Abstract
Steroid–resistant nephrotic syndrome (SRNS), a heterogeneous disorder of the renal glomerular filtration barrier, results in impairment of glomerular permselectivity. Inheritance of genetic SRNS may be autosomal dominant or recessive, with a subset of autosomal recessive SRNS presenting as congenital nephrotic syndrome (CNS). Mutations in 53 genes are associated with human SRNS, but these mutations explain ≤30% of patients with hereditary cases and only 20% of patients with sporadic cases. The proteins encoded by these genes are expressed in podocytes, and malfunction of these proteins leads to a universal end point of podocyte injury, glomerular filtration barrier disruption, and SRNS. Here, we identified novel disease–causing mutations in membrane–associated guanylate kinase, WW, and PDZ domain–containing 2 (MAGI2) through whole-exome sequencing of a deeply phenotyped cohort of patients with congenital, childhood–onset SRNS. Although MAGI2 has been shown to interact with nephrin and regulate podocyte cytoskeleton and slit diaphragm dynamics, MAGI2 mutations have not been described in human SRNS. We detected two unique frameshift mutations and one duplication in three patients (two families); two siblings shared the same homozygous frameshift mutation, whereas one individual with sporadic SRNS exhibited compound heterozygosity. Two mutations were predicted to introduce premature stop codons, and one was predicted to result in read through of the normal translational termination codon. Immunohistochemistry in kidney sections from these patients revealed that mutations resulted in lack of or diminished podocyte MAGI2 expression. Our data support the finding that mutations in the MAGI2 gene are causal for congenital SRNS.
Keywords: podocyte, genetic renal disease, nephrin, familial nephropathy, proteinuria, nephrotic syndrome
Steroid–resistant nephrotic syndrome (SRNS), a disorder of glomerular filtration, results in severe proteinuria, hypoalbuminaemia, and edema. Currently, 53 genes are implicated (Supplemental Table 1), but these account for only 20%–30% of patients with hereditary cases and only 10%–20% of patients with sporadic cases, supporting significant genetic heterogeneity.1–8 To date, all known SRNS–associated genes encode proteins expressed in podocytes (or associated basement membrane), polarized cells connected by highly specialized junctions called slit diaphragms. Correct podocyte morphology is essential for maintaining glomerular filtration barrier (GFB) integrity, and the podocyte actin cytoskeleton is tightly regulated by cell surface receptors, including the multiprotein complex at the slit diaphragm. Known SRNS gene mutations disrupt key cellular functions, resulting in podocyte injury and disruption of glomerular permselectivity. Typically, patients with a mutation in an SRNS–associated gene are less likely to respond to immunosuppressive treatment but have a reduced risk of disease recurrence after kidney transplant.9 SRNS may present at birth (congenital nephrotic syndrome [CNS]), usually with an early and severe phenotype, with about 80%10 of patients with cases ascribed to only six causal genes: NPHS1, NPHS2, LAMB2, WT1, PLCE1, and LMX1B,11 all key players in podocyte biology; the genetic cause of the remainder is unknown. The majority of patients with SRNS in childhood have an autosomal recessive mode of inheritance.
Results
Screening the Known Nephrotic Genes
We expanded the study originally described in the work by McCarthy et al.1 and performed whole-exome sequencing on a deeply phenotyped cohort of 187 patients with childhood SRNS (onset <18 years old; 11.8% familial and 7% consanguineous; 48.7% girls and 51.3% boys; 69.5% white and 30.5% South Asian, mixed race, African, and East Asian) collected via a United Kingdom–wide registry.
The cohort was first screened for the presence of disease-causing mutations in the 53 published genes known associate with SRNS (Supplemental Table 1, mapping statistics are presented in Supplemental Table 2). Our findings correlated with previous published studies,2,12 in that mutations in known SRNS genes were only detected in approximately 25% of patients. Assuming that mutations in the exome were present in a proportion of the remaining 75% of patients, variants detected in the whole exome were filtered to identify potential mutations in genes not previously directly associated with SRNS.
Membrane–Associated Guanylate Kinase, WW, and PDZ Domain–Containing 2 Mutations Identified by Whole-Exome Sequencing
After filtering and additional analysis of potentially pathogenic mutations in genes not previously directly associated with SRNS, we identified three novel, likely disease–causing frameshift mutations (Figure 1) in membrane–associated guanylate kinase, WW, and PDZ domain–containing 2 (MAGI2; MIM: 606382) in one patient with a sporadic case (180) (Supplemental Figure 1) and two patients with familial cases (175 and 175S) (Supplemental Figure 2) presenting with nonsyndromic congenital SRNS. Because the parents of patients 175 and 175S were consanguineous, autosomal recessive inheritance was considered the most likely mechanism of inheritance (Supplemental Figure 3).
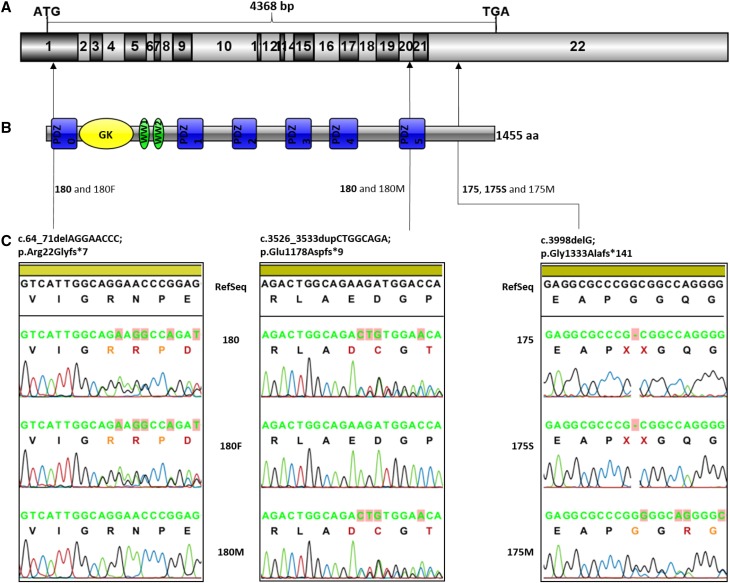
Figure 1.
MAGI2 mutations causing CNS. (A) The exon structure of MAGI2 cDNA (NM_012301.3); 22 coding exons with start and stop codons are indicated. (B) Domain structure of the MAGI2 protein. Six PDZ domains (PDZ0–PDZ5) are shown in blue, one guanylate kinase (GK) domain is shown in yellow, and two WW domains are shown in green. (C) Frameshift variants found in three patients with CNS. Individual 180 is a compound heterozygote: the variant in exon 1 was inherited from the father (180F), and the variant in exon 20 was inherited from the mother (180M). Individual 175 and her sister, 175S, are homozygous for a single-nucleotide deletion in exon 22. The mother of the siblings is heterozygous for the variant.
Whole-exome sequencing was performed on patients 175 and 180. Subsequent segregation analysis confirmed that two siblings (175 and 175S) shared the same homozygous frameshift deletion MAGI2:NM_012301:exon22:c.3998delG:p.(Gly1333Alafs*141). The patient with the sporadic case (180) exhibited compound heterozygosity: a deletion (paternal) resulting in a premature stop codon MAGI2:NM_012301:exon1:c.64_71delAGGAACCC:p.(Arg22Glyfs*7) together with a duplication (maternal) MAGI2:NM_012301:exon20:c.3526_3533dupCTGGCAGA:p.(Glu1178Aspfs*9). All three variants were absent in the ExAC database and had high CADD scores (34 [175 and 175S] and 35 [180]), supporting pathogenicity of these mutations. Translation of the mutated protein (ExPASy; http://web.expasy.org/translate) indicated that the two frameshift mutations located in exons 1 and 20 resulted in stop codons and premature termination of protein translation, and they were located in PDZ domains. The exon 22 frameshift deletion resulted in predicted read through of the normal termination (stop) codon, resulting in likely continued translation of the mRNA farther downstream into the 3′-untranslated region.
Patients 175, 175S, and 180 did not show any other significant rare or novel variants in other genes expressed in podocytes during exome screening, supporting MAGI2 as the most likely causative gene. Furthermore, there were no rare frameshift mutations at the same site as any of the MAGI2 mutations within 100 bp in Ensembl or other public databases, although one in-house control carried a heterozygous frameshift insertion in exon 20:c.3512_3513insTGTA:p.(Leu1171Phefs*27). Other MAGI2 variants were not detected in these or other control samples.
The MAGI2 homozygous frameshift deletion p.(Gly1333Alafs*141) was verified by Sanger sequencing, and this was the only homozygous variant present in both 175 and 175S. It was also present in the mother (175M) as a heterozygote but could not be verified in the father, because DNA was not available.
Of the eight rare/novel variants found in 175, two (RAD51D and NUP155) were absent in 175S, and two (GABRD and POLR2M) were only present as heterozygous variants. Furthermore, one variant (AKR1C1) was present as homozygous in the ExAC database, and one (IGLL5) was predicted to be tolerated by in silico tools, excluding both as likely candidates. Both heterozygous variants in SKOR1 found in 175 were inherited from the mother and thus, present on the same allele. Only one rare heterozygous potentially known SRNS gene variant was shared by the siblings: COL4A3 c.4421T>C:p.(Leu1474Pro); it was detected before enrollment into the study by the Bristol Genetics Laboratory (SRNS 37 gene panel; www.nbt.nhs.uk/severn-pathology/pathology-services/bristol-genetics-laboratory-bgl). This variant was seen over 300 times as a heterozygote in the ExAC database and is, therefore, of unlikely significance.
Because DNA from both parents was available for 180, we were able to verify that the novel MAGI2 mutations were in trans; p.(Arg22Glyfs*7) was inherited on a paternal allele, and p.(Glu1178Aspfs*9) was inherited on a maternal allele. The frameshift variations in MAGI2 were not present in the ExAC database. Six other nonsense variants in MAGI2 were present; however, each was seen only once and as a heterozygote. Similarly, one splice acceptor and two splice donor variants are also present in the ExAC database but again, only present as heterozygotes. If we include non-PASS variants, an additional nonsense and three frameshift variants are also present; however, again, all were seen only as heterozygotes.
Five other genes were also left after the filtering steps; however, variants in three genes were maternally inherited in cis, and insertions in MICALCL and ZIC5 were considered of unlikely significance due to the presence of similar insertions around this region in the ExAC database. MAGI2 remained the sole candidate gene for causing nephrotic syndrome in our patients. (Data are presented in Supplemental Table 3.)
There was no associated extrarenal phenotype in patient 175, whereas patient 175S had some minor cardiac (possibly unrelated) abnormalities. Patient 180 had associated polydactyly and a previous pyloric stenosis, although the patient did not show any other features of any characterized syndrome. The inheritance pattern in all three patients was compatible with autosomal recessive disease. All had presented with significant proteinuria and hypoalbuminaemia within the first 4 months of life compatible with a CNS. Patient 175 was diagnosed after renal biopsy with Finnish–type nephrotic syndrome, which is usually caused by NPHS1 mutations; however, the renal histology in early life may be relatively nonspecific in appearance, and no NPHS1 mutations were detected on screening the entire gene. Patient 175 had rapid disease progression and required a kidney transplant at age 3.5 years old; in contrast, the sibling, 175S, has followed a more benign course. Similarly, patient 180 has had persistent proteinuria for 9 years with only mild renal impairment to date. Phenotypic variability between different family members is not unusual in SRNS and has been previously described. Interestingly, the most severely affected patient (175) also carried a single heterozygous variant in LAMB2, resulting in c.4274G>C:p.(Gly1425Ala), the significance of which is unknown, but it was not present as a homozygote on the ExAC database and had an MAF of 0.000075. Disease modification due to bigenic heterozygosity or triallelic hits may occur but is unusual in SRNS.13 Phenotypic details are presented in Table 1.
Table 1.
Clinical features of the affected individuals with MAGI2 mutations
| Patient No. | Sex | Ethnicity | Familial/Sporadic | Age at Onset, mo | Consanguinity | Resistance to Steroids | First Native Biopsy | Second Native Biopsy | CKD Stage | Time to ESRF | Tx? | Disease Recurrence? | Extrarenal Phenotype | Treatment Used | Most Recent Serum Creatinine, μmol/L | Most Recent Albumin-to-Creatinine Ratio, mg/mmol | Length of Follow-Up, yr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 180 | M | W | Sporadic | 3 | No | Presumed | MCD (28 mo) | N/A | 2 | N/A | No | N/A | Pyloric stenosis, single–digit postaxial polydactyly, thrombocytosis | Enalapril, cyclosporin, tacrolimus, prednisolone, mycophenolate mofetil | 32 | 83 (Serum albumin 17 g/L) | 9.08 |
| 175 | F | W | Familial | 4 | Yes | Presumed | Late Finnish–type CNS (22 mo) | End stage of Finnish-type CNS (26 mo) | Tx | 1 yr, 9 mo | Yes | No | No | None | 73 | 3.2 | 9.58 |
| 175S | F | W | Familial | 1 | Yes | Presumed | N/A | N/A | 1 | N/A | N/A | N/A | Mild peripheral branch pulmonary stenosis, patent foramen ovale | Captopril | 24 | 2160 (Serum albumin 17 g/L) | 2.33 |
ESRF, end stage renal failure; Tx, transplanted; M, boy; W, white; MCD, minimal change disease; N/A, not applicable; F, girl.
Immunohistochemistry
Kidney biopsy sections (from individual 180 and an individual with minimal change disease), a nephrectomy section from individual 175, and an unsuitable for transplant normal human kidney section used for immunohistochemistry were formalin fixed and paraffin embedded (Figure 2).
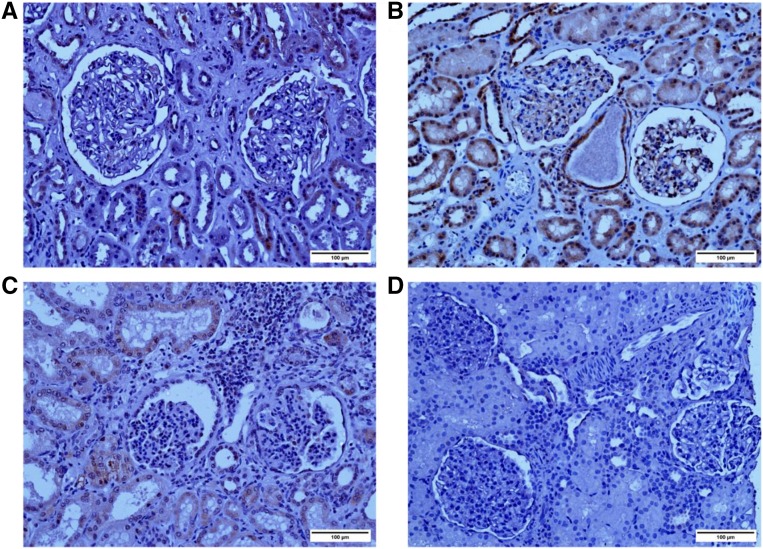
Figure 2.
MAGI2 expression is altered in patients with mutations. Immunohistochemical staining with human anti–MAGI2 antibody (Sigma-Aldrich) is shown. (A) A kidney that was not suitable for transplant was used as a control. (B) Biopsy specimen obtained from an individual with minimal change disease; MAGI2 staining is seen in the glomeruli at the periphery of capillary loops, consistent with podocyte localization. Staining in tubules is also seen. (C) Nephrectomy specimen obtained from patient 175 (homozygous mutation in MAGI2) shows weak but positive glomerular MAGI2 staining. (D) Biopsy specimen obtained from patient 180 (compound heterozygous mutation in MAGI2) shows complete lack of MAGI2. Original magnification, ×20. Scale bar, 100 μm.
Glomeruli from patient 175 showed marked lobulation, with fibrosis or global sclerosis, interstitial fibrosis, and diffuse inflammation. From patient 180, changes were milder (consistent with milder renal impairment), with two of 59 glomeruli showing global sclerosis and the rest looking normal on light microscopy. Electron microscopy (Supplemental Figure 4) from this specimen shows a normal mesangial area, normal glomerular basement membrane, and diffuse podocyte foot process effacement, similar to the reported mouse model.14
MAGI2 staining was absent in patient 180, consistent with an early truncation resulting in nontranslation of the MAGI2 protein through nonsense-mediated decay. MAGI2 staining was, however, weakly positive in podocytes of patient 175. The terminal position of the frameshift mutation (exon 22) and predicted read through into the 3′-untranslated region support weak expression of a translated but dysfunctional protein.
We observed MAGI2 staining in the tubules in control sections, which was absent in patient 180 with this antibody (Sigma-Aldrich). We, therefore, stained sections with a different antibody (Santa Cruz), which showed different degrees of tubular staining in all sections, weak glomerular staining in the normal kidney, weak/negative staining in patient 175, and negative staining in patient 180 (Supplemental Figure 5). The best fit explanation for the difference in tubular staining with the Santa Cruz antibody is that this is due to a component of nonspecific staining with this antibody, and therefore, we cannot be conclusive about the specificity of tubular staining.
Nephrin expression seemed to be decreased and localization was altered in both patients 175 and 180 compared with control (Figure 3), similar to what was observed in MAGI2 mouse models.15,16
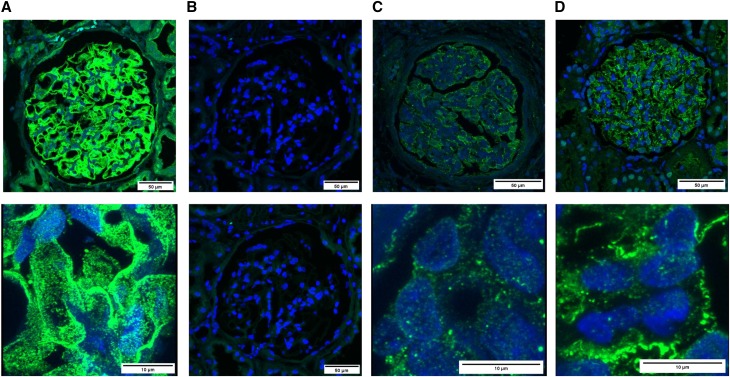
Figure 3.
Nephrin immunostaining is altered in patients with MAGI2 mutations. Nephrin immunostaining on paraffin–embedded renal tissue sections. Confocal microscopy images show nephrin staining in green and DAPI (4′,6-diamidino-2-phenylindole) nuclear staining in blue. (A) Positive control; normal human kidney. (B) Negative control (no nephrin antibody); normal human kidney. (C) Nephrectomy specimen obtained from patient 175 (homozygous mutation in MAGI2). (D) Biopsy specimen obtained from patient 180 (compound heterozygous mutation in MAGI2). Upper panels show the whole glomerulus (×40±3), and lower panels in A, C, and D show higher original magnification (×100±7). Lower panels show decreased/disrupted nephrin expression between normal and patient podocytes.
Discussion
We performed whole-exome sequencing using an Illumina platform on 187 patients with SRNS as an extension of a previous analysis of a cohort of childhood SRNS.1 Patients lacking mutations in the known 53 nephrotic genes were analyzed further, particularly those with CNS, which is generally autosomal recessive and a developmental disorder. The exome data were, therefore, analyzed to look for novel candidate genes. After filtering and additional analysis of potentially pathogenic mutations in genes expressed in podocytes but not previously directly associated with SRNS, we identified likely disease–causing frameshift mutations in MAGI2 in one patient with a sporadic case (180) and two patients with familial cases (175 and 175S).
Although a recently ascribed podocyte gene, MAGI2 mutations have not previously been directly associated with human SRNS. MAGI2 together with its paralogs MAGI1 and MAGI3 belong to the membrane–associated guanylate kinase (MAGUK) family of scaffolding proteins highly expressed in neurons and normally associated with neurologic function.17–19 MAGI proteins function as molecular scaffolds, coordinating signaling complexes by linking cell surface receptors to the cytoskeleton, but differ from other members of the MAGUK family by having a guanylate kinase domain at the N terminus as well as WW and PDZ domains in reverse orientation to MAGUK.20 MAGI2 is expressed in podocyte foot processes together with IQGAP1, CASK, spectrins, and α-actinin and is known to directly bind the cytosolic tail of nephrin, an essential component of the slit diaphragm, as part of the nephrin multiprotein complex.21 It also plays a role in RhoA-dependent regulation of the actin cytoskeleton, an important process in podocytes.22 Moreover, MAGI2 has recently been described as a WT1 target gene and required for podocyte development in both zebrafish23 and mice. Mice with MAGI2 deletion display disruption of the slit diaphragm formation, podocyte foot process effacement, and severe glomerular pathology compatible with human SRNS, with early death from nephrotic syndrome and kidney failure.16 In humans, expression data from microdissected renal biopsies (Nephromine; http://www.nephromine.org/) indicated that MAGI2 was one of the top deregulated genes in glomerular disease, with 3.2- and 6.2-fold decreases in FSGS and diabetic nephropathy, respectively, further underlining its potential importance in human SRNS.15
Aside from published evidence that MAGI2 is a component of the multiprotein complex at the slit diaphragm, MAGI2-deleted mice die soon after birth from podocyte injury, severe proteinuria, and end stage renal failure, indicating a profound developmental slit diaphragm defect.17,19,20,22 Furthermore, MAGI2 is a WT1 target during podocyte development and has been previously implicated in RhoA regulation/signaling known to play a critical role in actin cytoskeletal regulation in podocytes.22–25
In conclusion, we present genetic data that support MAGI2 mutations as a cause of congenital SRNS in humans. Although additional functional studies are required to establish exactly how MAGI2 mutations lead to human CNS, we propose that MAGI2 gene mutations should be added to gene panels when investigating SRNS and considered in patients with congenital-onset SRNS, particularly where mutation in other known genes is not found.
Concise Methods
Sequencing
Patients 175 and 180 underwent exome sequencing. DNA from peripheral blood was extracted with the Gentra Puregene Blood Kit (Qiagen). DNA libraries were prepared from 3 μg dsDNA using the SureSelect Human All Exon 50 Mb Kit (Agilent). Samples were multiplexed (four samples on each lane), and 100-bp paired end sequencing was performed on the Illumina HiSeq System. Sequence data were aligned to the hg19 human reference genome using Novoalign (Novocraft Technologies SDN BHD), and variants were called with SAMtools (Sequence Alignment/Map Tools)26 and annotated via multiple passes through Annovar.27 Initial exploration of datasets was performed using the Integrative Genome Browser (https://www.broadinstitute.org/igv).
Variant Filtering
(1) Only variants with a minor allele frequency (MAF)<0.01 were considered (1000 Genomes Project; http://www.1000genomes.org/; NHLBI Exome Sequencing Project [ESP]; http://evs.gs.washington.edu/EVS/; http://exac.broadinstitute.org/; KCL in–house dataset [King’s College London; in-house data from 5000 control individuals without kidney disease]). (2) Variants seen as a homozygote in the ExAC database (http://exac.broadinstitute.org/), ESP, or 1000 Genomes Project (http://www.internationalgenome.org/) were excluded from additional analysis. (3) For the novel/not previously described missense variants, information from Alamut Visual (http://interactive-biosoftware.com/alamut-visual) and UCSC (http://genome.ucsc.edu/) was used to check whether an amino acid is conserved. For filtering, the amino acid must be conserved, and the new amino acid must not be present in another multicellular organism. (4) Analysis of potential synonymous and splice site variants was performed using Alamut Visual 2.7 (SpliceSiteFinder-like, MaxEntScan, NNSPLICE, GeneSplicer, and Human Splicing Finder) to check if there was a consistent predicted splice effect across the majority of tools. MAGI2 frameshift mutations were examined for deleteriousness using CADD (http://cadd.gs.washington.edu). (5) Variants present within 50 bp and subsequently shown to be present in cis were excluded and not considered compound heterozygotes. (6) Any variants that did not meet criteria above were discarded. (7) All mutations of interest were confirmed by Sanger sequencing in probands and parents.
To ascertain additional annotative information about potential genes of interest, we also examined genomic, proteomic, transcriptomic, genetic, and functional information on all new candidates using databases, such as GeneCards (http://www.genecards.org/), Ensembl (http://ensembl.org), and Uniprot (http://www.uniprot.org), as well as the Human Protein Atlas (http://www.proteinatlas.org/) and literature searches to help decisions regarding potential pathogenicity of findings. Mouse Genome Informatics (http://www.informatics.jax.org/) was used for murine data and establishing whether SRNS disease phenotypes might occur in mice. Additional details are provided in Supplemental Tables 3 and 4.
Immunohistochemistry
Two MAGI2 antibodies were used.
MAGI2 (HPA013650; Sigma-Aldrich) was used 1:250. Heat–induced epitope retrieval was performed using a domestic stainless steel pressure cooker; 5 minutes were counted as soon as the cooker reached full pressure. (10 mM sodium citrate tribasic, pH 6), and 10% goat serum (Sigma-Aldrich) was used for blocking.
MAGI2 (sc-25664; Santa Cruz) was used 1:50. The heat–induced antigen retrieval method was used (boiled for 5 minutes; 10 mM sodium citrate tribasic, pH 6); 3% BSA and 3% goat serum were used for blocking.
Fluorescent Immunohistochemistry Staining of Paraffin–Embedded Tissue Sections
Nephrin (AF4269; R&D) was used 1:200. Donkey anti–sheep IgG (H+L) secondary antibody and Alexa Fluor 488 conjugate (A11015; Life Technologies) were used 1:300. Proteinase K was used as antigen retrieval (20 μg/ml); 1.5% BSA (A9647; Sigma-Aldrich) plus 5% donkey serum (D9663; Sigma-Aldrich) was used for blocking.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank Lauren Flanagan, Liz Bailey, and Hannah Leyland for their help with data and sample collection.
The research was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St. Thomas’ National Health Service (NHS) Foundation Trust and King’s College London. Funding sources were Kids Kidney Research, Nephrotic Syndrome Trust, NephCure, NIHR, and Guy’s and St. Thomas’ Hospital Charity. The United Kingdom Renal Rare Disease Registry is funded by Kidney Research UK and the British Kidney Patients’ Association.
The views expressed are those of the author(s) and are not necessarily those of the NHS, the NIHR, or the Department of Health.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016040387/-/DCSupplemental.
Contributor Information
Collaborators: Agnieszka Bierzynska, Katrina Soderquest, Philip Dean, Elizabeth Colby, Ruth Rollason, Caroline Jones, Carol D. Inward, Hugh J. McCarthy, Michael A. Simpson, Graham M. Lord, Maggie Williams, Gavin I. Welsh, Ania B. Koziell, and Moin A. Saleem
References
- 1.McCarthy HJ, Bierzynska A, Wherlock M, Ognjanovic M, Kerecuk L, Hegde S, Feather S, Gilbert RD, Krischock L, Jones C, Sinha MD, Webb NJ, Christian M, Williams MM, Marks S, Koziell A, Welsh GI, Saleem MA; RADAR the UK SRNS Study Group : Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 8: 637–648, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadowski CE, Lovric S, Ashraf S, Pabst WL, Gee HY, Kohl S, Engelmann S, Vega-Warner V, Fang H, Halbritter J, Somers MJ, Tan W, Shril S, Fessi I, Lifton RP, Bockenhauer D, El-Desoky S, Kari JA, Zenker M, Kemper MJ, Mueller D, Fathy HM, Soliman NA, Hildebrandt F; SRNS Study Group : A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 26: 1279–1289, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierzynska A, Soderquest K, Koziell A: Genes and podocytes - new insights into mechanisms of podocytopathy. Front Endocrinol (Lausanne) 5: 226, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyake N, Tsukaguchi H, Koshimizu E, Shono A, Matsunaga S, Shiina M, Mimura Y, Imamura S, Hirose T, Okudela K, Nozu K, Akioka Y, Hattori M, Yoshikawa N, Kitamura A, Cheong HI, Kagami S, Yamashita M, Fujita A, Miyatake S, Tsurusaki Y, Nakashima M, Saitsu H, Ohashi K, Imamoto N, Ryo A, Ogata K, Iijima K, Matsumoto N: Biallelic mutations in nuclear pore complex subunit NUP107 cause early-childhood-onset steroid-resistant nephrotic syndrome. Am J Hum Genet 97: 555–566, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun DA, Sadowski CE, Kohl S, Lovric S, Astrinidis SA, Pabst WL, Gee HY, Ashraf S, Lawson JA, Shril S, Airik M, Tan W, Schapiro D, Rao J, Choi WI, Hermle T, Kemper MJ, Pohl M, Ozaltin F, Konrad M, Bogdanovic R, Büscher R, Helmchen U, Serdaroglu E, Lifton RP, Antonin W, Hildebrandt F: Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat Genet 48: 457–465, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gee HY, Zhang F, Ashraf S, Kohl S, Sadowski CE, Vega-Warner V, Zhou W, Lovric S, Fang H, Nettleton M, Zhu JY, Hoefele J, Weber LT, Podracka L, Boor A, Fehrenbach H, Innis JW, Washburn J, Levy S, Lifton RP, Otto EA, Han Z, Hildebrandt F: KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest 125: 2375–2384, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebarasi L, Ashraf S, Bierzynska A, Gee HY, McCarthy HJ, Lovric S, Sadowski CE, Pabst W, Vega-Warner V, Fang H, Koziell A, Simpson MA, Dursun I, Serdaroglu E, Levy S, Saleem MA, Hildebrandt F, Majumdar A: Defects of CRB2 cause steroid-resistant nephrotic syndrome. Am J Hum Genet 96: 153–161, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Mutiis C, Pasini A, La Scola C, Pugliese F, Montini G: Nephrotic-range albuminuria as the presenting symptom of Dent-2 disease. Ital J Pediatr 41: 46, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jungraithmayr TC, Hofer K, Cochat P, Chernin G, Cortina G, Fargue S, Grimm P, Knueppel T, Kowarsch A, Neuhaus T, Pagel P, Pfeiffer KP, Schäfer F, Schönermarck U, Seeman T, Toenshoff B, Weber S, Winn MP, Zschocke J, Zimmerhackl LB: Screening for NPHS2 mutations may help predict FSGS recurrence after transplantation. J Am Soc Nephrol 22: 579–585, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machuca E, Benoit G, Nevo F, Tête MJ, Gribouval O, Pawtowski A, Brandström P, Loirat C, Niaudet P, Gubler MC, Antignac C: Genotype-phenotype correlations in non-Finnish congenital nephrotic syndrome. J Am Soc Nephrol 21: 1209–1217, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sweeney E, Fryer A, Mountford R, Green A, McIntosh I: Nail patella syndrome: A review of the phenotype aided by developmental biology. J Med Genet 40: 153–162, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trautmann A, Bodria M, Ozaltin F, Gheisari A, Melk A, Azocar M, Anarat A, Caliskan S, Emma F, Gellermann J, Oh J, Baskin E, Ksiazek J, Remuzzi G, Erdogan O, Akman S, Dusek J, Davitaia T, Özkaya O, Papachristou F, Firszt-Adamczyk A, Urasinski T, Testa S, Krmar RT, Hyla-Klekot L, Pasini A, Özcakar ZB, Sallay P, Cakar N, Galanti M, Terzic J, Aoun B, Caldas Afonso A, Szymanik-Grzelak H, Lipska BS, Schnaidt S, Schaefer F; PodoNet Consortium : Spectrum of steroid-resistant and congenital nephrotic syndrome in children: The PodoNet registry cohort. Clin J Am Soc Nephrol 10: 592–600, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koziell A, Grech V, Hussain S, Lee G, Lenkkeri U, Tryggvason K, Scambler P: Genotype/phenotype correlations of NPHS1 and NPHS2 mutations in nephrotic syndrome advocate a functional inter-relationship in glomerular filtration. Hum Mol Genet 11: 379–388, 2002 [DOI] [PubMed] [Google Scholar]
- 14.Balbas MD, Burgess MR, Murali R, Wongvipat J, Skaggs BJ, Mundel P, Weins A, Sawyers CL: MAGI-2 scaffold protein is critical for kidney barrier function. Proc Natl Acad Sci USA 111: 14876–14881, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefebvre J, Clarkson M, Massa F, Bradford ST, Charlet A, Buske F, Lacas-Gervais S, Schulz H, Gimpel C, Hata Y, Schaefer F, Schedl A: Alternatively spliced isoforms of WT1 control podocyte-specific gene expression. Kidney Int 88: 321–331, 2015 [DOI] [PubMed] [Google Scholar]
- 16.Ihara K, Asanuma K, Fukuda T, Ohwada S, Yoshida M, Nishimori K: MAGI-2 is critical for the formation and maintenance of the glomerular filtration barrier in mouse kidney. Am J Pathol 184: 2699–2708, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Hirao K, Hata Y, Ide N, Takeuchi M, Irie M, Yao I, Deguchi M, Toyoda A, Sudhof TC, Takai Y: A novel multiple PDZ domain-containing molecule interacting with N-methyl-D-aspartate receptors and neuronal cell adhesion proteins. J Biol Chem 273: 21105–21110, 1998 [DOI] [PubMed] [Google Scholar]
- 18.Shoji H, Tsuchida K, Kishi H, Yamakawa N, Matsuzaki T, Liu Z, Nakamura T, Sugino H: Identification and characterization of a PDZ protein that interacts with activin type II receptors. J Biol Chem 275: 5485–5492, 2000 [DOI] [PubMed] [Google Scholar]
- 19.Wood JD, Yuan J, Margolis RL, Colomer V, Duan K, Kushi J, Kaminsky Z, Kleiderlein JJ, Sharp AH, Ross CA: Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol Cell Neurosci 11: 149–160, 1998 [DOI] [PubMed] [Google Scholar]
- 20.Nagashima S, Kodaka M, Iwasa H, Hata Y: MAGI2/S-SCAM outside brain. J Biochem 157: 177–184, 2015 [DOI] [PubMed] [Google Scholar]
- 21.Lehtonen S, Ryan JJ, Kudlicka K, Iino N, Zhou H, Farquhar MG: Cell junction-associated proteins IQGAP1, MAGI-2, CASK, spectrins, and alpha-actinin are components of the nephrin multiprotein complex. Proc Natl Acad Sci USA 102: 9814–9819, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iida J, Ishizaki H, Okamoto-Tanaka M, Kawata A, Sumita K, Ohgake S, Sato Y, Yorifuji H, Nukina N, Ohashi K, Mizuno K, Tsutsumi T, Mizoguchi A, Miyoshi J, Takai Y, Hata Y: Synaptic scaffolding molecule alpha is a scaffold to mediate N-methyl-D-aspartate receptor-dependent RhoA activation in dendrites. Mol Cell Biol 27: 4388–4405, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong L, Pietsch S, Tan Z, Perner B, Sierig R, Kruspe D, Groth M, Witzgall R, Grone HJ, Platzer M, Englert C: Integration of cistromic and transcriptomic analyses identifies Nphs2, Mafb, and Magi2 as Wilms' Tumor 1 target genes in podocyte differentiation and maintenance. J Am Soc Nephrol 26: 2118–2128, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Ellis MJ, Gomez JA, Eisner W, Fennell W, Howell DN, Ruiz P, Fields TA, Spurney RF: Mechanisms of the proteinuria induced by Rho GTPases. Kidney Int 81: 1075–1085, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babayeva S, Zilber Y, Torban E: Planar cell polarity pathway regulates actin rearrangement, cell shape, motility, and nephrin distribution in podocytes. Am J Physiol Renal Physiol 300: F549–F560, 2011 [DOI] [PubMed] [Google Scholar]
- 26.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup : The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Li M, Hakonarson H: ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.