a derived allele frequency of 77.8% (95% CI: [66.7%, 88.9%]) in Tibetans and 44.9% (95% CI: [40.0%, 50.0%]) in Han Chinese, consistent with previous findings[14]. The CMS score is -60.43 for c.12C>G and -72.08 for c.380G>C, both ranked among the top 0.01% variants genome-wide. We also identified a novel rare EGLN1 variant (c.C358T, p.P120L) within one Tibetan individual, who also carries the c.380G>C but not c.12C>G variant. In our dataset this variant has a frequency of 1.85% (95% CI: [0.05%, 9.89%]) in Tibetans, 0.81% (95% CI: [0.02%, 4.41%]) in Han Chinese and is absent from other populations in the 1KG dataset.
Mapping regions with high Denisovan ancestry in the Tibetan genome
To evaluate whether any genomic regions with Denisovan introgression other than EPAS1 may contribute to Tibetan high-altitude adaptation, we first calculated D-statistics[46] to evaluate the overall level of Denisovan admixture in the Tibetan genome. When comparing Tibetans to Yoruba, we observed a D of 0.011 (95% CI: 0.0068 to 0.0163), indicating a significant excess of Denisovan admixture among Tibetans. However, we observed no significant difference in D between Tibetans and Han Chinese (D = -0.0007, 95% CI -0.0027 to 0.0021), demonstrating that the genome-wide level of Denisovan admixture does not differ substantially between Tibetans and Han Chinese, and is consistent with a previous report[47]. Using the Q statistic of Rogers and Bohlender[48], we estimated that the proportion of Denisovan admixture in Tibetans is 0.4% (95% CI: 0.2% to 0.6%, see Methods and S4 Fig). Briefly, Q is an f3 ratio estimator of admixture[48]. Given a sample from three populations X, Y and N, so that X and Y are more recently diverged from each other than either is from N, the calculation of Q uses a ratio of expectations to calculate the total admixture from N into Y, in a similar way as other f3-ratio estimators[46]. Q has the advantage of requiring only a single archaic individual, and uses external information about ghost admixture to correct bias due to admixture from an unsampled archaic population (Neanderthal).
To investigate the variation in Denisovan admixture across the genome among Tibetans, we first used an LD-based statistic S*[32] to identify introgressed regions originated from Denisovans. S* searches for haplotypes that share a substantial amount of similarity with the archaic genome and are present in the population of interest but not in the comparison population (see S1 Supplementary Methods). In our study, we used Han Chinese as the comparison population and Denisovan alleles as the archaic genome to search for Denisovan haplotypes absent among the Han Chinese genomes and present in at least one Tibetan genome (Supplementary Methods). In total, S* identified 660 50kb candidate regions using this procedure (S5 Fig; S7 Table). As an alternative approach, we also developed a new statistic, D*, which normalizes the D-statistics to enable the identification of admixed regions with calibrated Type I error (Methods). We used D* to compare Tibetans to Han Chinese for all 200kb genomic regions (S5 Fig). With this procedure, we identified 11 regions with significant proportions of Denisovan admixture after multiple-testing correction (FDR < 0.30). The region containing EPAS1 ranked second, with D* = 6.2 (p = 6.1 x 10−6). Six of 11 regions identified by D* overlapped with the 660 regions identified by S*, including the region containing EPAS1 (Table 4).
Table 4. 200-kb genome regions with higher Denisovan ancestry in Tibetan than in Han Chinese in both S* and D* tests.
| Chr | start | end | raw D-value | D* | p-value | q-value | Genes in the regions |
|---|---|---|---|---|---|---|---|
| 7 | 26800001 | 27000000 | 0.86 | 6.25 | 6.1E-06 | 2.7E-02 | SKAP2 |
| 2 | 46400001 | 46600000 | 0.56 | 6.15 | 6.1E-06 | 2.7E-02 | PRKCE,EPAS1 |
| 2 | 47600001 | 47800000 | 0.65 | 5.92 | 6.1E-06 | 2.7E-02 | MIR559,MSH2,EPCAM,KCNK12 |
| 5 | 200001 | 400000 | 0.52 | 4.99 | 6.1E-05 | 1.4E-01 | CCDC127,PDCD6,SDHA |
| 12 | 8800001 | 9000000 | 0.63 | 4.63 | 1.2E-04 | 1.7E-01 | MFAP5,RIMKLB,A2ML1 |
| 4 | 100400001 | 100600000 | 0.48 | 4.58 | 1.2E-04 | 1.7E-01 | TRMT10A,C4orf17,MTTP |
Next, to test for evidence of adaptive Denisovan introgression, we compared the location of the Denisovan DNA regions identified by either S* or D* to the regions with the highest CMS scores. The top candidate identified by this analysis is EPAS1. Since we were interested in identifying novel adaptive Denisovan introgression, we removed the EPAS1 region and evaluated the statistical significance of the observed number of overlaps. To calculate the p-value, we permuted the CMS scores of all 200kb genomic regions to generate the distribution of overlaps under the null hypothesis that no additional Denisovan-like DNA was positively selected in Tibetans. When we compared the Denisovan-like genomic regions to the top 0.2% of CMS regions, we found no overlapping region with D* and only one with S* (Chr2:42200001–42400000). In the top 1% of CMS regions, we identified no overlapping regions with D* and three overlapping regions (Chr2: 42200001–42400000; Chr1: 79200001–79400000; Chr1:1200001–1400000) with S*; however, the p-values from all tests were non-significant. Therefore, we find no evidence that additional Denisovan-like DNA has contributed to Tibetan high-altitude adaptation. We summarized the CMS scores of Denisovan variants that are present in Tibetans but uncommon among major population groups in Fig 3b.
Discussion
Our demographic history estimates from ∂a∂i suggest that population structure existed between the Tibetan and Han ancestral subpopulations as early as 44 to 58 kya, with high rates of admixture maintained between the two subpopulations until around 9 kya. This agrees well with the findings of Lu et al.[49] that the early colonization of the Tibetan plateau occurred between 62kya and 38kya, and the post-Last Glacial Maximum (LGM) arrival at the Tibetan plateau could be between 15kya and 9kya. Moreover, archaeological evidence [1,50] suggests that the Tibetan plateau was occupied during the Late Pleistocene, roughly coinciding with the time of Han-Tibetan separation in our demographic model. Interestingly, MSMC did not detect a decrease in relative cross coalescence rate between Han and Tibetans until 3 to 9 kya. We hypothesized that this discrepancy was due to an inability of MSMC to differentiate between panmixia and high rates of gene flow between Tibetan and Han Chinese populations between 9kya and 54kya. To test this hypothesis, we conducted coalescent simulations to evaluate the ability of MSMC to accurately estimate the Tibetan-Han divergence time under a model where the two populations diverged at 54 kya but experienced a large amount of gene-flow (6.8x10-4 and 9.0x10-4 per generation per chromosome) until 9 kya. As shown in S6 Fig, MSMC was indeed unable to detect the ancestral population split at 54 kya. The observed pattern of estimated relative cross-coalescence rate was consistent between the observed data and simulations based on the best-fitting ∂a∂i model, with no decrease in the relative cross-coalescence rate prior to 10 kya. Conversely, when we conducted coalescence simulation using the simplified demographic model predicted by MSMC (i.e., Han and Tibetan separated 7,000 years ago) and ran ∂a∂i, we found that ∂a∂i accurately recovered the simulated divergence date (S8 Table), providing support for the ∂a∂i model with an earlier divergence date but a high migration rate between the two populations until approximately 9 kya.
Our admixture analysis in the EPAS1 region confirmed findings from previous studies that a common Tibetan haplotype in this region contains excessive Denisovan-like DNA[16], but also showed that on a genome-wide level, the amount of Denisovan admixture in Tibetans (0.4%) is similar to that of Han Chinese. This suggests that at least one major introgression occurred from the archaic population to the common ancestor of Han and Tibetans. The high-altitude adaptive haplotype in the EPAS1 region may have been acquired prior to Han-Tibetan divergence, but was either lost or present at low frequencies in Han due to the lack of selection. This explanation agreed with the hypothesis proposed by Huerta-Sanchez et al[16], who posit that the EPAS1 haplotype may have been introduced prior to the separation of Han and Tibetans, considering the presence of the Tibetan EPAS1 haplotype in a single Han Chinese individual. Alternatively, it is possible that the low frequency of the Tibetan EPAS1 haplotype in China is the result of relatively recent migration from Tibet[16] and that the haplotype was originally introduced into Tibet after the Han-Tibetan divergence. This alternative explanation is supported by our 95% C.I. for the introduction of the EPAS1 haplotype into the ancestral Tibetan population (32 to 12 kya) which is later than the previous estimates for the date of Denisovan admixture into modern humans (44.0 to 54.0 kya)[51]. Other than the EPAS1 haplotype, we observed no evidence of other Denisovan-like DNA segments contributing to high-altitude adaptation in Tibetans, given that we did not detect significant overlaps between introgressed regions and the top 1% of CMS regions. However, we cannot rule out the possibility that with larger sample sizes, subtle adaptive introgression signals may be detected.
In addition to the previously reported 32.7kb haplotype block in EPAS1[16], we identified a much larger haplotype that contains both the EPAS1 genic region and at least 150kb 3’ of EPAS1. However, the extensive LD within the EPAS1 and its 3’ region prevented us from pinpointing the location of the adaptive mutation. Interestingly, the selected haplotype (Fig 4b; S3 Fig) contains many Denisovan-like and non-Denisovan-like variants, and many top candidate SNVs in our analysis, as well as the previously reported 3.4kb Tibetan enriched deletion, were absent from the high-coverage Denisovan genome. This observation is reflected in the estimated population divergence time of 952 to 238 kya between the population represented by the Denisovan reference genome and the archaic population that admixed with Tibetans (Supplementary Methods). This estimate is broadly consistent with previous evidence that the archaic Denisovan-like population that admixed with modern human populations separated from the population represented by the Denisovan reference genome between 276 kya and 403 kya[52]. Thus, the advantageous EPAS1 haplotype shows far greater similarity to Denisovans than would be expected in the absence of archaic admixture, but substantial mismatch should be expected given the apparent diversity of archaic populations outside of Africa[53].
Our CMS test combines five statistical tests of positive selection (iHS, XP-EHH, FST, ΔiHH and ΔDAF), offering a more robust performance across a wide variety of scenarios compared to any single test[28]; this allowed us to identify novel candidate genes contributing to adaptations on the Tibetan plateau. One of the candidate gene for adaptive selection is VDR, a gene encoding nuclear hormone receptor for 1, 25 dihydroxyvitamin D3 and functions in Vitamin D metabolism. Vitamin D is a secosteroid nutrient that plays an important role in calcium homeostasis and bone mineralization, and mainly comes from two sources: 1) through skin exposure to sunlight and 2) food, such as fish, milk and egg yolk. Deficiency in vitamin D leads to impairment of bone mineralization and skeletal deformities, known as rickets in children and osteomalacia and osteoporosis in adults [54]. Low level of vitamin D has also been associated with cancers, diabetes, autoimmune diseases, hypertension, and infectious disease[35,55]. In a study involving vitamin D status in a cohort of 63 Tibetans [54], the proportion of nomadic Tibetans with vitamin D deficiency (25(OH)D <75 nmol/L) was 100% with 80% of people having severe deficiency (25(OH)D < 30nmol/L); the proportion of non-nomadic Tibetans with vitamin D deficiency ranges from 40% to 83%. Consistently, in another study, 61% of Tibetan children suffer from rickets and 51% have stunted growth[56]. Such a high prevalence of vitamin D deficiency may be explained by the traditional Tibetan diet consisting of barley, yak meat and butter tea which are poor sources of vitamin D, and clothing habits in cold temperatures which allow for minimal skin exposure to the sunlight [54]. Therefore, we hypothesize that VDR gene is positively selected to compensate for the lack of vitamin D, the mechanism of which remains to be determined. Previously, it has been shown that genomic regions bound by VDR were under adaptive selection in the human genome[57].
A second promising candidate identified in our CMS test is PTGIS, a gene associated with three hypoxia-response GO terms (0071456, 0036294,0071453) and regulated by hypoxia-inducible factor 1 (HIF-1). PTGIS encodes prostaglandin I2 synthase, which participates in the synthesis of prostanoid. Previous studies have shown that the expression of PTGIS is induced by hypoxic conditions in human lung fibroblast cells and cancer cell lines[58], and can activate vascular endothelial growth factor. Therefore, it is possible that a Tibetan-unique genomic variant may induce vasodilation and angiogenesis in response to hypoxia by altering the expression of PTGIS.
The CMS test also identified KCTD12 as a new high-altitude adaptation candidate. Previously, it has been shown in B16 murine melanoma cells that KCTD12 is up-regulated in hypoxic conditions[59], and thus it may play a role in the hypoxic responses. This gene is predominantly expressed in fetal organs, suggesting a potentially important role in early development, but dramatically lower levels in adult tissue including brain and lung[60]. KCTD12 is differentially expressed in human pulmonary endothelial cells upon 48 hours of hypoxia exposure[59] and noted as one of 40 CpG sites with the greatest difference in methylation levels between highland and lowland Oromo Ethiopians[61].
Our functional annotation of Tibetan genomic variants identified a frequent protein-coding SNV in ALPP, a gene associated with pregnancy losses[25]. Since Tibetans tend to have higher birth weight in high altitude environments compared to lowland native populations living at similar altitudes[62], it is possible that the nonsynonymous variant in ALPP may affect the birth weight, development, and pregnancy outcomes in Tibetans. For every 1000 meters of altitude, birth weight decreases an average of 100 grams due to restricted fetal growth[63–66], with a three-fold increase in the number of infants born small for gestational age (SGA) at high altitude[67]. Weights of infants born at high altitudes to mothers of highland origin, however, are greater than those of lowland origin. This is shown specifically in infants born to Bolivian versus European women in the Andes, whose birth weights are about 300g higher in the former group, while no difference is reported by ethnicity at low altitude[67]. Whether these putatively adaptive markers play roles in this process remains to be determined.
In summary, we performed a comprehensive genomic analysis on whole-genome sequence data from 27 Tibetan individuals. Our analyses detected evidence of population structure between the ancestral Han and Tibetan subpopulations beginning between 44 and 58 kya, although admixture rates between the two subpopulations remained high until around 9 kya. The Denisovan EPAS1 haplotype introgressed into the Tibetan population between 12 and 32 kya, and positive adaptive pressure on this haplotype began between 7 and 28 kya. We summarized the dates of important demographic events in Fig 5. The Our CMS test identified novel candidate genes for high-altitude adaptation including KCTD12, VDR and PTGIS, and also generated a list of candidate variants within the EPAS1 gene region. We estimated that 0.4% of the Tibetan genome are introgressed DNA from Denisovans, although EPAS1 is probably the only introgressed locus that was influenced by strong positive selection in Tibetans. Our study provided a rich genomic resource of the Tibetan population and generated hypotheses for future positive selection tests.
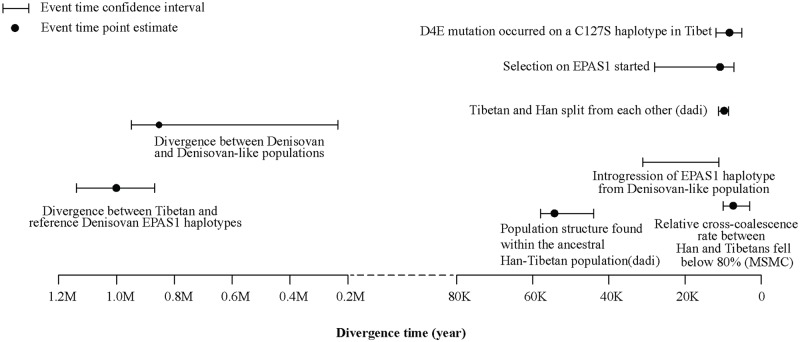
Fig 5. Timeline of important evolutionary events in the demographic history of Tibetans.
The horizontal axis represents the estimated time of events (in years) in the past. The age of the EGLN1 D4E mutation is based on previous estimates.
Materials and methods
Sequencing samples and variants calling
Our study subjects included 27 Tibetan individuals recruited from Tibetans living in San Diego, California (n = 3), Salt Lake City, Utah (n = 5), and the United Kingdom (n = 19). The sample type and place of origin information are listed in S9 Table. Whole-genome sequencing and variant-calling was performed by CG (v2.0 for the first 17 samples and v2.5 for the remaining 10 samples). Variants with genotyping quality less than 30 (i.e., more than 1 calling error among 1000 variants) were converted to missing genotype calls, and sites with more than 5% missing genotype rate were removed. To identify additional variants enriched for higher genotyping error rates, we compared 62 genomes that had been sequenced in both CG public genome data[33,68] and 1KG Project[18] Phase I data. We identified 652,899 SNVs with more than 5% discordant genotype calls between these two datasets, and removed them from all further analyses. We then performed haplotype phasing and imputation using Shapeit2[69]. After previous steps, 10,405,415 SNVs remained for subsequent analyses.
Whole-genome positive selection scan using Composite of Multiple Signals (CMS) test
The CMS test combines the signals from five different tests (ΔiHH, iHS, XP-EHH, FST and ΔDAF) to create a single test statistic[28]. On each variant, CMS calculates the posterior probability of the variant being selected in a naïve Bayes framework by taking the product of the posterior probabilities from each of the five tests. A higher CMS score is consistent with a stronger signal of positive selection. Our implementation of the CMS test closely follows the recommendations given in the previous report[28]. Briefly, we first performed coalescence simulations using cosi[34] based on the demographic model estimated by ∂a∂i. To simulate genetic data under various scenarios of selective sweeps, we used the following selection coefficients (s): 0.02, 0.03 and 0.04; the following start times of selection: 0, 100, 200, 300 and 400 generations ago (the last one represents the oldest selection start time that cosi may simulate within our model); and the following non-reference allele frequencies at the conclusion of the sweep: 0.2, 0.4, 0.6 and 0.8. In addition, we also simulated the scenario where no selective sweep had occurred, which corresponded to the null model in the CMS test. In each situation, we performed 1000 simulations. We calculated the CMS scores for all SNVs that passed our quality-control procedures. To calculate their p-values, we also ran the CMS test over the null model that we simulated above. The p-value for a certain CMS score was calculated as the proportion of SNVs in the null model that has the same or higher CMS scores.
Testing for Denisovan admixture using the D statistic
The D statistic tests whether the amount of archaic admixture in one population exceeds that of another population by examining variant allele frequency spectrum in the case population, control population, an archaic population and an outgroup population[70]. In our study, these were selected as Tibetans, Han Chinese/Yoruba, Denisovan and chimpanzee. Let , , , and , be the allele frequency of the i-th variant in the case, control, archaic and outgroup populations, respectively. D is defined as:
| (1) |
We estimated the confidence interval of D using bootstrap sampling with 200 replicates. Specifically, we divided the whole genome into 1MB blocks, and then randomly sampled with replacement the same number of 1MB blocks as in the actual data, each time calculating a sample D statistic. The 2.5% and 97.5% quantiles of the 200 D statistics values were used as the two end points of the 95% confidence interval.
Identifying regions of Denisovan admixture using the D* statistic
We developed the D* statistic as a complement to S* to identify local genomic regions with excess archaic admixture in a population. We first intended to use the D statistic to identify 200kb genomic regions with archaic admixture in the case population, but noticed that the variance of the D-statistic was high if the number of archaic variants in a target region was small. This sensitivity of the D-statistic to regions of low archaic gene flow has been noted previously, and alternatives to the D-statistic have been suggested[71]. To illustrate this sensitivity, consider the extreme scenario where only one archaic variant exists in a 200kb genomic region, and this variant was present in only one case genome and absent from all control genomes. In this situation, D will be the largest possible value Eq (1) despite the fact that the evidence supporting an archaic admixture in this region is poor. This property of D makes it unsuitable for prioritizing candidate introgression regions, since the majority of regions with high D values will be those with few archaic variants in modern human genomes.
Previous work has suggested using direct estimators of the quantity of gene flow as an alternative to a method like D-statistics due to the sensitivity of the statistic to population history and other exogenous parameters[71]. However, D-statistics and the related f-statistics vary only in magnitude as a result of these biases, and will only deviate from zero, in expectation, as a result of gene flow[46]. Normalization of D-statistics preserves their desirable characteristics, while eliminating a source of bias.
To normalize D-statistics, we calculated the following statistic (U) for each 200kb genomic region:
| (2) |
As can be seen, U is the denominator of the D in Eq (1) and is negatively correlated with the standard deviation associated with D (S7 Fig). To create a normalized D-statistic (D*) that can be used to compare admixture signals across regions, we sort all 200-kb genomics regions according to their U values and then divide the regions into 20 equal-sized groups. Then D* of a 200kb region is equal to its D value divided by the within-group standard deviation. We estimated the significance of a given D* by comparing it to the null distribution of D* generated by coalescence simulation.
Estimating the proportion of Denisovan admixture
Because only one Denisovan genome is available, we use the statistic Q of Rogers and Bohlender (Equation 11 of [48]), interpreting the primary source of introgression as Denisovan rather than Neanderthal. The expectation of Q depends not only on the fraction, mD, of Denisovan admixture but also on mN, the fraction of ghost admixture from another archaic such as Neanderthal. Equating observed and expected values defines mD as an implicit function of mN. To evaluate this function, we assumed the parameter values in table 3 of [48]. The result is shown as a solid black line in S4 Fig. We repeated this process, swapping the roles of Neanderthal and Denisovan, to estimate mN as a function of mD. This result is shown as a solid red line in S4 Fig. The intersection provides a simultaneous estimate of mD and mN.
Availability of data and material
The database of Genotypes and Phenotypes (dbGAP) accession number for the whole-genome sequencing data reported in this paper is phs001338.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board at University of Utah and at the University of California, San Diego, by the Berkshire Clinical Research Ethics Committee, UK, and by the Western Institutional Review Board (WIRB). Informed consent was obtained from all participants.
Supporting information
Each circle in the plot represents one genome. A) The 1st and 2nd principal components; B) The 3rd and 4th principal components; C) the 5th and 6th principal components; D) The 1st and 2nd principal components in the PCA of Chinese and Tibetans only.
(DOCX)
Each column represents one individual and different colors represent different ancestral population; each panel represents one K value. The reported ethnicities were listed on the top of each panel.
(DOCX)
The x-axis shows the position on chromosome 2, in Hg19 coordinate. The y-axis shows the FST value. Red color indicates Denisovan-like alleles. Only variants with MAF no less than 5% in either Han or Tibetan were shown. A) Chr2: 44000000–49000000. The EPAS1 region is indicated by the two vertical dotted lines. B) Chr2:46500000–46800000. The two blue vertical dotted lines indicate chr2: 46567916–46600661 (the 32.7kb region reported previously). The two red vertical dotted lines indicate chr2: 46675505–46714553, a second region with high FST and many Denisovan-like variants.
(DOCX)
Key: solid black, mD given mN; solid red, mN given mD; dashed, 95% confidence regions based on moving-blocks bootstrap; circle, simultaneous estimate.
(DOCX)
The x-axis represents the chromosome number. Each dot represents one 200kb genomic region identified by S*.
(DOCX)
The red curve shows the relative cross-coalescence rate based on the whole genome sequencing data; the grey curves show the relative cross-coalescence rate based on 20 coalescence simulations from our best-fitting dadi model with a true Han-Tibetan divergence time of 54 kya but with high rates of gene flow until 9 kya (see Fig 2).
(DOCX)
We sorted all the 200-kb genomic regions by their U values and calculated the standard deviations of the D-statistics within each percentile of U. Each dot represents the D and U for one percentile. The target population is Tibetan and the background population is Yoruba.
(DOCX)
(DOCX)
Panel A, B and C corresponds to ∂a∂i model A, B and C. In each panel, the first two plots show the observed and model predicted SFS heatmaps, respectively; the third plot shows the residual heatmap; the fourth plot shows a histogram of the residuals. Panel D shows the one-dimensional SFS for Han Chinese (left) and Tibetans (right) separately. Within each combination of population and model, the top plot shows the frequencies of variants for each minor allele count, with the red line showing the expected frequencies predicted by the model and blue line showing the observed frequencies; the bottom plot shows the standardized residuals of frequencies within each minor allele count category, assuming the frequencies are Poisson-distributed.
(DOCX)
(DOCX)
(XLSX)
(XLSX)
The start and end positions are in hg19 coordinates.
(XLSX)
(XLSX)
Red-color indicates SNVs present in the Denisovan genome.
(XLSX)
(XLSX)
(XLSX)
We first simulated a 50MB genomic region (with msms) under the MSMC demographic model, and then used dadi to estimate the Han-Tibetan divergence time. The actual demographic parameters are in the Simulated columns, and the dadi estimates are in the Estimated column.
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
An allocation of computer time on the University of Texas MD Anderson Research Computing High Performance Computing (HPC) facility is gratefully acknowledged.
Data Availability
The whole genome-sequencing data are available at The database of Genotypes and Phenotypes (dbGaP) under accession number phs001338.
Funding Statement
This work was supported by NIH R00 HL118215. LBJ was supported by NIH grants GM59290, GM104390, and GM118335. NP was supported by the Wellcome Trust (award number 089457/Z/09/Z, PI: Peter Robbins, DPhil). RB was supported by NCI Award Numbers R25CA057730 (PI: Shine Chang, PhD) and CA016672 (PI: Ronald Depinho, MD). AMC and GLC were supported by an Investigators Programme grant from Science Foundation Ireland (12/IP/1727). GG, JCR and MEB were supported by the University of Luxembourg - Institute for Systems Biology Program and by NIH grant P50 GM076547. ARR was supported by NSF grant BCS 1638840. JTP was supported by 1I01 CX001372-01A2. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aldenderfer M (2011) Peopling the Tibetan plateau: insights from archaeology. High Alt Med Biol 12: 141–147. 10.1089/ham.2010.1094 [DOI] [PubMed] [Google Scholar]
- 2.Gilbert-Kawai ET, Milledge JS, Grocott MP, Martin DS (2014) King of the mountains: Tibetan and Sherpa physiological adaptations for life at high altitude. Physiology (Bethesda) 29: 388–402. [DOI] [PubMed] [Google Scholar]
- 3.Qin Z, Yang Y, Kang L, Yan S, Cho K, et al. (2010) A mitochondrial revelation of early human migrations to the Tibetan Plateau before and after the last glacial maximum. Am J Phys Anthropol 143: 555–569. 10.1002/ajpa.21350 [DOI] [PubMed] [Google Scholar]
- 4.Zhao M, Kong QP, Wang HW, Peng MS, Xie XD, et al. (2009) Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau. Proc Natl Acad Sci U S A 106: 21230–21235. 10.1073/pnas.0907844106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheinfeldt LB, Soi S, Thompson S, Ranciaro A, Woldemeskel D, et al. (2012) Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol 13: R1 10.1186/gb-2012-13-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simonson TS, McClain DA, Jorde LB, Prchal JT (2012) Genetic determinants of Tibetan high-altitude adaptation. Hum Genet 131: 527–533. 10.1007/s00439-011-1109-3 [DOI] [PubMed] [Google Scholar]
- 7.Petousi N, Croft QP, Cavalleri GL, Cheng HY, Formenti F, et al. (2014) Tibetans living at sea level have a hyporesponsive hypoxia-inducible factor system and blunted physiological responses to hypoxia. J Appl Physiol (1985) 116: 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bigham AW, Lee FS (2014) Human high-altitude adaptation: forward genetics meets the HIF pathway. Genes Dev 28: 2189–2204. 10.1101/gad.250167.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simonson TS, Wei G, Wagner HE, Wuren T, Qin G, et al. (2015) Low haemoglobin concentration in Tibetan males is associated with greater high-altitude exercise capacity. J Physiol 593: 3207–3218. 10.1113/JP270518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, et al. (2010) Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A 107: 11459–11464. 10.1073/pnas.1002443107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bigham A, Bauchet M, Pinto D, Mao X, Akey JM, et al. (2010) Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet 6: e1001116 10.1371/journal.pgen.1001116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simonson TS, Yang Y, Huff CD, Yun H, Qin G, et al. (2010) Genetic evidence for high-altitude adaptation in Tibet. Science 329: 72–75. 10.1126/science.1189406 [DOI] [PubMed] [Google Scholar]
- 13.Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, et al. (2010) Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329: 75–78. 10.1126/science.1190371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenzo FR, Huff C, Myllymaki M, Olenchock B, Swierczek S, et al. (2014) A genetic mechanism for Tibetan high-altitude adaptation. Nat Genet 46: 951–956. 10.1038/ng.3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song D, Li LS, Arsenault PR, Tan Q, Bigham AW, et al. (2014) Defective Tibetan PHD2 binding to p23 links high altitude adaption to altered oxygen sensing. J Biol Chem 289: 14656–14665. 10.1074/jbc.M113.541227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huerta-Sanchez E, Jin X, Asan, Bianba Z, Peter BM, et al. (2014) Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512: 194–197. 10.1038/nature13408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao Hu, Simonson T, Glusman G, Roach J, Cavalleri G, et al. (2013) Insights on the evolutionary history of Tibetans from whole-genome sequence data. American Society of Human Genetics 2013 Annual Meeting. Boston, MA.
- 18.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudson RR, Slatkin M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weir BS, Cockerham CC (1984) Estimating F-Statistics for the Analysis of Population-Structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- 22.Wuren T, Simonson TS, Qin G, Xing J, Huff CD, et al. (2014) Shared and unique signals of high-altitude adaptation in geographically distinct Tibetan populations. PLoS One 9: e88252 10.1371/journal.pone.0088252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelis M, Esko T, Magi R, Zimprich F, Zimprich A, et al. (2009) Genetic structure of Europeans: a view from the North-East. PLoS One 4: e5472 10.1371/journal.pone.0005472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu H, Huff CD, Moore B, Flygare S, Reese MG, et al. (2013) VAAST 2.0: improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet Epidemiol 37: 622–634. 10.1002/gepi.21743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vatin M, Bouvier S, Bellazi L, Montagutelli X, Laissue P, et al. (2014) Polymorphisms of human placental alkaline phosphatase are associated with in vitro fertilization success and recurrent pregnancy loss. Am J Pathol 184: 362–368. 10.1016/j.ajpath.2013.10.024 [DOI] [PubMed] [Google Scholar]
- 26.Schiffels S, Durbin R (2014) Inferring human population size and separation history from multiple genome sequences. Nat Genet 46: 919–925. 10.1038/ng.3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD (2009) Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet 5: e1000695 10.1371/journal.pgen.1000695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossman SR, Shlyakhter I, Karlsson EK, Byrne EH, Morales S, et al. (2010) A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327: 883–886. 10.1126/science.1183863 [DOI] [PubMed] [Google Scholar]
- 29.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, et al. (2007) Genome-wide detection and characterization of positive selection in human populations. Nature 449: 913–918. 10.1038/nature06250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding Q, Hu Y, Xu S, Wang J, Jin L (2014) Neanderthal introgression at chromosome 3p21.31 was under positive natural selection in East Asians. Mol Biol Evol 31: 683–695. 10.1093/molbev/mst260 [DOI] [PubMed] [Google Scholar]
- 31.Ding Q, Hu Y, Xu S, Wang CC, Li H, et al. (2014) Neanderthal origin of the haplotypes carrying the functional variant Val92Met in the MC1R in modern humans. Mol Biol Evol 31: 1994–2003. 10.1093/molbev/msu180 [DOI] [PubMed] [Google Scholar]
- 32.Vernot B, Akey JM (2014) Resurrecting surviving Neandertal lineages from modern human genomes. Science 343: 1017–1021. 10.1126/science.1245938 [DOI] [PubMed] [Google Scholar]
- 33.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, et al. (2010) Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327: 78–81. 10.1126/science.1181498 [DOI] [PubMed] [Google Scholar]
- 34.Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, et al. (2005) Calibrating a coalescent simulation of human genome sequence variation. Genome Res 15: 1576–1583. 10.1101/gr.3709305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner CL, Greer FR, American Academy of Pediatrics Section on B, American Academy of Pediatrics Committee on N (2008) Prevention of rickets and vitamin D deficiency in infants, children, and adolescents. Pediatrics 122: 1142–1152. 10.1542/peds.2008-1862 [DOI] [PubMed] [Google Scholar]
- 36.Bodnar LM, Platt RW, Simhan HN (2015) Early-pregnancy vitamin D deficiency and risk of preterm birth subtypes. Obstet Gynecol 125: 439–447. 10.1097/AOG.0000000000000621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. (2002) The human genome browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwenk J, Metz M, Zolles G, Turecek R, Fritzius T, et al. (2010) Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature 465: 231–235. 10.1038/nature08964 [DOI] [PubMed] [Google Scholar]
- 39.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, et al. (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, et al. (2014) RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 42: D756–763. 10.1093/nar/gkt1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE (2002) Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci U S A 99: 5271–5276. 10.1073/pnas.082121399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki M, Kobayashi-Osaki M, Tsutsumi S, Pan X, Ohmori S, et al. (2013) GATA factor switching from GATA2 to GATA1 contributes to erythroid differentiation. Genes Cells 18: 921–933. 10.1111/gtc.12086 [DOI] [PubMed] [Google Scholar]
- 44.Lou H, Lu Y, Lu D, Fu R, Wang X, et al. (2015) A 3.4-kb Copy-Number Deletion near EPAS1 Is Significantly Enriched in High-Altitude Tibetans but Absent from the Denisovan Sequence. Am J Hum Genet 97: 54–66. 10.1016/j.ajhg.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, et al. (2012) A high-coverage genome sequence from an archaic Denisovan individual. Science 338: 222–226. 10.1126/science.1224344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, et al. (2012) Ancient admixture in human history. Genetics 192: 1065–1093. 10.1534/genetics.112.145037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeong C, Alkorta-Aranburu G, Basnyat B, Neupane M, Witonsky DB, et al. (2014) Admixture facilitates genetic adaptations to high altitude in Tibet. Nat Commun 5: 3281 10.1038/ncomms4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogers AR, Bohlender RJ (2015) Bias in estimators of archaic admixture. Theor Popul Biol 100C: 63–78. 10.1016/j.tpb.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 49.Lu D, Lou H, Yuan K, Wang X, Wang Y, et al. (2016) Ancestral Origins and Genetic History of Tibetan Highlanders. Am J Hum Genet 99: 580–594. 10.1016/j.ajhg.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu H (2016) Colonization of the Tibetan Plateau, permanent settlement, and the spread of agriculture: Reflection on current debates on the prehistoric archeology of the Tibetan Plateau. Archaeological Research in Asia 5: 12–15. [Google Scholar]
- 51.Sankararaman S, Mallick S, Patterson N, Reich D (2016) The Combined Landscape of Denisovan and Neanderthal Ancestry in Present-Day Humans. Curr Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, et al. (2014) The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505: 43–49. 10.1038/nature12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, et al. (2016) Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norsang G, Ma L, Dahlback A, Zhuoma C, Tsoja W, et al. (2009) The vitamin D status among Tibetans. Photochem Photobiol 85: 1028–1031. 10.1111/j.1751-1097.2009.00552.x [DOI] [PubMed] [Google Scholar]
- 55.Holick MF, Chen TC (2008) Vitamin D deficiency: a worldwide problem with health consequences. Am J Clin Nutr 87: 1080S–1086S. [DOI] [PubMed] [Google Scholar]
- 56.Harris NS, Crawford PB, Yangzom Y, Pinzo L, Gyaltsen P, et al. (2001) Nutritional and health status of Tibetan children living at high altitudes. N Engl J Med 344: 341–347. 10.1056/NEJM200102013440504 [DOI] [PubMed] [Google Scholar]
- 57.Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, et al. (2010) A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res 20: 1352–1360. 10.1101/gr.107920.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Ikeda R, Che XF, Ooyama A, Yamamoto M, et al. (2013) VEGF expression is augmented by hypoxiainduced PGIS in human fibroblasts. Int J Oncol 43: 746–754. 10.3892/ijo.2013.1994 [DOI] [PubMed] [Google Scholar]
- 59.Zhou L, Wang LM, Song HM, Shen YQ, Xu WJ, et al. (2013) Expression profiling analysis of hypoxic pulmonary disease. Genet Mol Res 12: 4162–4170. 10.4238/2013.October.7.2 [DOI] [PubMed] [Google Scholar]
- 60.Resendes BL, Kuo SF, Robertson NG, Giersch AB, Honrubia D, et al. (2004) Isolation from cochlea of a novel human intronless gene with predominant fetal expression. J Assoc Res Otolaryngol 5: 185–202. 10.1007/s10162-003-4042-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alkorta-Aranburu G, Beall CM, Witonsky DB, Gebremedhin A, Pritchard JK, et al. (2012) The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet 8: e1003110 10.1371/journal.pgen.1003110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tripathy V, Gupta R (2005) Birth weight among Tibetans at different altitudes in India: are Tibetans better protected from IUGR? Am J Hum Biol 17: 442–450. 10.1002/ajhb.20400 [DOI] [PubMed] [Google Scholar]
- 63.Jensen GM, Moore LG (1997) The effect of high altitude and other risk factors on birthweight: independent or interactive effects? Am J Public Health 87: 1003–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krampl E, Lees C, Bland JM, Espinoza Dorado J, Moscoso G, et al. (2000) Fetal biometry at 4300 m compared to sea level in Peru. Ultrasound Obstet Gynecol 16: 9–18. 10.1046/j.1469-0705.2000.00156.x [DOI] [PubMed] [Google Scholar]
- 65.Lichty JA, Ting RY, Bruns PD, Dyar E (1957) Studies of babies born at high altitudes. I. Relation of altitude to birth weight. AMA J Dis Child 93: 666–669. [DOI] [PubMed] [Google Scholar]
- 66.Unger C, Weiser JK, McCullough RE, Keefer S, Moore LG (1988) Altitude, low birth weight, and infant mortality in Colorado. JAMA 259: 3427–3432. [PubMed] [Google Scholar]
- 67.Julian CG, Vargas E, Armaza JF, Wilson MJ, Niermeyer S, et al. (2007) High-altitude ancestry protects against hypoxia-associated reductions in fetal growth. Arch Dis Child Fetal Neonatal Ed 92: F372–377. 10.1136/adc.2006.109579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, et al. (2015) A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delaneau O, Marchini J, Zagury JF (2012) A linear complexity phasing method for thousands of genomes. Nat Methods 9: 179–181. [DOI] [PubMed] [Google Scholar]
- 70.Durand EY, Patterson N, Reich D, Slatkin M (2011) Testing for ancient admixture between closely related populations. Mol Biol Evol 28: 2239–2252. 10.1093/molbev/msr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin SH, Davey JW, Jiggins CD (2015) Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol 32: 244–257. 10.1093/molbev/msu269 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each circle in the plot represents one genome. A) The 1st and 2nd principal components; B) The 3rd and 4th principal components; C) the 5th and 6th principal components; D) The 1st and 2nd principal components in the PCA of Chinese and Tibetans only.
(DOCX)
Each column represents one individual and different colors represent different ancestral population; each panel represents one K value. The reported ethnicities were listed on the top of each panel.
(DOCX)
The x-axis shows the position on chromosome 2, in Hg19 coordinate. The y-axis shows the FST value. Red color indicates Denisovan-like alleles. Only variants with MAF no less than 5% in either Han or Tibetan were shown. A) Chr2: 44000000–49000000. The EPAS1 region is indicated by the two vertical dotted lines. B) Chr2:46500000–46800000. The two blue vertical dotted lines indicate chr2: 46567916–46600661 (the 32.7kb region reported previously). The two red vertical dotted lines indicate chr2: 46675505–46714553, a second region with high FST and many Denisovan-like variants.
(DOCX)
Key: solid black, mD given mN; solid red, mN given mD; dashed, 95% confidence regions based on moving-blocks bootstrap; circle, simultaneous estimate.
(DOCX)
The x-axis represents the chromosome number. Each dot represents one 200kb genomic region identified by S*.
(DOCX)
The red curve shows the relative cross-coalescence rate based on the whole genome sequencing data; the grey curves show the relative cross-coalescence rate based on 20 coalescence simulations from our best-fitting dadi model with a true Han-Tibetan divergence time of 54 kya but with high rates of gene flow until 9 kya (see Fig 2).
(DOCX)
We sorted all the 200-kb genomic regions by their U values and calculated the standard deviations of the D-statistics within each percentile of U. Each dot represents the D and U for one percentile. The target population is Tibetan and the background population is Yoruba.
(DOCX)
(DOCX)
Panel A, B and C corresponds to ∂a∂i model A, B and C. In each panel, the first two plots show the observed and model predicted SFS heatmaps, respectively; the third plot shows the residual heatmap; the fourth plot shows a histogram of the residuals. Panel D shows the one-dimensional SFS for Han Chinese (left) and Tibetans (right) separately. Within each combination of population and model, the top plot shows the frequencies of variants for each minor allele count, with the red line showing the expected frequencies predicted by the model and blue line showing the observed frequencies; the bottom plot shows the standardized residuals of frequencies within each minor allele count category, assuming the frequencies are Poisson-distributed.
(DOCX)
(DOCX)
(XLSX)
(XLSX)
The start and end positions are in hg19 coordinates.
(XLSX)
(XLSX)
Red-color indicates SNVs present in the Denisovan genome.
(XLSX)
(XLSX)
(XLSX)
We first simulated a 50MB genomic region (with msms) under the MSMC demographic model, and then used dadi to estimate the Han-Tibetan divergence time. The actual demographic parameters are in the Simulated columns, and the dadi estimates are in the Estimated column.
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
The whole genome-sequencing data are available at The database of Genotypes and Phenotypes (dbGaP) under accession number phs001338.
The database of Genotypes and Phenotypes (dbGAP) accession number for the whole-genome sequencing data reported in this paper is phs001338.