Abstract
Parathyroid hormone (PTH) is a primary calcium regulatory hormone. Elevated serum PTH concentrations in primary and secondary hyperparathyroidism have been associated with bone disease, hypertension, and in some studies, cardiovascular mortality. Genetic causes of variation in circulating PTH concentrations are incompletely understood. We performed a genome-wide association study of serum PTH concentrations among 29,155 participants of European ancestry from 13 cohort studies (n=22,653 and n=6502 in discovery and replication analyses, respectively). We evaluated the association of single nucleotide polymorphisms (SNPs) with natural log-transformed PTH concentration adjusted for age, sex, season, study site, and principal components of ancestry. We discovered associations of SNPs from five independent regions with serum PTH concentration, including the strongest association with rs6127099 upstream of CYP24A1 (P=4.2 × 10−53), a gene that encodes the primary catabolic enzyme for 1,25-dihydroxyvitamin D and 25-dihydroxyvitamin D. Each additional copy of the minor allele at this SNP associated with 7% higher serum PTH concentration. The other SNPs associated with serum PTH concentration included rs4074995 within RGS14 (P=6.6 × 10−17), rs219779 adjacent to CLDN14 (P=3.5 × 10−16), rs4443100 near RTDR1 (P=8.7 × 10−9), and rs73186030 near CASR (P=4.8 × 10−8). Of these five SNPs, rs6127099, rs4074995, and rs219779 replicated. Thus, common genetic variants located near genes involved in vitamin D metabolism and calcium and renal phosphate transport associated with differences in circulating PTH concentrations. Future studies could identify the causal variants at these loci, and the clinical and functional relevance of these variants should be pursued.
Keywords: parathyroid hormone, mineral metabolism, human genetics, genome-wide association study
Parathyroid hormone (PTH) is the major calcium regulatory hormone in humans. The physiologic role of PTH is to maintain serum calcium concentration by (1) stimulating calcium release from bone, (2) enhancing renal calcium reabsorption, and (3) catalyzing the production of 1,25-dihydroxyvitamin D (1,25[OH]2D), the active form of vitamin D, from 25-hydroxyvitamin D (25[OH]D).1–6 Higher circulating PTH concentrations are associated with increased risk of fracture, hypertension, left ventricular hypertrophy, and in some studies with cardiovascular mortality.7–11
PTH is regulated in response to circulating calcium concentrations by the calcium-sensing receptor, which is highly expressed in parathyroid tissue.12–14 PTH concentration is further modulated by calcitriol, which interacts with DNA regulatory elements via the vitamin D nuclear receptor to suppress PTH, and is an established treatment for the secondary hyperparathyroidism seen in CKD.15,16 Additional causes of secondary hyperparathyroidism include calcium deficiency, vitamin D deficiency, vitamin D resistance, hypercalciuria, hyperphosphatemia, and vitamin D nuclear receptor abnormalities.17,18
A genetic basis for variability in PTH concentrations is suggested by rare Mendelian disorders that cause primary hyper- and hypoparathyroidism.19–27 Further, a classic twin study estimated that 54%–65% of variation in PTH concentration (broad-sense heritability) could be due to genetic/familial factors.28 Additionally, mutations in cell cycle and cell regulatory genes have been described in surgically removed parathyroid adenomas29,30; however, the contribution of genetic changes to the initial development of nonadenomatous parathyroid disease is unclear.
To investigate the role of common genetic variants (single nucleotide polymorphisms, SNPs) on variation in PTH concentration we conducted a genome wide association study (GWAS) meta-analysis of serum PTH concentrations in 27,561 individuals from 13 cohort studies.
Results
Genome-Wide Associations for Serum PTH Concentrations in Cohorts of European Ancestry
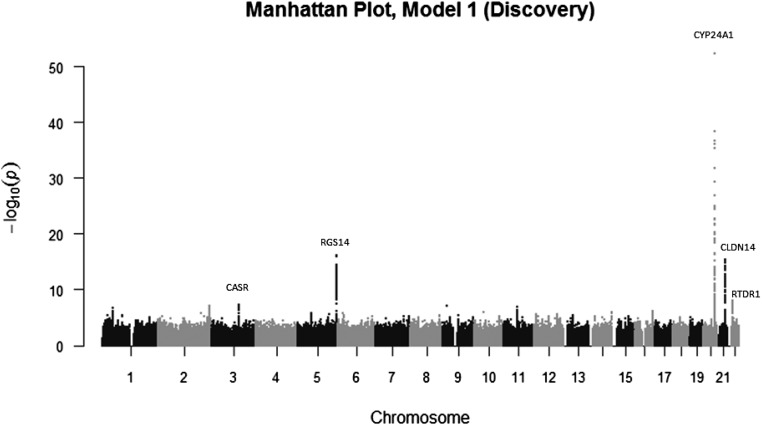
A total of 22,653 participants from ten cohort studies were available for the discovery meta-analysis.31–40 The majority of studies were community-based or population-based cohort studies of healthy adults from the United States and Europe. (Table 1) There was little evidence for population stratification at study level (median genomic inflation factor, λ=1.02) or meta-analysis level (λ=1.03). Inspection of the quantile-quantile plot suggested an excess of association signals beyond those expected (Supplemental Figure 1). SNPs in five independent regions (chromosomes 3, 5, 20, 21, and 22) exceeded the threshold of genome-wide significance for association with PTH concentration (Figure 1).
Table 1.
Characteristics of study participants
| Variables | ARIC (Whites) | CHS (Whites) | DCCT | Indiana | MESA (Whites) | MrOS | NESDA | OGP–Talana | SHIP-1 | SHIP-Trend | GOOD | LURIC | TwinsUK | Amish |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cohort information | Discovery Cohorts | Replication Cohorts | ||||||||||||
| Study design | Community-based | Community-based | RCT | Community-based | Community-based | Community-based | Community (19%), general practice (54%), and secondary mental health care (27%) | Population- and family-based | Population-based | Population-based | Population-based | Hospital-based case-control study | Twins registry | Community-based |
| Overall sample size with PTH and genotype | 8135 | 1782 | 278 | 994 | 2455 | 1785 | 2393 | 696 | 3176 | 977 | 911 | 2729 | 1982 | 621 |
| Mean age, yr (SD) | 57.1 (5.7) | 73.8 (4.8) | 32.4 (6.9) | 36.4 (8.5) | 62.6 (10.3) | 73.7 (5.8) | 42.3 (13.0) | 49.2 (20.8) | 54.3 (15.2) | 50.1 (13.7) | 18.9 (0.6) | 62.9 (10.6) | 48.2 (13.2) | 53.5 (13.9) |
| Men, n (%) | 3747 (46.1) | 680 (30.3) | 154 (55.4) | 0 (0) | 1164 (47.4) | 1785 (100) | 813 (34.0) | 284 (41.3) | 1526 (48.0) | 432 (44.2) | 911 (100) | 1879 (68.9) | 44 (2.22) | 265 (42.7) |
| Season of PTH measurement | ||||||||||||||
| Winter, n (%) | 1973 (24.3) | 564 (25.1) | 82 (29.5) | 198 (19.9) | 682 (27.8) | 350 (19.6) | 628 (26.2) | 696 (100) | 773 (24.3) | 184 (18.8) | 13 (1.4) | 593 (21.7) | 646 (32.6) | 183 (29.5) |
| Spring, n (%) | 2410 (29.6) | 515 (23.0) | 71 (25.5) | 249 (25.1) | 658 (26.8) | 468 (26.2) | 578 (24.2) | 0 (0) | 900 (28.3) | 293 (30.0) | 336 (36.9) | 549 (20.1) | 645 (32.5) | 183 (29.5) |
| Summer, n (%) | 1984 (24.4) | 650 (29.0) | 56 (20.1) | 268 (26.9) | 522 (21.3) | 515 (28.9) | 592 (24.7) | 0 (0) | 678 (21.4) | 268 (27.4) | 215 (23.6) | 754 (27.6) | 414 (20.9) | 187 (30.1) |
| Fall, n (%) | 1768 (21.7) | 514 (22.9) | 69 (24.8) | 279 (28.1) | 593 (24.2) | 452 (25.3) | 595 (24.9) | 0 (0) | 825 (26.0) | 232 (23.8) | 347 (38.1) | 833 (30.5) | 277 (14.0) | 68 (11.0) |
| BMI, kg/m2 (SD) | 27.3 (5.0) | 26.5 (4.6) | 25.9 (3.5) | 26.3 (6.0) | 27.7 (5.1) | 27.5 (3.7) | 25.6 (5.0) | 26.3 (4.9) | 28.0 (4.9) | 27.3 (4.6) | 22.3 (3.1) | 27.5 (4.1) | 26.0 (4.9) | 28.0 (5.1) |
| eGFR (MDRD), ml/min per 1.73 m2 (SD) | 87.8 (13.2) | 79.6 (18.7) | 106.3 (13.4) | NA | 76.0 (17.1) | 77.7 (16.7) | 103.7 (15.7) | 77.6 (21.5) | 89.2 (21.0) | 96.0 (18.0) | 100.1 (14.5) | 82.1 (18.3) | 83.6 (16.2) | 90.2 (19.8) |
| Mean Serum PTH, pg/ml (SD) | 40.4 (14.6) | 56.3 (28.8) | 30.6 (13.3) | 29.5 (13.2) | 40.6 (16.8) | 32.8 (12.1) | 55.7 (27.8) | 39.4 (25.1) | 37.6 (19.0) | 34.7 (20.7) | 39.3 (14.7) | 32.9 (17.9) | 34.5 (18.1) | 57.2 (19.6) |
Indiana, Indiana Sisters Cohort; NESDA, Netherlands Study of Depression and Anxiety; OGP–Talana, Ogliastra Genetic Park–Talana Study; GOOD, Gothenburg Osteoporosis and Obesity Determinants; Amish, Amish Family Osteoporosis Study; MDRD, Modification of Diet in Renal Disease.
Figure 1.
Graphical summary of genome-wide association results. Manhattan plot of the strength of association of SNPs ln-transformed PTH, on the basis of association with 22,653 participants for autosomal SNPs and 17,865 for X-chromosome (discovery stage).
The SNP having the strongest association with PTH concentration was rs6127099 (P=4.2 × 10−53; Supplemental Figure 2A, Table 2). This SNP lies 38 kbp upstream of CYP24A1 (cytochrome P450, family 24, subfamily A, polypeptide 1) on chromosome 20q13.2. Each additional copy of the rs6127099 T allele is associated with approximately 7% higher PTH concentration, after adjustment for age, sex, the first ten principal components of ancestry (where available), geographic site (where applicable), and season of PTH measurement (model 1). The CYP24A1 gene encodes the major enzyme responsible for catabolizing calcitriol to water-soluble 1,24,25-trihydroxyvitamin D and 25(OH)D to 24,25-dihydroxyvitamin D (24,25[OH]2D) for excretion.41 Further adjustment for body mass index (BMI) and eGFR (model 2) did not alter the magnitude of the association (7% higher PTH concentration after adjustment; P=2.2 × 10−43).
Table 2.
Associations of top single nucleotide polymorphisms with ln-transformed serum PTH concentrations
| SNP | Nearest Gene | Chr | Position | PTH-Increasing Allele | Other Allele | PTH-Increasing Allele Frequencya | Model 1 | Model 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Discovery (n=22,653) | Replication (n=6502) | Discovery + Replication (n=29,165) | Discovery (n=22,653) | Replication (n=4908b) | ||||||||||||
| βc (SEM) | P Value | β (SEM) | P Value | β (SEM) | P Value | β (SEM) | P Value | β (SEM) | P Value | |||||||
| rs6127099 | CYP24A1 | 20 | 52,731,402 | T | A | 0.34 | 0.07 (0.003) | 4.2 × 10−53 | 0.07 (0.011) | 9.1 × 10−11 | 0.07 (0.003) | 2.4 × 10−72 | 0.07 (0.005) | 2.2 × 10−43 | 0.08 (0.012) | 2.3 × 10−11 |
| rs4074995 | RGS14 | 5 | 176,797,343 | G | A | 0.71 | 0.03 (0.003) | 6.6 × 10−17 | 0.05 (0.012) | 4.3 × 10−4 | 0.03 (0.003) | 3.3 × 10−23 | 0.03 (0.005) | 7.7 × 10−12 | 0.05 (0.012) | 9.3 × 10−5 |
| rs219779 | CLDN14 | 21 | 37,833,751 | G | A | 0.75 | 0.04 (0.003) | 3.5 × 10−16 | 0.05 (0.011) | 4.3 × 10−5 | 0.04 (0.003) | 8.9 × 10−22 | 0.03 (0.005) | 1.6 × 10−11 | 0.05 (0.012) | 1.6 × 10−4 |
| rs4443100 | RTDR1 | 22 | 23,372,864 | G | C | 0.32 | 0.02 (0.003) | 8.7 × 10−9 | 0.01 (0.01) | 0.50 | 0.02 (0.003) | 4.1 × 10−11 | 0.02 (0.005) | 3.0 × 10−7 | 0.01 (0.012) | 0.20 |
| rs73186030 | CASR | 3 | 122,013,465 | T | C | 0.14 | 0.03 (0.003) | 4.8 × 10−8 | 0.03 (0.01) | 0.02 | 0.03 (0.004) | 1.2 × 10−9 | 0.03 (0.006) | 6.4 × 10−7 | 0.03 (0.015) | 0.04 |
In aggregate, rs6127099, rs4074995, rs219779, rs4443100, and rs73186030 together accounted for 4.2% of circulating PTH variation. Only top SNP from each region shown. Model 1 includes age, sex, season of PTH measurement, geographic site (if applicable), and first ten principal components of ancestry (if available). Model 2 additionally adjusts for BMI, eGFR, and eGFR-squared. Top SNPs were defined as the most significant SNP (lowest P value) within a 500 kb window. Chr, chromosome.
Allele frequency data from 1000 Genome Phase 1 genotype data (European Super Population [EUR]).
Results for model 2 were not available for the Amish Family Osteoporosis Study.
β-estimates are interpreted as the absolute difference in ln(PTH) per PTH-increasing allele, e.g., +0.07 is a 0.07-unit higher ln(PTH) per T allele. The exponentiated β coefficients are interpreted as the relative difference in PTH concentration per T allele, e.g., for rs6127099, every allele is associated with a e0.07-fold, or 1.073-fold higher, or 7.3% higher PTH concentration per T allele.
rs4074995 was associated with PTH concentrations (P=6.6 × 10−17, Table 2) and is located on chromosome 5q35.3. This SNP is intronic within RGS14 (regulator of G-protein signaling 14).42 However, rs4074995 is in strong linkage disequilibrium (LD) with SNPs in the directly adjacent gene, SLC34A1 (solute carrier family 34 [type 2 sodium/phosphate cotransporter], member 1). SLC34A1 is a strong candidate gene for PTH concentration, as it encodes the kidney-specific sodium-phosphate type 2a transporter (Npt2a) responsible for phosphate reabsorption in the proximal tubule.43
The third SNP robustly associated with serum PTH concentrations was rs219779 (P=3.5 × 10−16, Table 2), located in a transcript (AP000695.6) on chromosome 21q22.13, adjacent to CLDN14 (Claudin 14). Each copy of the rs219779 G allele was associated with approximately 4% higher PTH concentration, after full adjustment. CLDN14 is a candidate gene for serum PTH concentration, as it plays a major role in tight junction-specific obliteration of the intercellular space, through calcium-independent cell-adhesion activity.44
rs4443100 was associated with PTH concentration and is located approximately 20 kb 3′ of RTDR1 (RSPH14, radial spoke head 14 homolog) on chromosome 22q11.23 (P=8.7 × 10−9, Table 2). This gene encodes a protein with no known function but is located in a region deleted in pediatric rhabdoid tumors of the kidney.45
rs73186030 was also significantly associated with PTH concentration (P=4.8 × 10−8, Table 2), and is located approximately 8 kb 3′ of CASR (calcium-sensing receptor) on chromosome 3q13.33. The CaSR protein is a G protein–coupled receptor that is expressed primarily in the parathyroid gland and the ascending loop of the kidney. This receptor senses small changes in circulating calcium concentration and couples this information to intracellular signaling pathways that modify PTH secretion and renal cation handling, thus this protein plays an essential role in maintaining mineral ion homeostasis.46,47
Analyses of the CYP24A1 region, conditioning on rs6127099, identified one additional locus which was independently associated with circulating PTH (rs35194449; PTH-increasing allele: T [frequency=0.24]; effect estimate: 4% higher PTH per additional copy of T-allele; pc=1.8 × 10−10; r2 between SNP and rs6127099 = 0.00). Conditional analyses of the remaining four regions did not identify any secondary association signals, indicating no additional independently-associated SNPs after conditioning on the region’s lead SNP.
In aggregate, the loci (rs6127099, rs35194449, rs4074995, rs219779, rs4443100, and rs73186030) explained 4.5% of the variance in circulating PTH.
Replication in Four Cohorts of European Ancestry
A total of 6502 individuals were available for the replication meta-analysis of model 1 results, and 4908 for model 2. Regression coefficients for each of the top five SNPs in the discovery sample were in the same direction in the replication sample (Table 2). Replication associations were considered significant at the Bonferroni-corrected P=0.01 level for three of five SNPs meta-analyzed in replication. Post hoc power calculations of SNPs rs6127099, rs4074995, rs219779, rs4443100, and rs73186030 for replication analyses with 6502 individuals revealed 99%, 77%, 96%, 36%, and 31% power, respectively, to detect the β coefficients from the discovery analyses, with a type 1 error α=0.01. Primary regression coefficients and interpretation of our results were not affected by additional adjustment for BMI and eGFR in sensitivity analyses (Table 1).
Replication in Populations of African Ancestry
In black populations, data were available for four of the five SNPs. Three of the four SNPs had β coefficients that were direction-consistent with the primary analysis and one SNP (rs4074995) was significantly (P<0.01) associated (Table 3). rs219779 and rs4443100 showed the most allelic differentiation between individuals of African and European ancestry. The frequencies of PTH-increasing alleles differed across the five Super Populations assessed in the 1000 Genomes Project (Supplemental Table 5).
Table 3.
Associations of top single nucleotide polymorphisms with ln-transformed serum PTH concentrations among individuals of black descent (n=4279)
| SNP | Nearest Gene | Chr | Position | PTH-Increasing Allele | Other Allele | PTH-Increasing Allele Frequencya | β (SEM)b | P Value | Fstc |
|---|---|---|---|---|---|---|---|---|---|
| rs6127099 | CYP24A1 | 20 | 52,731,402 | T | A | 0.21 | +0.03 (0.0136) | 0.0363 | 0.020 |
| rs4074995 | RGS14 | 5 | 176,797,343 | G | A | 0.92 | −0.04 (0.0160) | 0.0059 | 0.057 |
| rs219779 | CLDN14 | 21 | 37,833,751 | G | A | 0.69 | −0.01 (0.0096) | 0.5337 | 0.102 |
| rs4443100 | RTDR1 | 22 | 23,372,864 | C | G | 0.80 | −0.02 (0.0106) | 0.05366 | 0.100 |
| rs73186030 | CASR | 3 | 122,013,465 | T | C | 0.01 | NA | NA | 0.055 |
Chr, chromosome; Fst, fixation index.
MAF data from 1000 Genome Phase 1 genotype data (African Super Population [AFR]).
Effect estimate adjusted for age, sex, season of PTH measurement, geographic site (if applicable), and first ten principal components of ancestry (if available) and are interpreted as the absolute difference in ln(PTH) per minor allele, e.g., +0.03 is a 0.03-unit higher ln(PTH) per minor allele. The exponentiated β coefficients are interpreted as the relative difference in PTH concentration per T allele, e.g., for rs6127099, every T allele is associated with a e0.03-fold, or 1.03-fold higher, or 3% higher PTH concentration per minor allele.
Fst is an estimate of genetic differentiation calculated using the variance in allele frequencies among European and African samples from the 1000 Genomes and standardized according to the mean allele frequency in the combined sample.73
Expression Quantitative Trait Locus and Functional Prediction
For each of the replicated SNPs in European populations, we identified all proxy SNPs with r2>0.8 in the 1000 Genomes Pilot 1 pairwise LD data, yielding a total of 120 SNPs. We then queried each of these SNPs in the expression quantitative trait locus (eQTL) database of the Phenotype-Genotype Integrator and the Genotype-Tissue Expression project.48–50 rs4074995 was associated with gene expression at RGS14 (strongest association in transformed fibroblast cells, P=1.0 × 10−28), F12 in esophagus mucosa (P=1.2 × 10−7), and FGFR4 in tibial nerve tissue (P=6.8 × 10−7), whereas rs219779 was associated with expression at AP000695.4 in skeletal muscle tissue (P=9.1 × 10−6). Associations between SNPs and SLC34A1 and CLDN14 expression could not be analyzed, as no information for these genes was included in either database. Furthermore, neither kidney nor parathyroid tissue eQTL data were available to be queried.
Associations with Bone Mineral Density
Each of the three replicated SNPs was associated (P<0.01) with femoral neck bone density in the Genetic Factors of OSteroporosis (GEFOS) consortium.51 Each additional copy of the PTH-increasing allele at rs6127099 (CYP24A1), rs4074995 (RGS14/SLC34A1), and rs219779 (CLDN14) was associated with a +0.030 (SEM=0.009, P=5 × 10−4), +0.023 (SEM=0.008, P=8 × 10−3), and −0.022 (SEM=0.008, P=9 × 10−3) difference in bone mineral density (BMD) SD.
Associations with Mineral Metabolism Biomarkers
rs6127099 (CYP24A1) is significantly associated with circulating concentrations of PTH, and circulating fibroblast growth-factor 23 (FGF-23), serum phosphorus, and urine concentrations of calcium and phosphate normalized to urine creatinine (Table 4). rs4443100 (RTDR1) and rs219779 (CLDN14) are associated with PTH concentrations and with FGF-23 concentrations. rs4074995 (RGS14/SLC34A1) is also associated with FGF-23 and phosphorus concentrations, and 25(OH)D and calcium concentrations. Serum concentrations of 24,25(OH)2D were not associated with any of the most associated SNPs identified in the discovery/replication GWAS meta-analysis (Table 4).
Table 4.
Associations of top single nucleotide polymorphisms with mineral metabolism measurements
| SNP and Nearest Gene | PTH, pg/ml | FGF23, pg/ml | 24,25D, ng/ml | 25D, ng/ml | Serum Calcium, mg/dl | Serum Phosphorus, mg/dl | Urine Calcium-to-Creatinine Ratio, mg/mg | Urinary FePO4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Mean (SD) | N | Mean (SD) | N | Mean(SD) | N | Mean (SD) | N | Mean (SD) | N | Mean (SD) | N | Mean (SD) | N | Mean (SD) | |
| rs6127099 | ||||||||||||||||
| CYP24A1 | ||||||||||||||||
| AA | 5164 | 38.88 (10.5) | 5164 | 44.17 (11.3) | 1244 | 4.54 (2.7) | 5164 | 27.93 (6.7) | 6638 | 9.44 (0.24) | 6638 | 3.57 (0.3) | 1244 | 0.102 (0.07) | 1244 | 12.71 (5.7) |
| AT | 3906 | 41.76 (11.1) | 3906 | 42.24 (12.9) | 1009 | 4.51 (2.7) | 3906 | 27.17 (6.6) | 5041 | 9.44 (0.24) | 5041 | 3.57(0.3) | 1009 | 0.102 (0.07) | 1009 | 12.62 (5.3) |
| TT | 703 | 44.08 (11.8) | 703 | 40.83 (10.2) | 202 | 4.69 (2.8) | 703 | 27.16 (6.1) | 907 | 9.45 (0.26) | 907 | 3.55 (0.3) | 202 | 0.090 (0.06) | 202 | 11.78 (5.3) |
| P-for-trenda | <0.001 | <0.001 | 0.46 | 0.004 | 0.33 | 0.02 | 0.03 | 0.03 | ||||||||
| rs4074995 | ||||||||||||||||
| RGS14 | ||||||||||||||||
| AA | 830 | 37.22 (9.19) | 830 | 40.79 (9.97) | 202 | 4.66 (2.8) | 830 | 28.02 (6.6) | 1053 | 9.45 (0.25) | 1053 | 3.52 (0.3) | 202 | 0.105 (0.07) | 202 | 12.44 (5.0) |
| GA | 4173 | 40.08 (11.1) | 4173 | 42.78 (12.3) | 968 | 4.49 (2.6) | 4173 | 27.59 (6.6) | 5416 | 9.44 (0.24) | 5416 | 3.56 (0.3) | 968 | 0.101 (0.07) | 968 | 12.70 (5.4) |
| GG | 5587 | 41.23 (11.1) | 5587 | 43.69 (11.7) | 1285 | 4.55 (2.7) | 5587 | 27.44 (6.6) | 7109 | 9.44 (0.24) | 7109 | 3.58 (0.3) | 1285 | 0.101 (0.07) | 1285 | 12.54 (5.7) |
| P-for-trend | <0.001 | <0.001 | 0.59 | 0.02 | <0.001 | <0.001 | 0.46 | 0.81 | ||||||||
| rs219779 | ||||||||||||||||
| CLDN14 | ||||||||||||||||
| AA | 645 | 39.12 (11.2) | 645 | 44.46 (12.4) | 168 | 4.53 (2.5) | 645 | 27.63 (6.5) | 828 | 9.45 (0.23) | 828 | 3.58 (0.3) | 168 | 0.087 (0.06) | 168 | 12.09 (5.2) |
| AG | 3823 | 39.91 (10.6) | 3823 | 42.98 (11.7) | 907 | 4.55 (2.7) | 3823 | 27.41 (6.6) | 4858 | 9.44 (0.24) | 4858 | 3.57 (0.3) | 907 | 0.104 (0.07) | 907 | 12.56 (5.3) |
| GG | 5878 | 40.89 (11.0) | 5878 | 43.03 (12.1) | 1380 | 4.53 (2.7) | 5878 | 27.62 (6.6) | 7647 | 9.45 (0.24) | 7647 | 3.57 (0.3) | 1380 | 0.101 (0.07) | 1380 | 12.68 (5.7) |
| P-for-trend | 0.001 | 0.004 | 1.00 | 0.99 | 0.65 | 0.39 | 0.02 | 0.19 | ||||||||
| rs4443100 | ||||||||||||||||
| RTDR1 | ||||||||||||||||
| CC | 4898 | 39.90 (10.7) | 4898 | 43.60 (13.1) | 1188 | 4.48 (2.6) | 4898 | 27.56 (6.5) | 6246 | 9.46 (0.24) | 6246 | 3.57 (0.3) | 1188 | 0.099 (0.07) | 1188 | 12.62 (5.4) |
| CG | 4406 | 40.86 (11.2) | 4406 | 42.73 (11.2) | 1014 | 4.61 (2.7) | 4406 | 27.73 (6.7) | 5680 | 9.42 (0.25) | 5680 | 3.57 (0.3) | 1014 | 0.102 (0.07) | 1014 | 12.52 (5.8) |
| GG | 994 | 41.74 (11.3) | 994 | 42.64 (10.1) | 253 | 4.52 (2.8) | 994 | 26.87 (6.5) | 1340 | 9.43 (0.23) | 1340 | 3.58 (0.3) | 253 | 0.107 (0.07) | 253 | 12.80 (5.1) |
| P-for-trend | <0.001 | 0.02 | 0.83 | 0.003 | 0.001 | 0.20 | 0.11 | 0.64 | ||||||||
| rs73186030 | ||||||||||||||||
| CASR | ||||||||||||||||
| CC | 7817 | 40.00 (10.9) | 7817 | 43.05 (12.3) | 1746 | 4.54 (2.7) | 7817 | 27.61 (6.7) | 9916 | 9.41 (0.24) | 9916 | 3.59 (0.3) | 1746 | 0.101 (0.07) | 1746 | 12.45 (5.6) |
| CT | 2456 | 41.76 (11.3) | 2456 | 43.32 (11.4) | 636 | 4.52 (2.5) | 2456 | 27.37 (6.5) | 3264 | 9.51 (0.24) | 3264 | 3.53 (0.3) | 636 | 0.102 (0.07) | 636 | 12.94 (5.5) |
| TT | 212 | 42.15 (10.4) | 212 | 42.98 (9.68) | 73 | 4.60 (2.6) | 212 | 27.17 (5.5) | 283 | 9.55 (0.21) | 283 | 3.53 (0.3) | 73 | 0.103 (0.07) | 73 | 13.03 (4.8) |
| P-for-trend | 0.01 | 0.94 | 0.85 | 0.35 | <0.001 | 0.001 | 0.81 | 0.38 | ||||||||
24,25D, 24,25-dihydroxyvitamin D; 25D, 25-hydroxyvitamin D; FePO4, Fractional excretion of phosphorus.
P-for-trend obtained from Wald test of linear regression coefficient for number of copies of the minor allele.
Discussion
In this first reported GWAS of PTH concentration, we identified five loci, located on chromosomes 3, 5, 20, 21, and 22, that were associated with variation in PTH concentrations. Upon replication, three of the five loci were associated with PTH concentration after Bonferroni correction. The strongest association was observed for rs6127099, located 38 kbp upstream of CYP24A1, encoding the primary catabolic enzyme for 1,25(OH)2D. Other significant associations in discovery and replication included SNPs located near genes encoding the renal type 2a sodium-phosphate cotransporter and claudin 14. Taken together, these findings provide the first human evidence linking PTH concentrations and common polymorphisms near genes involved with vitamin D and calcium mineral metabolism.
The cytochrome p450 enzyme CYP24A1 is critically important for maintaining serum 1,25(OH)2D concentrations within a tight physiologic range and preventing vitamin D toxicity by catalyzing the conversion of 1,25(OH)2D to 1,24,25(OH)3D for subsequent excretion.41 Inactivating mutations of CYP24A1 cause severe infantile hypercalcemia, hypercalcemic syndrome in adults, and nephrolithiasis, with potent suppression of PTH.52–58 We found the T allele of rs6127099 to be associated with greater serum PTH concentrations, suggesting that the associated haplotype may confer increased CYP24A1 activity, accelerated 1,25(OH)2D catabolism, and disinhibition of PTH levels. The association of the T allele of rs6127099 with lower serum concentrations of FGF-23, which is upregulated by 1,25(OH)2D, is also consistent with increased CYP24A1 activity and accelerated 1,25(OH)2D catabolism. CYP24A1 also converts 25(OH)D to 24,25(OH)2D; however, we detected only small differences in serum 25(OH)D and no differences in serum 24,25(OH)2D concentrations according to the number of rs6127099 minor alleles. This suggests that environmental factors (diet and sunlight) play a more important role than genetic variation for circulating 25(OH)D.59 Of note, rs6127099 is in near perfect linkage (or LD) with a variant (i.e., rs1570669) which has been associated with lower circulating calcium and with BMD at the lumbar spine.60 Differences in 1,25(OH)2D catabolism could have important clinical implications for the therapeutic approach to parathyroid disorders; future studies are needed to test whether rs6127099 is associated with variation in the response to vitamin D treatment.
rs4074995 is located within RGS14, which encodes a G-protein signal-regulating protein, and is in LD with multiple SNPs within the gene coding for type 2a sodium phosphate cotransporter (Npt2a, SLC34A1) expressed in the proximal tubule of the kidney.61 In a previous GWAS, as in ours, rs4074995 was found to be associated with serum phosphate concentrations.62 Hypothetically, this variant could lead to greater ion leakage in the sodium channel, leading to reduced need for phosphaturic hormones. Not surprisingly, we did not detect differences in the spot urinary excretion of calcium or phosphate according to the number of rs4074995 minor alleles; these parameters are highly influenced by dietary intake. The functional relevance of this polymorphism to renal phosphate transport and PTH regulation awaits further elucidation.
CLDN14 encodes a tight junction protein expressed in the thick ascending limb of the kidney that inhibits calcium and magnesium reabsorption, and is regulated by the CaSR.44 The CaSR, located at the surface of parathyroid cells, mediates their detection of small changes in blood ionized calcium concentration, and subsequent modification of PTH release.44,63,64 Located in a CLDN14 intron, rs219779 is in LD with several other SNPs within this gene. The rs219779 allele was associated with significantly lower urinary calcium excretion and circulating PTH, consistent with enhanced CLDN14 function. The common variant rs219780, 400 bp upstream of, and in moderate LD with, rs219779 (r2=0.78) has been associated with kidney stones and BMD.65 Further studies are needed to identify causal variants in this region that affect renal calcium handling and modulate PTH.
Perhaps surprisingly, common SNPs near CYP24A1, SLC34A1, and CLDN14 were more strongly associated with serum PTH concentrations than those located near or within CASR, the gene encoding the primary receptor involved in PTH regulation. This may have been due to differences in minor allele frequency (MAF), however, as the CASR locus had the lowest MAF of the discovered loci. Post hoc power calculations of the replication analysis of rs73186030 to detect the same β coefficient as the discovery analyses showed 31% power with a type 1 error α=0.01. Nevertheless, rs73186030, which is in LD with several SNPs within CASR, was associated with significant differences in serum PTH in combined discovery and replication analyses. Rare genetic disruptions of the CASR gene cause familial hypocalciuric hypercalcemia and autosomal dominant hypoparathyroidism with hypocalcemia, and rs1801725, a common polymorphism within CASR, was previously identified to be associated with serum calcium concentrations in a GWAS.60,63,66 The rs73186030 identified here is in strong LD with rs1801725 (R2=0.925, D′=1.00), and was associated with differences in serum calcium and phosphate concentrations.
Strengths of our study include the large and diverse sample, the population-based settings, and the comprehensive set of common genetic variants and mineral metabolites examined. Potential limitations include a restriction to common variants only and discovery efforts in an exclusively European American sample. The top SNPs identified in this study were located near genes encoding proteins that are typically expressed in the kidneys and parathyroid tissue and are therefore unlikely to be related to gene expression in circulating white blood cells or fibroblasts.49,50 Follow-up expression work in renal and parathyroid tissue is required and may shed additional light on the functional relevance of the identified polymorphisms.
Although the observed variants were associated with mild differences in PTH concentrations, it is possible that the effects may be more pronounced among individuals with mineral metabolism disturbances, such as those with CKD. It will be crucial to test these results among individuals with CKD, in whom the biologic implications and potential treatment options are most relevant.
In summary, we demonstrate that common genetic variants are associated with circulating PTH concentrations in adults using meta-analysis of study-specific GWAS from thirteen large population-based studies. Follow-up studies are needed to identify potential causal genetic loci in or near the identified candidate genes. Candidate genes may be explored in more comprehensive metabolic studies of PTH metabolism and in translational animal models that will shed new light on the mechanisms and clinical implications of PTH in calcium homeostasis and its treatment.
Concise Methods
Subjects
Discovery Study Populations
Ten cohorts contributed to the discovery meta-analysis, by providing study-specific genome-wide analyses of PTH concentrations, for a total of 22,653 individuals of European ancestry (Table 1). Contributing studies included the Atherosclerosis Risk in Communities Study (ARIC; number of individuals of European ancestry, n=8135),31 the Cardiovascular Health Study (CHS; n=1782),32 the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC; n=278),33 the Indiana Sisters Study (n=994),34 the Multi-Ethnic Study of Atherosclerosis (MESA; n=2446),35 the Osteoporotic Fractures in Men Study (MrOS; n=1785),36 the Netherlands Study of Depression and Anxiety (n=2393),37 the Ogliastra Genetic Park–Talana Study (n=687),39 the Study of Health in Pomerania (SHIP; n=3176),38 and the SHIP-Trend Study (n=977).40
Replication Study Populations
In a second stage, we followed up SNPs showing significant association with PTH (P<5 × 10−8) and performed meta-analysis of β-estimates and SEM across discovery and replication stages. The replication analyses encompassed 1982 subjects from the TwinsUK Study,67 911 subjects from the Gothenburg Osteoporosis and Obesity Determinants Study,68 2988 participants from the Ludwigshafen Risk and Cardiovascular Health (LURIC) study,69 and 621 participants from the Amish Family Osteoporosis Study70 (Table 1).
For all study populations, we excluded participants who had an eGFR<30 ml/min per 1.73 m2 calculated from serum creatinine, because of the possibility that the strong influence of kidney disease may overwhelm potentially subtler influences of SNPs on circulating PTH concentrations. All participants provided written informed consent. The study was approved by the institutional ethics committee of each study site and was conducted according to Declaration of Helsinki principles.
The meta-analysis was extended to include the evaluation of the X chromosome across seven cohorts from the discovery GWAS (MESA, ARIC, the Indiana Sisters Study, DCCT/EDIC, MrOS, SHIP, and SHIP-TREND), for a total of 17,865 participants.
Genotyping and Imputation
Genome-wide commercial arrays were used by all cohorts as described in Supplemental Table 1. Genotype calling for SNPs on the X-chromosome was performed separately in men and women. Quality control methods implemented by each study are described in the Supplemental Material. Imputation to the Phase 1 or 2 1000 Genomes panel was conducted using either MACH, IMPUTE, or BIMBAM with quality control metrics as shown in Supplemental Table 2. Imputation results were summarized as an “allele dosage” (a fractional value between 0 and 2), defined as the expected number of copies of the minor allele at that SNP.
Measurement of PTH
Circulating PTH concentrations were measured using second or third generation PTH immunoassays (Supplemental Table 3). We natural-log–transformed PTH as the dependent variable in analyses to evaluate proportionate differences in serum PTH concentrations, minimizing differences in mean PTH concentrations to yield comparable interpretation of coefficients across assays.
Statistical Analyses
Individual centers performed GWAS analyses using linear regression of natural-log–transformed PTH concentrations as the dependent variable, and genotypes (SNP allelic dosage) as predictors, under an additive genetic model. The exponentiated β coefficients from these models can be interpreted as the proportionate (or fold-) difference in serum PTH concentration associated with each additional copy of the minor allele at a given SNP, holding other model covariates constant.
Covariates included in model 1 were age, sex, and season of PTH measurement. If applicable, cohorts included geographic study site, and, where available, the first ≤10 principal components of ancestry to adjust for population stratification (Supplemental Table 2). PTH concentrations are strongly associated with eGFR and BMI. To detect PTH loci independent of these pathways and to diminish associations with effects modulated through these factors, model 2 added adjustment for BMI (calculated as weight in kilograms divided by height in meters-squared) and both linear and quadratic terms for the eGFR, calculated using the creatinine-based Chronic Kidney Disease Epidemiology Collaboration equation.71 Both discovery and replication analyses used the same adjustment models.
Genomic control parameters were estimated for each cohort and appropriate genomic control correction was applied to input statistics before performing meta-analysis to correct for residual cryptic relatedness or population stratification.72 Study-specific genomic control (λGC) estimates are shown in Supplemental Table 2.
Genetic differentiation was estimated using the Weir unbiased estimator of the fixation index, calculated using the variance in allele frequencies among European and African samples from the 1000 Genomes and standardized according to the mean allele frequency in the combined sample.73
Conditional analyses were performed using the Genome-wide Complex Traits Analysis tool, version 1.25.3,74 to test whether multiple independent risk alleles existed at any of the genome-wide significant loci, using a stepwise selection procedure and summary-level statistics from the meta-analysis (combined discovery and replication, model 1).
Post hoc power analyses were performed using the Quanto software.75
Meta-Analysis of Discovery Data
Fixed-effects inverse variance–weighted meta-analysis was conducted within each cohort. This approach takes directionality into account by aligning study results according to the same effect allele. Study-specific effect estimates and SEMs were combined using METAL.76
Analyses of the X-chromosome were carried out separately in men and women, and the studies were meta-analyzed separately by sex using an inverse-variance model with fixed effects. The sex-specific meta-analysis results were then combined using a sample-size weighted model.77
Variants with imputation quality <0.3 or a MAF <0.05 were excluded from each dataset before meta-analysis. The following exclusions were also applied: call rate <97%, Hardy–Weinberg equilibrium P value <10−5, duplicate error or Mendelian inconsistency, heterozygote frequency approximately 0, or SNP not found in dbSNP Build 142. SNPs were further excluded from analysis if the ratio of the variance of the allele dosage to the variance expected under Hardy–Weinberg equilibrium was <0.01. In the X-chromosome analyses, all exclusions and filters were applied to sex-stratified SNP-level data. In total, 8,020,965 autosomal SNPs and 538,222 markers on the X-chromosome were meta-analyzed in stage 1. Quantile-quantile plots and Manhattan plots were produced using R and regional association plots were created using SNAP.78
Selection of SNPs for Replication and Combined Discovery and Replication Analysis
SNPs exhibiting statistically significant association with PTH concentration (P<5 × 10−8) were evaluated for replication in four independent cohorts. For genetic regions that contained multiple SNPs significantly associated with PTH concentration, we selected the most significant SNP (lowest P value) within a 500 kb window for replication. Four of the top five SNPs found to be significantly associated with PTH concentration in individuals of European ancestry were imputed, with the following mean ratios of observed variance of the allele dosage to the expected binomial variance at Hardy–Weinberg equilibrium: rs6127099, oevar=0.91; rs219779, oevar=1.03; rs4443100, oevar=1.00; rs73186030, oevar=1.00. The rs4074995 SNP was directly genotyped.
We performed an inverse variance–weighted fixed-effects meta-analysis across replication results using METAL76 and obtained two-sided P values for the final effect estimates.
Proportion of Phenotypic Variance Explained
The proportion of variance (PVE) in circulating PTH levels explained by each top novel locus, jointly across all discovery cohorts, was estimated as:
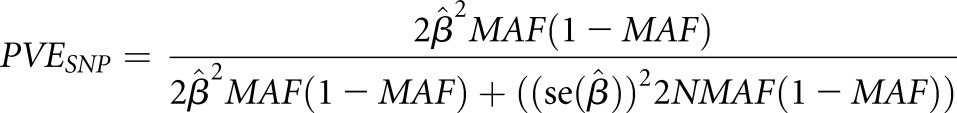 |
where 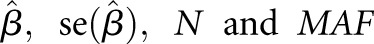 are the effect size estimate of each minor allele on the relative concentration of PTH, SEM of the effect size, sample size, and MAF for the SNP, respectively.79
are the effect size estimate of each minor allele on the relative concentration of PTH, SEM of the effect size, sample size, and MAF for the SNP, respectively.79
Follow-Up in African Ancestry Cohorts
The five top SNPs found to be significantly associated with PTH concentration in individuals of European ancestry were evaluated for replication among black individuals from three cohorts: ARIC (n=2464), MESA (n=1510), and CHS (n=305). Four of the SNPs were available in black participants and meta-analyzed. These analyses were performed using the same quality control and imputation exclusions, and meta-analysis was performed using METAL.76
eQTL and Functionality Prediction
For each of the SNPs replicated in the European ancestry populations, we identified all proxy SNPs with r2>0.8 in the 1000 Genomes Pilot 1 pairwise LD data using the SNP Annotation and Proxy Search online database (SNAP, Broad). For each SNP, we queried the eQTL database of the Phenotype-Genotype Integrator (http://www.ncbi.nlm.nih.gov/gap/phegeni).
Associations with BMD
We conducted look-ups for femoral bone density in the GEFOS dataset.51 BMD is used in clinical practice for the diagnosis of osteoporosis and bone density at the femoral neck is predictive of fracture risk. BMD was measured in all cohorts at the femoral neck using dual-energy x-ray absorptiometry following standard manufacturer protocols (General Electric Lunar Corp., Madison, WI or Hologic Inc., Bedford, MA).51
Associations with Mineral Metabolism Traits
For the SNPs associated with serum PTH concentrations, we examined their association with related mineral metabolism biomarkers where available, including levels of FGF-23 (ARIC and MESA), 25(OH)D (ARIC and MESA), 24,25(OH)2D (MESA), serum calcium and phosphate (ARIC, LURIC, and MESA), and urine calcium and phosphorus (MESA). Details of these laboratory measurements are provided in Supplemental Tables 4 and 5.
Disclosures
B.K. reports receiving consulting fees and grant support from Amgen Inc. (Thousand Oaks, CA). I.H.d.B. reports receiving research support from Abbvie (Chicago, IL), MedTronic (Minneapolis, MN), and Abbott (Chicago, IL), and consulting fees from Amgen Inc., Bayer (Leverkusen, Germany), and Janssen (Beerse, Belgium). B.M.P. serves on the Data and Safety Monitoring Board of a clinical trial of a device funded by the manufacturer (Zoll LifeCor Corp.; Pittsburgh, PA) and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson (New Brunswick, NJ).
Supplementary Material
Acknowledgments
The authors wish to thank the investigators, staff, and participants of the individual participating studies for their valuable contributions.
The Multi-Ethnic Study of Atherosclerosis (MESA) and the MESA Single Nucleotide Polymorphism Health Association Resource (SHARe) project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, UL1-TR-000040, and DK063491. Support is provided by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071258, R01HL071259, by the National Center for Research Resources, Grant UL1RR033176, and the National Center for Advancing Translational Sciences, Grant UL1TR000124. Industry contributors have had no role in the DCCT/EDIC study but have provided free or discounted supplies or equipment to support participants’ adherence to the study: Abbott Diabetes Care (Alameda, CA), Animas (Westchester, PA), Bayer Diabetes Care (North America Headquarters, Tarrytown, NY), Becton Dickinson (Franklin Lakes, NJ), Eli Lilly (Indianapolis, IN), Extend Nutrition (St. Louis, MO), Insulet Corporation (Bedford, MA), Lifescan (Milpitas, CA), Medtronic Diabetes (Minneapolis, MN), Nipro Home Diagnostics (Ft. Lauderdale, FL), Nova Diabetes Care (Billerica, MA), Omron (Shelton, CT), Perrigo Diabetes Care (Allegan, MI), Roche Diabetes Care (Indianapolis, IN), and Sanofi-Aventis (Bridgewater, NJ). The DCCT/EDIC has been supported by cooperative agreement grants (1982–1993, 2012–2017), and contracts (1982–2012) with the Division of Diabetes Endocrinology and Metabolic Diseases of the National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) (current grant numbers U01 DK094176 and U01 DK094157), and through support by the National Eye Institute, the National Institute of Neurologic Disorders and Stroke, the General Clinical Research Centers Program (1993–2007), and Clinical Translational Science Center Program (2006–present), Bethesda, MD. Trial Registration: clinicaltrials.gov NCT00360815 and NCT00360893. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by NHLBI contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367, and R01HL086694, National Human Genome Research Institute Grant U01HG004402, and National Institutes of Health (NIH) contract HHSN268200625226C. PTH measurements were supported by R01HL103706. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research. For the Gothenburg Osteoporosis and Obesity Determinants Study, financial support was received from the Swedish Research Council, the Swedish Foundation for Strategic Research, the Avtal om Läkarutbildning och Forskning (ALF-LUA) research grant in Gothenburg, the Lundberg Foundation, the Torsten and Ragnar Söderberg’s Foundation, the Västra Götaland Foundation, the Göteborg Medical Society, the Novo Nordisk foundation, and the European Commission grant HEALTH-F2-2008-201865-GEFOS. The work within the Indiana Sisters cohort was supported by NIH Grant R01AG041517. Genotyping services were provided by Center for Inherited Disease Research, which is fully funded through a federal contract from the NIH to the Johns Hopkins University (contract HHSN268200782096C). This research was supported in part by the Intramural Research Program of the NIH, National Library of Medicine. The Osteoporotic Fractures in Men Study is supported by NIH funding. The following institutes provide support: the National Institute on Aging (NIA), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Center for Advancing Translational Sciences, and NIH Roadmap for Medical Research under the following grant numbers: U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160, and UL1 TR000128. C.M.N.’s work is supported by NIAMS: K01AR062655. For the Netherlands Study of Depression and Anxiety, funding was obtained from the Netherlands Organization for Scientific Research (Geestkracht program grant 10-000-1002), the Center for Medical Systems Biology (CSMB, NWO Genomics), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL), VU University’s Institutes for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam, University Medical Center Groningen, Leiden University Medical Center, and the NIH (R01D0042157-01A, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995). Part of the genotyping and analyses were funded by the Genetic Association Information Network of the Foundation for the NIH. Computing was supported by BiG Grid, the Dutch e-Science Grid, which is financially supported by The Netherlands Organisation for Scientific Research. Funding for the Amish studies was provided by R01 AR46838 and P30 DK072488.The Study of Health in Pomerania is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania, and the network ‘Greifswald Approach to Individualized Medicine’ funded by the Federal Ministry of Education and Research (grant 03IS2061A). Genome-wide data have been supported by the Federal Ministry of Education and Research (grant no. 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg–West Pomerania. The University of Greifswald is a member of the ‘Center of Knowledge Interchange’ program of the Siemens Aktiengesellschaft and the Caché Campus program of the InterSystems GmbH. E.S. was supported by NIH/NIDDK grant K24DK106414. For the Ludwigshafen Risk and Cardiovascular Health study, the genotyping was funded by the Seventh Framework Program AtheroRemo (grant agreement number 201668) of the European Union and the analyses were supported by the Seventh Framework Program RiskyCAD (grant agreement number 305739). The TwinsUK study was funded by the Wellcome Trust, European Community’s Seventh Framework Programme (FP7/2007-2013). The study also receives support from the National Institute for Health Research–funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ National Health Service Foundation Trust in partnership with King’s College London. Single nucleotide polymorphism genotyping was performed by The Wellcome Trust Sanger Institute and National Eye Institute via NIH/Center for Inherited Disease Research. The Cardiovascular Health Study (CHS) research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268200960009C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and NHLBI grants U01HL080295, R01HL087652, R01HL085251, R01HL105756, R01HL103612, and R01HL120393 with additional contribution from the National Institute of Neurological Disorders and Stroke. Additional support was provided through R01AG023629 from the NIA. A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. Infrastructure for the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium is supported in part by the NHLBI grant R01HL105756.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
A complete list of participants in the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) Research Group is presented in the Supplemental Material published online for the article in N Engl J Med 372:1722–1733, 2015.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016010069/-/DCSupplemental.
References
- 1.Brown EM: Four-parameter model of the sigmoidal relationship between parathyroid hormone release and extracellular calcium concentration in normal and abnormal parathyroid tissue. J Clin Endocrinol Metab 56: 572–581, 1983 [DOI] [PubMed] [Google Scholar]
- 2.Kumar R: Vitamin D and calcium transport. Kidney Int 40: 1177–1189, 1991 [DOI] [PubMed] [Google Scholar]
- 3.Portale AA, Halloran BP, Morris RC Jr: Physiologic regulation of the serum concentration of 1,25-dihydroxyvitamin D by phosphorus in normal men. J Clin Invest 83: 1494–1499, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Divieti P, John MR, Jüppner H, Bringhurst FR: Human PTH-(7-84) inhibits bone resorption in vitro via actions independent of the type 1 PTH/PTHrP receptor. Endocrinology 143: 171–176, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Hruska KA, Teitelbaum SL: Renal osteodystrophy. N Engl J Med 333: 166–174, 1995 [DOI] [PubMed] [Google Scholar]
- 6.Sherrard DJ, Hercz G, Pei Y, Maloney NA, Greenwood C, Manuel A, Saiphoo C, Fenton SS, Segre GV: The spectrum of bone disease in end-stage renal failure--an evolving disorder. Kidney Int 43: 436–442, 1993 [DOI] [PubMed] [Google Scholar]
- 7.Sugimoto T, Tanigawa T, Onishi K, Fujimoto N, Matsuda A, Nakamori S, Matsuoka K, Nakamura T, Koji T, Ito M: Serum intact parathyroid hormone levels predict hospitalisation for heart failure. Heart 95: 395–398, 2009 [DOI] [PubMed] [Google Scholar]
- 8.Hagström E, Hellman P, Larsson TE, Ingelsson E, Berglund L, Sundström J, Melhus H, Held C, Lind L, Michaëlsson K, Arnlöv J: Plasma parathyroid hormone and the risk of cardiovascular mortality in the community. Circulation 119: 2765–2771, 2009 [DOI] [PubMed] [Google Scholar]
- 9.Näppi S, Saha H, Virtanen V, Limnell V, Sand J, Salmi J, Pasternack A: Left ventricular structure and function in primary hyperparathyroidism before and after parathyroidectomy. Cardiology 93: 229–233, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Walker MD, Fleischer J, Rundek T, McMahon DJ, Homma S, Sacco R, Silverberg SJ: Carotid vascular abnormalities in primary hyperparathyroidism. J Clin Endocrinol Metab 94: 3849–3856, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverberg SJ, Clarke BL, Peacock M, Bandeira F, Boutroy S, Cusano NE, Dempster D, Lewiecki EM, Liu JM, Minisola S, Rejnmark L, Silva BC, Walker MD, Bilezikian JP: Current issues in the presentation of asymptomatic primary hyperparathyroidism: Proceedings of the Fourth International Workshop. J Clin Endocrinol Metab 99: 3580–3594, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown EM: Extracellular Ca2+ sensing, regulation of parathyroid cell function, and role of Ca2+ and other ions as extracellular (first) messengers. Physiol Rev 71: 371–411, 1991 [DOI] [PubMed] [Google Scholar]
- 13.Nemeth EF, Scarpa A: Cytosolic Ca2+ and the regulation of secretion in parathyroid cells. FEBS Lett 203: 15–19, 1986 [DOI] [PubMed] [Google Scholar]
- 14.Brent GA, LeBoff MS, Seely EW, Conlin PR, Brown EM: Relationship between the concentration and rate of change of calcium and serum intact parathyroid hormone levels in normal humans. J Clin Endocrinol Metab 67: 944–950, 1988 [DOI] [PubMed] [Google Scholar]
- 15.Delmez JA, Tindira C, Grooms P, Dusso A, Windus DW, Slatopolsky E: Parathyroid hormone suppression by intravenous 1,25-dihydroxyvitamin D. A role for increased sensitivity to calcium. J Clin Invest 83: 1349–1355, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slatopolsky E, Weerts C, Thielan J, Horst R, Harter H, Martin KJ: Marked suppression of secondary hyperparathyroidism by intravenous administration of 1,25-dihydroxy-cholecalciferol in uremic patients. J Clin Invest 74: 2136–2143, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rejnmark L, Vestergaard P, Heickendorff L, Mosekilde L: Determinants of plasma PTH and their implication for defining a reference interval. Clin Endocrinol (Oxf) 74: 37–43, 2011 [DOI] [PubMed] [Google Scholar]
- 18.Streeten EA, Rogstad AS, Flammer KM, Zarbalian K, Ryan K, Horwitz M, Holick MF, Shelton J: Reduced parathyroid hormone-stimulated 1,25-dihydroxyvitamin d production in vitamin d sufficient postmenoposual women with low bone mass and idiopathic secondary hyperparathyroidism. Endocr Pract 19: 91–99, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marx SJ, Simonds WF, Agarwal SK, Burns AL, Weinstein LS, Cochran C, Skarulis MC, Spiegel AM, Libutti SK, Alexander HR Jr, Chen CC, Chang R, Chandrasekharappa SC, Collins FS: Hyperparathyroidism in hereditary syndromes: Special expressions and special managements. J Bone Miner Res 17[Suppl 2]: N37–N43, 2002 [PubMed] [Google Scholar]
- 20.Mitnick PD, Goldfarb S, Slatopolsky E, Lemann J Jr, Gray RW, Agus ZS: Calcium and phosphate metabolism in tumoral calcinosis. Ann Intern Med 92: 482–487, 1980 [DOI] [PubMed] [Google Scholar]
- 21.Frishberg Y, Ito N, Rinat C, Yamazaki Y, Feinstein S, Urakawa I, Navon-Elkan P, Becker-Cohen R, Yamashita T, Araya K, Igarashi T, Fujita T, Fukumoto S: Hyperostosis-hyperphosphatemia syndrome: A congenital disorder of O-glycosylation associated with augmented processing of fibroblast growth factor 23. J Bone Miner Res 22: 235–242, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Scambler PJ: The 22q11 deletion syndromes. Hum Mol Genet 9: 2421–2426, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Daw SC, Taylor C, Kraman M, Call K, Mao J, Schuffenhauer S, Meitinger T, Lipson T, Goodship J, Scambler P: A common region of 10p deleted in DiGeorge and velocardiofacial syndromes. Nat Genet 13: 458–460, 1996 [DOI] [PubMed] [Google Scholar]
- 24.Lichtner P, König R, Hasegawa T, Van Esch H, Meitinger T, Schuffenhauer S: An HDR (hypoparathyroidism, deafness, renal dysplasia) syndrome locus maps distal to the DiGeorge syndrome region on 10p13/14. J Med Genet 37: 33–37, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parvari R, Hershkovitz E, Kanis A, Gorodischer R, Shalitin S, Sheffield VC, Carmi R: Homozygosity and linkage-disequilibrium mapping of the syndrome of congenital hypoparathyroidism, growth and mental retardation, and dysmorphism to a 1-cM interval on chromosome 1q42-43. Am J Hum Genet 63: 163–169, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly TE, Blanton S, Saif R, Sanjad SA, Sakati NA: Confirmation of the assignment of the Sanjad-Sakati (congenital hypoparathyroidism) syndrome (OMIM 241410) locus to chromosome lq42-43. J Med Genet 37: 63–64, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trump D, Dixon PH, Mumm S, Wooding C, Davies KE, Schlessinger D, Whyte MP, Thakker RV: Localisation of X linked recessive idiopathic hypoparathyroidism to a 1.5 Mb region on Xq26-q27. J Med Genet 35: 905–909, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter D, De Lange M, Snieder H, MacGregor AJ, Swaminathan R, Thakker RV, Spector TD: Genetic contribution to bone metabolism, calcium excretion, and vitamin D and parathyroid hormone regulation. J Bone Miner Res 16: 371–378, 2001 [DOI] [PubMed] [Google Scholar]
- 29.Thomopoulou GE, Tseleni-Balafouta S, Lazaris AC, Koutselini H, Kavantzas N, Davaris PS: Immunohistochemical detection of cell cycle regulators, Fhit protein and apoptotic cells in parathyroid lesions. Eur J Endocrinol 148: 81–87, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Szijan I, Orlow I, Dalamon V, Vergani P, Danilowicz K, Mezzadri N, Cordon-Cardo C, Bruno OD: Alterations in the retinoblastoma pathway of cell cycle control in parathyroid tumors. Oncol Rep 7: 421–425, 2000 [DOI] [PubMed] [Google Scholar]
- 31.The Atherosclerosis Risk in Communities (ARIC) Study: Design and objectives. The ARIC investigators. Am J Epidemiol 129: 687–702, 1989 [PubMed] [Google Scholar]
- 32.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, O’Leary DH, Psaty B, Rautaharju P, Tracy RP, Weiler PG; The Cardiovascular Health Study group : The Cardiovascular Health Study: Design and rationale. Ann Epidemiol 1: 263–276, 1991 [DOI] [PubMed] [Google Scholar]
- 33.The DCCT Research Group : The Diabetes Control and Complications Trial (DCCT). Design and methodologic considerations for the feasibility phase. Diabetes 35: 530–545, 1986 [PubMed] [Google Scholar]
- 34.Peacock M, Koller DL, Hui S, Johnston CC, Foroud T, Econs MJ: Peak bone mineral density at the hip is linked to chromosomes 14q and 15q. Osteoporos Int 15: 489–496, 2004 [DOI] [PubMed] [Google Scholar]
- 35.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR Jr, Kronmal R, Liu K, Nelson JC, O’Leary D, Saad MF, Shea S, Szklo M, Tracy RP: Multi-Ethnic Study of Atherosclerosis: Objectives and design. Am J Epidemiol 156: 871–881, 2002 [DOI] [PubMed] [Google Scholar]
- 36.Orwoll E, Blank JB, Barrett-Connor E, Cauley J, Cummings S, Ensrud K, Lewis C, Cawthon PM, Marcus R, Marshall LM, McGowan J, Phipps K, Sherman S, Stefanick ML, Stone K: Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study--a large observational study of the determinants of fracture in older men. Contemp Clin Trials 26: 569–585, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Penninx BW, Beekman AT, Smit JH, Zitman FG, Nolen WA, Spinhoven P, Cuijpers P, De Jong PJ, Van Marwijk HW, Assendelft WJ, Van Der Meer K, Verhaak P, Wensing M, De Graaf R, Hoogendijk WJ, Ormel J, Van Dyck R; NESDA Research Consortium : The Netherlands Study of Depression and Anxiety (NESDA): Rationale, objectives and methods. Int J Methods Psychiatr Res 17: 121–140, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.John U, Greiner B, Hensel E, Lüdemann J, Piek M, Sauer S, Adam C, Born G, Alte D, Greiser E, Haertel U, Hense HW, Haerting J, Willich S, Kessler C: Study of Health In Pomerania (SHIP): A health examination survey in an east German region: Objectives and design. Soz Praventivmed 46: 186–194, 2001 [DOI] [PubMed] [Google Scholar]
- 39.Biino G, Palmas MA, Corona C, Prodi D, Fanciulli M, Sulis R, Serra A, Fossarello M, Pirastu M: Ocular refraction: Heritability and genome-wide search for eye morphometry traits in an isolated Sardinian population. Hum Genet 116: 152–159, 2005 [DOI] [PubMed] [Google Scholar]
- 40.Völzke H: [Study of Health in Pomerania (SHIP). Concept, design and selected results]. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 55: 790–794, 2012 [DOI] [PubMed] [Google Scholar]
- 41.Jones G, Prosser DE, Kaufmann M: 25-Hydroxyvitamin D-24-hydroxylase (CYP24A1): Its important role in the degradation of vitamin D. Arch Biochem Biophys 523: 9–18, 2012 [DOI] [PubMed] [Google Scholar]
- 42.Martin-McCaffrey L, Willard FS, Pajak A, Dagnino L, Siderovski DP, D’Souza SJ: RGS14 is a microtubule-associated protein. Cell Cycle 4: 953–960, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Tenenhouse HS: Regulation of phosphorus homeostasis by the type iia na/phosphate cotransporter. Annu Rev Nutr 25: 197–214, 2005 [DOI] [PubMed] [Google Scholar]
- 44.Gong Y, Renigunta V, Himmerkus N, Zhang J, Renigunta A, Bleich M, Hou J: Claudin-14 regulates renal Ca⁺⁺ transport in response to CaSR signalling via a novel microRNA pathway. EMBO J 31: 1999–2012, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biegel JA, Tan L, Zhang F, Wainwright L, Russo P, Rorke LB: Alterations of the hSNF5/INI1 gene in central nervous system atypical teratoid/rhabdoid tumors and renal and extrarenal rhabdoid tumors. Clin Cancer Res 8: 3461–3467, 2002 [PubMed] [Google Scholar]
- 46.Brown EM, MacLeod RJ: Extracellular calcium sensing and extracellular calcium signaling. Physiol Rev 81: 239–297, 2001 [DOI] [PubMed] [Google Scholar]
- 47.Pearce SH, Williamson C, Kifor O, Bai M, Coulthard MG, Davies M, Lewis-Barned N, McCredie D, Powell H, Kendall-Taylor P, Brown EM, Thakker RV: A familial syndrome of hypocalcemia with hypercalciuria due to mutations in the calcium-sensing receptor. N Engl J Med 335: 1115–1122, 1996 [DOI] [PubMed] [Google Scholar]
- 48.Ramos EM, Hoffman D, Junkins HA, Maglott D, Phan L, Sherry ST, Feolo M, Hindorff LA: Phenotype-Genotype Integrator (PheGenI): Synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur J Hum Genet 22: 144–147, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.GTEx Consortium : The Genotype-tissue expression (GTEx) project. Nat Genet 45: 580–585, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.GTEx Consortium : Human genomics. The Genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348: 648–660, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, Koller DL, Li G, Liu CT, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao SM, Yerges-Armstrong LM, Zheng HF, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, Garcia-Giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim GS, Kooperberg C, Koromila T, Kruk M, Laaksonen M, Lacroix AZ, Lee SH, Leung PC, Lewis JR, Masi L, Mencej-Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi ML, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, González-Macías J, Kähönen M, Karlsson M, Khusnutdinova E, Koh JM, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren Ö, Lorenc RS, Marc J, Mellström D, Obermayer-Pietsch B, Olmos JM, Pettersson-Kymmer U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Slagboom PE, Tang NL, Urreizti R, Van Hul W, Viikari J, Zarrabeitia MT, Aulchenko YS, Castano-Betancourt M, Grundberg E, Herrera L, Ingvarsson T, Johannsdottir H, Kwan T, Li R, Luben R, Medina-Gómez C, Palsson ST, Reppe S, Rotter JI, Sigurdsson G, van Meurs JB, Verlaan D, Williams FM, Wood AR, Zhou Y, Gautvik KM, Pastinen T, Raychaudhuri S, Cauley JA, Chasman DI, Clark GR, Cummings SR, Danoy P, Dennison EM, Eastell R, Eisman JA, Gudnason V, Hofman A, Jackson RD, Jones G, Jukema JW, Khaw KT, Lehtimäki T, Liu Y, Lorentzon M, McCloskey E, Mitchell BD, Nandakumar K, Nicholson GC, Oostra BA, Peacock M, Pols HA, Prince RL, Raitakari O, Reid IR, Robbins J, Sambrook PN, Sham PC, Shuldiner AR, Tylavsky FA, van Duijn CM, Wareham NJ, Cupples LA, Econs MJ, Evans DM, Harris TB, Kung AW, Psaty BM, Reeve J, Spector TD, Streeten EA, Zillikens MC, Thorsteinsdottir U, Ohlsson C, Karasik D, Richards JB, Brown MA, Stefansson K, Uitterlinden AG, Ralston SH, Ioannidis JP, Kiel DP, Rivadeneira F: Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 44: 491–501, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molin A, Baudoin R, Kaufmann M, Souberbielle JC, Ryckewaert A, Vantyghem MC, Eckart P, Bacchetta J, Deschenes G, Kesler-Roussey G, Coudray N, Richard N, Wraich M, Bonafiglia Q, Tiulpakov A, Jones G, Kottler ML: CYP24A1 mutations in a cohort of hypercalcemic patients: Evidence for a recessive trait. J Clin Endocrinol Metab 100: E1343–E1352, 2015 [DOI] [PubMed] [Google Scholar]
- 53.Schlingmann KP, Kaufmann M, Weber S, Irwin A, Goos C, John U, Misselwitz J, Klaus G, Kuwertz-Bröking E, Fehrenbach H, Wingen AM, Güran T, Hoenderop JG, Bindels RJ, Prosser DE, Jones G, Konrad M: Mutations in CYP24A1 and idiopathic infantile hypercalcemia. N Engl J Med 365: 410–421, 2011 [DOI] [PubMed] [Google Scholar]
- 54.Streeten EA, Zarbalian K, Damcott CM: CYP24A1 mutations in idiopathic infantile hypercalcemia. N Engl J Med 365: 1741–1742 2011. [DOI] [PubMed] [Google Scholar]
- 55.Nesterova G, Malicdan MC, Yasuda K, Sakaki T, Vilboux T, Ciccone C, Horst R, Huang Y, Golas G, Introne W, Huizing M, Adams D, Boerkoel CF, Collins MT, Gahl WA: 1,25-(OH)2D-24 hydroxylase (CYP24A1) deficiency as a cause of nephrolithiasis. Clin J Am Soc Nephrol 8: 649–657, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dauber A, Nguyen TT, Sochett E, Cole DE, Horst R, Abrams SA, Carpenter TO, Hirschhorn JN: Genetic defect in CYP24A1, the vitamin D 24-hydroxylase gene, in a patient with severe infantile hypercalcemia. J Clin Endocrinol Metab 97: E268–E274, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tebben PJ, Milliner DS, Horst RL, Harris PC, Singh RJ, Wu Y, Foreman JW, Chelminski PR, Kumar R: Hypercalcemia, hypercalciuria, and elevated calcitriol concentrations with autosomal dominant transmission due to CYP24A1 mutations: Effects of ketoconazole therapy. J Clin Endocrinol Metab 97: E423–E427, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacobs TP, Kaufman M, Jones G, Kumar R, Schlingmann KP, Shapses S, Bilezikian JP: A lifetime of hypercalcemia and hypercalciuria, finally explained. J Clin Endocrinol Metab 99: 708–712, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, Peltonen L, Cooper JD, O’Reilly PF, Houston DK, Glazer NL, Vandenput L, Peacock M, Shi J, Rivadeneira F, McCarthy MI, Anneli P, de Boer IH, Mangino M, Kato B, Smyth DJ, Booth SL, Jacques PF, Burke GL, Goodarzi M, Cheung CL, Wolf M, Rice K, Goltzman D, Hidiroglou N, Ladouceur M, Wareham NJ, Hocking LJ, Hart D, Arden NK, Cooper C, Malik S, Fraser WD, Hartikainen AL, Zhai G, Macdonald HM, Forouhi NG, Loos RJ, Reid DM, Hakim A, Dennison E, Liu Y, Power C, Stevens HE, Jaana L, Vasan RS, Soranzo N, Bojunga J, Psaty BM, Lorentzon M, Foroud T, Harris TB, Hofman A, Jansson JO, Cauley JA, Uitterlinden AG, Gibson Q, Järvelin MR, Karasik D, Siscovick DS, Econs MJ, Kritchevsky SB, Florez JC, Todd JA, Dupuis J, Hyppönen E, Spector TD: Common genetic determinants of vitamin D insufficiency: A genome-wide association study. Lancet 376: 180–188, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Seaghdha CM, Wu H, Yang Q, Kapur K, Guessous I, Zuber AM, Köttgen A, Stoudmann C, Teumer A, Kutalik Z, Mangino M, Dehghan A, Zhang W, Eiriksdottir G, Li G, Tanaka T, Portas L, Lopez LM, Hayward C, Lohman K, Matsuda K, Padmanabhan S, Firsov D, Sorice R, Ulivi S, Brockhaus AC, Kleber ME, Mahajan A, Ernst FD, Gudnason V, Launer LJ, Mace A, Boerwinckle E, Arking DE, Tanikawa C, Nakamura Y, Brown MJ, Gaspoz JM, Theler JM, Siscovick DS, Psaty BM, Bergmann S, Vollenweider P, Vitart V, Wright AF, Zemunik T, Boban M, Kolcic I, Navarro P, Brown EM, Estrada K, Ding J, Harris TB, Bandinelli S, Hernandez D, Singleton AB, Girotto G, Ruggiero D, d’Adamo AP, Robino A, Meitinger T, Meisinger C, Davies G, Starr JM, Chambers JC, Boehm BO, Winkelmann BR, Huang J, Murgia F, Wild SH, Campbell H, Morris AP, Franco OH, Hofman A, Uitterlinden AG, Rivadeneira F, Völker U, Hannemann A, Biffar R, Hoffmann W, Shin SY, Lescuyer P, Henry H, Schurmann C, Munroe PB, Gasparini P, Pirastu N, Ciullo M, Gieger C, März W, Lind L, Spector TD, Smith AV, Rudan I, Wilson JF, Polasek O, Deary IJ, Pirastu M, Ferrucci L, Liu Y, Kestenbaum B, Kooner JS, Witteman JC, Nauck M, Kao WH, Wallaschofski H, Bonny O, Fox CS, Bochud M, Consortium S; SUNLIGHT Consortium; GEFOS Consortium : Meta-analysis of genome-wide association studies identifies six new Loci for serum calcium concentrations. PLoS Genet 9: e1003796, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shachaf C, Skorecki KL, Tzukerman M: Role of AP2 consensus sites in regulation of rat Npt2 (sodium-phosphate cotransporter) promoter. Am J Physiol Renal Physiol 278: F406–F416, 2000 [DOI] [PubMed] [Google Scholar]
- 62.Kestenbaum B, Glazer NL, Köttgen A, Felix JF, Hwang SJ, Liu Y, Lohman K, Kritchevsky SB, Hausman DB, Petersen AK, Gieger C, Ried JS, Meitinger T, Strom TM, Wichmann HE, Campbell H, Hayward C, Rudan I, de Boer IH, Psaty BM, Rice KM, Chen YD, Li M, Arking DE, Boerwinkle E, Coresh J, Yang Q, Levy D, van Rooij FJ, Dehghan A, Rivadeneira F, Uitterlinden AG, Hofman A, van Duijn CM, Shlipak MG, Kao WH, Witteman JC, Siscovick DS, Fox CS: Common genetic variants associate with serum phosphorus concentration. J Am Soc Nephrol 21: 1223–1232, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hendy GN, D’Souza-Li L, Yang B, Canaff L, Cole DE: Mutations of the calcium-sensing receptor (CASR) in familial hypocalciuric hypercalcemia, neonatal severe hyperparathyroidism, and autosomal dominant hypocalcemia. Hum Mutat 16: 281–296, 2000 [DOI] [PubMed] [Google Scholar]
- 64.Chen RA, Goodman WG: Role of the calcium-sensing receptor in parathyroid gland physiology. Am J Physiol Renal Physiol 286: F1005–F1011, 2004 [DOI] [PubMed] [Google Scholar]
- 65.Thorleifsson G, Holm H, Edvardsson V, Walters GB, Styrkarsdottir U, Gudbjartsson DF, Sulem P, Halldorsson BV, de Vegt F, d’Ancona FC, den Heijer M, Franzson L, Christiansen C, Alexandersen P, Rafnar T, Kristjansson K, Sigurdsson G, Kiemeney LA, Bodvarsson M, Indridason OS, Palsson R, Kong A, Thorsteinsdottir U, Stefansson K: Sequence variants in the CLDN14 gene associate with kidney stones and bone mineral density. Nat Genet 41: 926–930, 2009 [DOI] [PubMed] [Google Scholar]
- 66.Zajickova K, Vrbikova J, Canaff L, Pawelek PD, Goltzman D, Hendy GN: Identification and functional characterization of a novel mutation in the calcium-sensing receptor gene in familial hypocalciuric hypercalcemia: Modulation of clinical severity by vitamin D status. J Clin Endocrinol Metab 92: 2616–2623, 2007 [DOI] [PubMed] [Google Scholar]
- 67.Spector TD, Williams FM: The UK Adult Twin Registry (TwinsUK). Twin Res Hum Genet 9: 899–906, 2006 [DOI] [PubMed] [Google Scholar]
- 68.Lorentzon M, Swanson C, Andersson N, Mellström D, Ohlsson C: Free testosterone is a positive, whereas free estradiol is a negative, predictor of cortical bone size in young Swedish men: The GOOD study. J Bone Miner Res 20: 1334–1341, 2005 [DOI] [PubMed] [Google Scholar]
- 69.Winkelmann BR, März W, Boehm BO, Zotz R, Hager J, Hellstern P, Senges J; LURIC Study Group (LUdwigshafen RIsk and Cardiovascular Health) : Rationale and design of the LURIC study--a resource for functional genomics, pharmacogenomics and long-term prognosis of cardiovascular disease. Pharmacogenomics 2[Suppl 1]: S1–S73, 2001 [DOI] [PubMed] [Google Scholar]
- 70.Brown LB, Streeten EA, Shuldiner AR, Almasy LA, Peyser PA, Mitchell BD: Assessment of sex-specific genetic and environmental effects on bone mineral density. Genet Epidemiol 27: 153–161, 2004 [DOI] [PubMed] [Google Scholar]
- 71.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) : A new equation to estimate glomerular filtration rate. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Devlin B, Roeder K: Genomic control for association studies. Biometrics 55: 997–1004, 1999 [DOI] [PubMed] [Google Scholar]
- 73.Weir BS, Cockerham CC: Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370, 1984 [DOI] [PubMed] [Google Scholar]
- 74.Yang J, Lee SH, Goddard ME, Visscher PM: GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet 88: 76–82, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gauderman WJ, Morrison JM: Quanto. 1.2.4 ed., 2009 Available at: http://biostats.usc.edu/Quanto.html. Accessed May 10, 2016
- 76.Willer CJ, Li Y, Abecasis GR: METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Magi R, Lindgren CM, Morris AP: Meta-analysis of sex-specific genome-wide association studies. Genet Epidemiol 34: 846–853, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI: SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24: 2938–2939, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shim H, Chasman DI, Smith JD, Mora S, Ridker PM, Nickerson DA, Krauss RM, Stephens M: A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS One 10: e0120758, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.