Abstract
Steroid-resistant nephrotic syndrome is characterized by podocyte dysfunction. Drosophila garland cell nephrocytes are podocyte-like cells and thus provide a potential in vivo model in which to study the pathogenesis of nephrotic syndrome. However, relevant pathomechanisms of nephrotic syndrome have not been studied in nephrocytes. Here, we discovered that two Drosophila slit diaphragm proteins, orthologs of the human genes encoding nephrin and nephrin-like protein 1, colocalize within a fingerprint-like staining pattern that correlates with ultrastructural morphology. Using RNAi and conditional CRISPR/Cas9 in nephrocytes, we found this pattern depends on the expression of both orthologs. Tracer endocytosis by nephrocytes required Cubilin and reflected size selectivity analogous to that of glomerular function. Using RNAi and tracer endocytosis as a functional read-out, we screened Drosophila orthologs of human monogenic causes of nephrotic syndrome and observed conservation of the central pathogenetic alterations. We focused on the coenzyme Q10 (CoQ10) biosynthesis gene Coq2, the silencing of which disrupted slit diaphragm morphology. Restoration of CoQ10 synthesis by vanillic acid partially rescued the phenotypic and functional alterations induced by Coq2-RNAi. Notably, Coq2 colocalized with mitochondria, and Coq2 silencing increased the formation of reactive oxygen species (ROS). Silencing of ND75, a subunit of the mitochondrial respiratory chain that controls ROS formation independently of CoQ10, phenocopied the effect of Coq2-RNAi. Moreover, the ROS scavenger glutathione partially rescued the effects of Coq2-RNAi. In conclusion, Drosophila garland cell nephrocytes provide a model with which to study the pathogenesis of nephrotic syndrome, and ROS formation may be a pathomechanism of COQ2-nephropathy.
Keywords: nephrocyte, Drosophila, nephrotic syndrome, COQ2, garland cell, SRNS
Steroid resistant nephrotic syndrome (SRNS) represents a common cause of CKD.1 In about 30% of cases mutation of a single gene can be identified as the cause2–4 and more than 30 causative genes have been discovered.5 The Drosophila nephrocyte forms slit diaphragms across membrane invaginations called labyrinthine channels. Nephrocytes have been suggested to be molecularly, ultrastructurally, and functionally analogous to mammalian podocytes.6–8 Thus they offer the opportunity for in vivo study of podocytopathies in a genetically highly-tractable model organism. There are two distinct nephrocyte populations: the pericardial nephrocytes along the heart tube and the garland cell nephrocytes (GCN) in a garland-like ring around the esophagus. In contrast to pericardial nephrocytes, GCN can be identified anatomically after brief dissection. The pericardial nephrocytes were first described as a screening tool for the discovery of renal disease genes9 and proposed as a model for protein uptake in the proximal tubulus.10 Mammalian Neph proteins partially rescue the Drosophila orthologs in GCN11 and pericardial nephrocytes were shown to be impaired by a high-glucose diet.12 Moreover, some novel human monogenic SRNS genes were studied in pericardial nephrocytes.13–15 But nephrocytes have not been tested systematically as a model for SRNS. The conservation of mechanisms involved in the pathogenesis of SRNS such as actin dysregulation, a role of the extracellular matrix (ECM), or CoQ10 deficiency is unclear.
Results
Slit Diaphragm Proteins Show a Fingerprint-Like Pattern
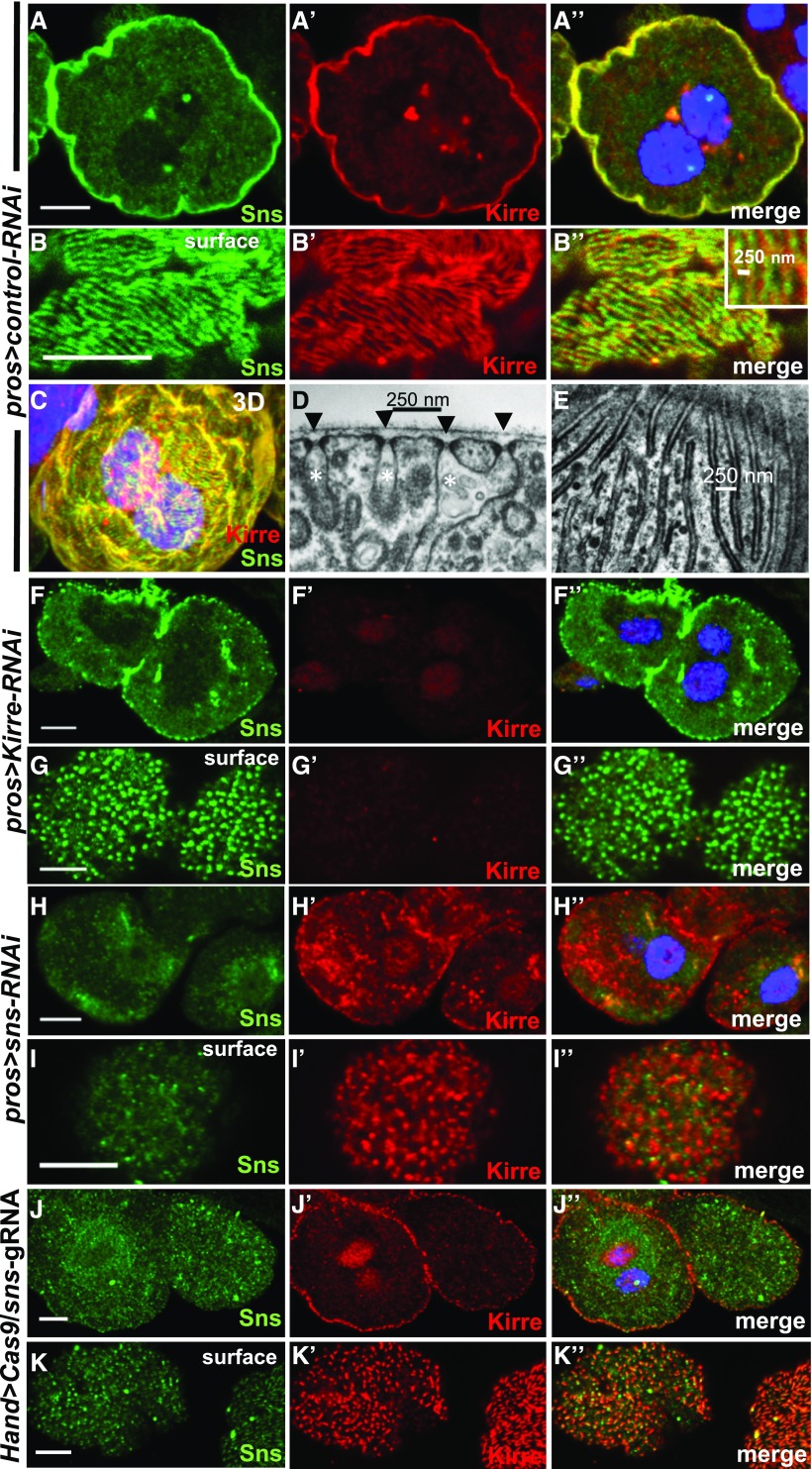
The slit diaphragm proteins sns and kirre, orthologs of NPHS1 and KIRREL, were shown to be a prerequisite for slit diaphragm formation in nephrocytes6,7 and immunofluorescence previously showed localization at the cell membrane.6,7,16 Using established Sns17 and Kirre18 antibodies we confirmed the published findings6,7,16 (Figure 1, A–A’’). To study subcellular localization in more detail we recorded sections tangential to the cell surface with high magnification. This revealed a pattern of Sns and Kirre in parallel lines reminiscent of a fingerprint (Figure 1, B–B’’). This pattern covered the entire cell (3D-projection in Figure 1C). The distance between parallel lines ranged from 250 to 500 nm (inset Figure 1B’’) corresponding to the distance between two slit diaphragms observed by transmission electron microscopy (TEM)(Figure 1D). We compared tangential sections of GCN in TEM with the tangential sections obtained by confocal microscopy and observed an analogous picture of slit diaphragms/labyrinthine channels as parallel lines with a distance of 250–500 nm (Figure 1E). This suggests that Sns and Kirre localize to the slit diaphragms. This is in accordance with the published findings using immunogold-labeling in TEM.6,7 The fingerprint-like pattern can be detected as early as first instar larvae and is present throughout larval development and adulthood (Supplemental Figures 1, A–G’’). Adult GCN were described as degenerated9 but testing the same genetic background we found adult GCN to maintain typical ultrastructure and endocytic activity (Supplemental Figure 2, A–C). Functionality is further supported by previous reports.19,20
Figure 1.
Slit diaphragm proteins localize in a fingerprint pattern analogous to slit diaphragm morphology in TEM and cluster to puncta upon loss of an interaction partner. (A–A’’) Equatorial cross section of GCN costained for the nephrin ortholog Sns (green) and the KIRREL/NEPH1 ortholog Kirre (red). Slit diaphragm proteins localize at the cell periphery. (B–B’’) Surface section of the same nephrocyte as in (A) reveals a fingerprint-like pattern. Distance between lines is 250–500 nm. (C) 3D reconstruction from a series of confocal sections of the same cell indicates that the visible circumference of the cell is densely covered by Sns/Kirre that are arranged in a fine linear pattern. (D) EM image of detail from a section cutting through nephrocyte surface perpendicularly shows cross-section of slit diaphragms (black arrow heads) and labyrinthine channels (white asterisks). Distance between two slit diaphragms is approximately 250–500 nm. (E) Planar section of a nephrocyte near the surface through the level of slit diaphragms/labyrinthine channels corresponds to lines in confocal image and confirms typical distance and pattern. Slit diaphragms are located between parallel lines of higher electrodensity. (F–F’’) Knockdown of kirre abolishes signal from Kirre-antibody whereas Sns remains detectable. (G–G’’) Surface section from GCN expressing kirre-RNAi shows Sns protein in a punctate staining pattern although the fingerprint-like pattern is lost. (H–H’’) Silencing of Sns results in strong reduction of cortical Sns signal whereas Kirre protein partially retains its localization at the periphery of the cell. (I–I’’) Surface section of GCN expressing sns-RNAi reveals loss of fingerprint-like pattern and strongly reduced Sns signal. Kirre staining pattern is punctate. Sns and Kirre seem mutually dependent and cluster in puncta upon loss of one interaction partner. (J–J’’) Shown is a GCN from an animal expressing two guide RNAs directed against sns ubiquitously and UAS-Cas9 under the hand-GAL4 driver in nephrocytes. The specific Sns signal at the periphery of the cell is strongly reduced although Kirre protein still is expressed. (K–K’’) CRISPR/Cas9-mediated loss-of-function of sns results in a punctate staining pattern of Kirre on the cell surface. The cell on the right shows residual amounts of Sns protein with an intermediate pattern of Kirre in dashed lines. Hand-GAL4>Cas9/+ (lacking gRNA expression) served as negative control and is shown elsewhere (Supplemental Figure 1, E–F’’). All scale bars represent 500 nm in electron microscopy images and 5 µm in confocal images unless otherwise indicated.
We wanted to evaluate Sns localization upon loss of kirre6 that is known to abrogate labyrinthine channels and slit diaphragms. Sns protein seems mostly retained at the membrane upon kirre knockdown in the equatorial sections (Figure 1F, Kirre control Figure 1F’) but tangential sections now revealed a punctate pattern (Figure 1G, kirre control Figure 1G’). Localization of Sns protein thus seems to be dependent on the presence of its binding partner Kirre. Conversely, we tested Kirre localization upon silencing of sns and observed a punctate staining pattern of Kirre (Figure 1, H and I’, Sns control in Figure 1, H and I). In conclusion, removal of the slit diaphragm protein Sns resulted in a punctate pattern of the unsilenced binding partner Kirre and vice versa. To evaluate this further we employed a conditional somatic CRISPR/Cas9 technique. This approach is useful for the study of nephrocytes as the classic Flp/FRT-based mosaic analysis is prevented in these nondividing cells. Transgenic expression of UAS-Cas9 in nephrocytes via Hand-GAL4 combined with ubiquitous expression of two guide RNAs led to strong reduction of Sns protein (Figure 1, J and K, negative control Supplemental Figure 1, E–F’’). Loss of Sns again ensued a punctate staining pattern of Kirre (Figure 1, J’ and K’) thus confirming our previous observations in this independent approach. Some cells displayed low residual levels of Sns protein (Figure 1K, cell on the right) resulting in an intermediate staining pattern of Kirre in dashed lines instead of puncta (Figure 1K’). This suggests that localization of Kirre is affected proportionally to the amount of Sns protein. The fingerprint pattern requires correct stoichiometry of slit diaphragm proteins.
Tracer Uptake Is Mediated by Receptor-Mediated Endocytosis via Cubilin/Amnionless
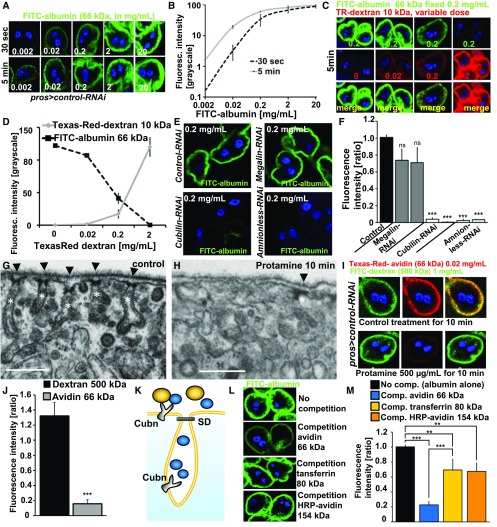
We then explored endocytosis of tracers like dextrans as an assay for nephrocyte function that was previously described.6,7,9 Dextrans are regarded as fluid-phase tracers. We hypothesized that tracer uptake is a read-out of fluid-phase endocytosis as a function of surface area. Fluid-phase endocytosis is generally linear whereas saturability is a hallmark of receptor-mediated endocytosis. We chose a modified approach by exposing third instar larvae to FITC-albumin and analyzed the dose-response relationship in (Figure 2, A and B). Fluorescence increased proportionally to an increasing dose of FITC-albumin below a threshold of 0.2 mg/ml. Further increase of the dose of FITC-albumin beyond this threshold resulted in a disproportionally slower increase of fluorescence reflected in the declining slope of the dose-response curve (Figure 2B, black). Increasing the incubation time tenfold lowered this threshold accordingly in a left-shifted curve (Figure 2B, gray). These findings suggest that tracer uptake occurs in a saturable process and thereby point toward receptor-mediated endocytosis. We then tested for receptor competition, another hallmark of receptor-mediated endocytosis. Keeping a saturated dose of FITC-albumin constant under an increasing dose of a Texas-Red–dextran with a molecular mass of 10 kD we observed a proportionate decrease of FITC-albumin uptake under increasing concentrations of 10 kD Texas-Red–dextran (Figure 2, C and D). High doses of Texas-Red–dextran entailed minimal uptake of FITC-albumin. FITC-albumin uptake thus also exhibits receptor competition which is characteristic for receptor-mediated endocytosis. The protein scavengers Cubilin and Amnionless have been shown to be required for the function of pericardial10 nephrocytes and GCN.21 Knockdown of the ortholog of the alternative protein scavenger Megalin did not result in a significant reduction, whereas knockdown of Cubilin or Amnionless seemed to abrogate uptake of FITC-albumin (0.2 mg/ml) almost entirely (Figure 2, E and F). This implies Cubilin/Amnionless as the receptor complex.
Figure 2.
Tracer endocytosis characteristics of GCN support a model of size-dependent filtration across slits and consecutive uptake via Cubilin/Amnionless coreceptors. (A) GCNs rapidly endocytose FITC-albumin. Fluorescence intensity increases with higher dose or incubation time (30 seconds versus 5 minutes). (B) Quantitation of fluorescence intensity after FITC-albumin exposure at two different incubation times using ImageJ shows linear increase until reaching saturation at higher concentrations. Values are presented as a mean±SD from the three brightest cells each from two independent experiments. (C) Simultaneous incubation with two small tracers, FITC-albumin (66 kD) and Texas-Red–dextran (10 kD), for 5 minutes. Increasing the dose of Texas-Red–dextran 10 kD reduces the uptake of FITC-albumin, whose applied concentration was kept constant. This suggests competition for uptake between both dyes. (D) Quantitation of fluorescence for coincubation of FITC-albumin at constant dose and variable concentrations of Texas-Red–dextran (10 kD). Intensity is recorded as grayscale (0–255) using ImageJ. FITC-albumin uptake is decreasing proportionally to the applied concentrations and increasing uptake of Texas-Red–dextran 10 kD. Offering Texas-Red–dextran in a tenfold mass excess reduces uptake of FITC-albumin to minimal levels. Values are presented as a mean±SD, n=3 per individual tracer dosage. (E) Representative images of fluorescence after incubation with FITC-albumin for 30 seconds at 0.2 mg/ml for control experiments used for normalization (GFP-RNAi) and RNAis for protein scavenger receptors. Cubilin-RNAi and Amnionless-RNAi showed strong reduction of the fluorescent signal. (F) Quantitation of FITC-fluorescence of (E) using ImageJ upon silencing of Megalin, Cubilin, and Amnionless from three independent experiments using two independent RNAis each shows a strong reduction for Cubilin and Amnionless, whereas Megalin has no significant effect. Values are presented as a mean±SD, n=3–4 per genotype. (G) Control treatment of nephrocytes for 10 minutes with PBS showed normal slit diaphragms (arrow heads) and labyrinthine channel morphology (white asterisks). (H) Pretreatment of nephrocytes with protamine 500 µg/ml for 10 minutes leads to a partial loss of labyrinthine channels and slit diaphragms. (I) Exposing cells to PBS alone followed by simultaneous exposure to the large tracer FITC-dextran 500 kD (1 mg/ml) and a small tracer Texas-Red–avidin 66 kD (0.02 mg/ml) resulted in strong uptake of both tracers (upper panel). The same experiment after protamine exposure for 10 min (500 µg/ml) showed strong reduction of avidin uptake whereas the large tracer FITC-dextran was not reduced by the treatment. (J) Quantitation of changes shown in (I) by protamine treatment normalized to mock-treatment for Texas-Red–avidin and FITC-dextran 500 kD shows reduction of 66 kD tracer endocytosis by loss of labyrinthine channels whereas the uptake of FITC-dextran 500 kD is increased in a nonsignificant manner. This suggests that uptake of the small tracer Texas-Red–dextran depends on slit diaphragms and labyrinthine channel surface but uptake of the large tracer FITC-dextran 500 kD is not affected by protamine-induced alteration of ultrastructure. n=5 per treatment. (K) Schematic illustrating hypothesis that competition between tracers depends on passage of the slit diaphragm which thus can serve as a read-out for filtration characteristics. Tracers which pass the slit diaphragm (blue) may approach all of the available receptors inside and outside the labyrinthine channels and thus are able to out-compete other tracers. Tracers whose mass prevents passage of the slit diaphragm (yellow) on the other hand cannot compete for receptors located inside the labyrinthine channels which results in incomplete competition. (L) Shown are representative images of control nephrocytes after incubation with 3 µM FITC-albumin for 5 minutes. The image on top shows FITC-albumin uptake without competition. Competition of FITC-albumin with a tenfold molar excess of tracers of increasing size are shown in a descending order. The tracer avidin (66 kD) competes strongly, whereas the larger tracers transferrin (80 kD) and HRP-avidin (avidin compound whose mass increased to 154 kD by conjugation of two molecules of peroxidase) show mild reduction of FITC-albumin endocytosis. This suggests partial competition and thus implies inability of the two larger tracers to pass the slit diaphragm. (M) Quantitation of experiments from (I). n=3–4 per intervention.
Tracer Experiments Are Compatible with Size-Selective Filtration in Nephrocytes
Size-selective filtration of hemolymph proteins before entering the labyrinthine channels analogous to size-selective filtration in the glomerulus has previously been proposed.6,7,9,10 We hypothesized that tracer endocytosis is not merely a function of Cubilin-mediated uptake but also reflects size-selective filtration. Therefore, we treated GCN with protamine sulfate, which rapidly induces foot-process–effacement in podocytes.22 Protamine at 500 µg/ml for 10 minutes induced partial loss of labyrinthine channels in nephrocytes compared with control treatment (Figure 2, G and H). Protamine hence perturbs nephrocyte ultrastructure including the slit diaphragms. We reasoned that uptake of a large tracer, whose size prevents the passage through the slit diaphragm, would be unaffected by protamine-induced loss of labyrinthine channels. On the other hand, endocytosis of a smaller tracer may occur within the labyrinthine channels after slit diaphragm passage. This uptake should be reduced by the loss of labyrinthine channel surface area. We applied 500 kD FITC-dextran (1 mg/ml) as a tracer that is unlikely to pass the slit diaphragm and Texas-Red–conjugated avidin of 66 kD (0.02 mg/ml) as a tracer that is expected to pass the slit diaphragm. In mock-treated nephrocytes (10 minutes PBS only) both tracers show robust endocytosis after 5 minutes (Figure 2I, upper panel). Then we recorded uptake of both tracers after treatment with protamine (Figure 2I, lower panel). Assessing the effect of protamine compared with mock treatment we noticed no reduction of 500 kD FITC-dextran but a strong reduction of uptake of the smaller Texas-Red–avidin (Figure 2J). The loss of labyrinthine channels/slit diaphragms therefore is reflected by the reduced uptake of the smaller tracer whereas endocytosis of 500 kD dextran appears to be independent of disturbed ultrastructure. This suggests that tracer endocytosis also reflects size-selective filtration and a 66 kD tracer can be useful to study nephrocyte function.
Receptor Competition Experiments Suggest a Filtration Cut-Off around 66–80 kD
The size cut-off for glomerular filtration in mammals is approximately 70 kD23,24 and we hypothesized that filtration in GCN has a similar cut-off. This is supported by findings in pericardial nephrocytes that suggested a size cut-off around 70 kD.9 To test this in GCN we employed the phenomenon of receptor competition. We reasoned that a tracer whose mass exceeds the filtration cut-off cannot compete for receptors located within the labyrinthine channels. As a smaller tracer can still be taken up inside the labyrinthine channels, this should result in partial competition (schematic in Figure 2K). We exposed uptake of 66 kD FITC-albumin to a tenfold molar competition of tracers with increasing size: 66 kD Texas-Red–avidin, 80 kD tracer Texas-Red–transferrin, and a 154 kD HRP-avidin (Figure 2, L and M). The smaller avidin showed strong reduction of FITC-albumin endocytosis whereas transferrin and the avidin, whose size had been modified by HRP-conjugation, reduced FITC-albumin endocytosis only about 30% compared with uptake without competition (Figure 2M). This suggests incomplete competition as predicted. Transferrin, a known ligand of cubilin,25 and avidin seem to compete for the same receptors as they are effectively out-competed by an excess of FITC-albumin in the reverse experiment (Supplemental Figure 3, A–D). These data are in accordance with a filtration cut-off between 66 and 80 kD analogous to the mammalian glomerular slit diaphragm.
Silencing Orthologs of Human Genes Defective in Monogenic SRNS Impairs Nephrocyte Tracer Uptake
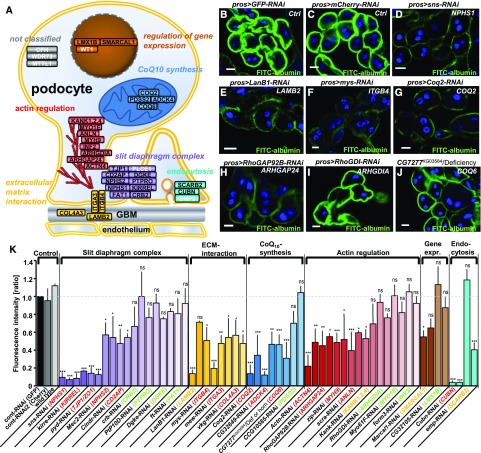
More than 30 genes have been identified as monogenic causes of SRNS in humans5 (Figure 3A). We hypothesized that the functional modules responsible for monogenic SRNS may be conserved in GCN. Analysis of a set of 36 established human and mouse genes using a combination of BLAST analysis and query of online databases (www.ensembl.org and the diopt tool26) rendered 29 putative Drosophila orthologs (Figure 3A, Supplemental Table 1). We silenced the Drosophila genes in GCN by 2–3 independent RNAis. Nephrocyte function was assessed by FITC-albumin endocytosis and the fluorescent signal was quantified and normalized to a control experiment performed in parallel (Figure 3K, Supplemental Figure 4, Supplemental Table 2). We categorized genes as “likely functionally-relevant” (red) if at least two RNAi lines impaired tracer uptake significantly, and “negative” if two RNAis had no significant effect (green). If an observed tracer impairment was not confirmed by a second RNAi and no further RNAi line was available, the result would be considered “undetermined” (yellow). Efficient knockdown was confirmed using immunofluorescence for the orthologs of CRB2, ITGA3, and ITGB4 (Supplemental Figure 5). Hence we identified five orthologs of genes related to the slit diaphragm complex (NPHS1, KIRREL, TJP1/ZO1, NPHS2, CD2AP), three orthologs connected to the ECM (ITGA3, ITGAB3, COL4A3), four orthologs involved in actin regulation (ACTN4, ARHGAP24, MYH9, ANLN), and one endocytosis factor (CUBN) as likely relevant for nephrocyte function. The orthologs of LAMB2, SMARCAL1, SCARB2, and the Drosophila KANK remain undetermined (Figure 3K, Supplemental Figure 4, Supplemental Table 2). A functional significance was also observed for the orthologs of the CoQ10 synthesis genes COQ2 and ADCK4. The single RNAi line targeting the ortholog of COQ6 had no effect but a p element insertion allele (CG7277KG03584) impairs tracer uptake significantly when being homozygous or hemizygous with a corresponding deficiency (Figure 3K). Neither allele nor deficiency affect tracer endocytosis heterozygous with wild type (Supplemental Figure 4, Supplemental Table 2) Thus a functional role for the ortholog of COQ6 seems likely. TUNEL colabeling suggested impairment of tracer uptake to be independent of apoptosis for sns-RNAi and Coq2-RNAi (Supplemental Figure 6, A–C).
Figure 3.
Endocytosis assay in Drosophila GCN shows reduced uptake of FITC-albumin in the majority of orthologs of mammalian genes that if mutated cause nephrosis. (A) Known monogenic causes of SRNS in humans or mice are shown within their protein interaction complexes and subcellular localization in the podocyte. Cellular functions are classified as: slit diaphragm complex, ECM interaction proteins, actin regulators, regulators of gene expression, proteins that play a role in endocytosis, and CoQ10 synthesis genes. Gene names in white letters indicate that no ortholog was identified. (B–J) Shown are examples of GCN that were exposed to FITC-albumin for 30 seconds for a selection of genotypes. All scale bars represent 10 µm. The human ortholog is denoted in white. (B) Nephrocytes expressing control RNAis against mCherry (B) or GFP (C) exhibit strong FITC-albumin uptake whereas cells expressing an RNAi directed against the nephrin ortholog sns result only in minimal uptake (D). Reduced uptake is also observed upon knockdown of the orthologs of LAMB2 (E), ITGB4 (F), COQ2 (G), and ARHGAP24 (H). Knockdown of the ortholog of ARHGDIA on the other hand shows uptake similar to control levels (I). An allele of the ortholog of COQ6 hemizygous over a corresponding deficiency shows reduced uptake as well (J). (K) Quantitation of fluorescent intensity from FITC-albumin uptake is shown. The human ortholog is denoted in brackets. Genes are grouped into functional complexes denoted on top (compare with figure in A). The human ortholog is colored in red when the uptake was significantly reduced in two RNAis and in green when no significant difference to control (mCherry) was observed (P<0.05) for two RNAis. Genes where significant reduction of tracer uptake was observed with one RNAi but did not confirm in the only available second RNAi are denoted as undetermined (yellow). Wild type nephrocytes and GCN expressing an RNAi directed against mCherry showed uptake that is comparable to GFP-RNAi. Knocking down orthologs of genes involved in the slit diaphragm complex, ECM interaction, CoQ10 synthesis, and actin regulation resulted in impaired tracer uptake. The p element insertion allele of the ortholog of COQ6 is quantified homozygous and hemizygous over a short deficiency both resulting in reduced uptake. Values are presented as mean±SD of the ratio to a control experiment (GFP-RNAi) done in parallel, n=3–4 per genotype. Statistical significance was calculated using ANOVA and Dunnet post hoc analysis. ACTN4, actinin, α4; ADCK4, aarf domain-–containing kinase 4; ANLN, actin-binding protein anillin; ARHGAP24, rho gtpase–activating protein 24; ARHGDIA, rho gdp–dissociation inhibitor α; CD2AP, cd2-associated protein; COL4A3, collagen Type IV, α-3; COQ2, coq2 homolog (s. cerevisiae); COQ6, coq6 homolog (s. cerevisiae); CRB2, crumbs homolog 2 (drosophila); CUBN, cubilin; DGKE, diacylglycerol kinase, ε, 64-kd; FAT1, fat tumor suppressor homolog 1 (drosophila); INF2, inverted formin 2; ITGA3, integrin α-3; ITGB4, integrin β-4; KANK1, kn motif- and ankyrin repeat domain–containing protein 1; KANK2, kn motif– and ankyrin repeat domain-containing protein 2; KANK4, kn motif– and ankyrin repeat domain–containing protein 4; KIRREL, kin of ire–like; LAMB2, laminin β-2; LMX1B, lim homeobox transcription factor 1 β; MYH9, myosin heavy chain 9, nonmuscle; MYO1E, myosin 1e; NPHS1, nephrin; NPHS2, podocin; PDSS2, prenyl diphosphate synthase, subunit 2; PTPRO, protein-tyrosine phosphatase, receptor type o; SCARB2, scavenger receptor class b, member 2; SMARCAL1, swi/snf-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a-like protein 1; TJP1, tight junction protein 1.
Taken together, 16 of 29 orthologous genes significantly affected nephrocyte function across the functional modules of slit diaphragm complex, ECM interactors, CoQ10 synthesis, and actin regulation. This suggests that the central pathomechanisms of monogenic human SRNS are reflected by GCN.
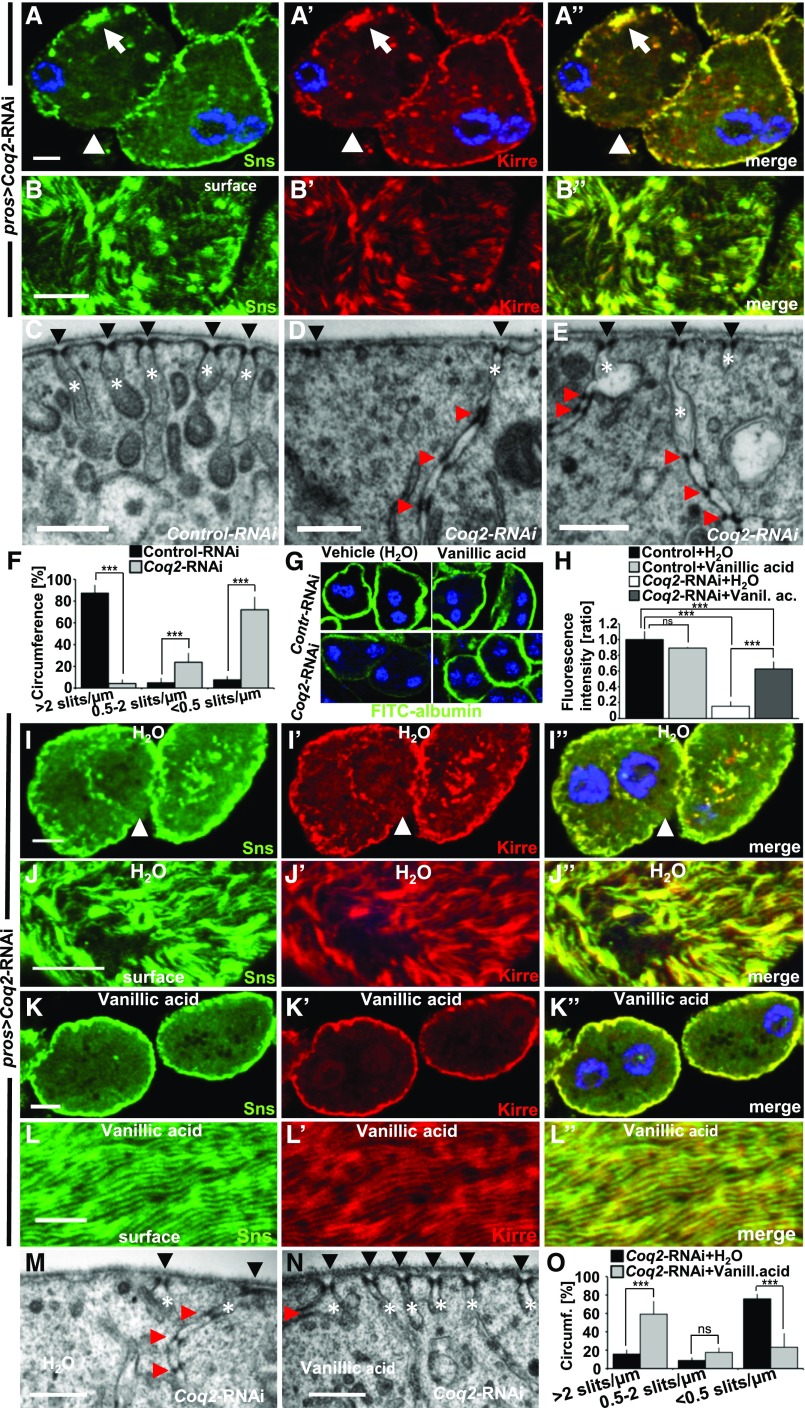
Knockdown of Coq2 Results in Mislocalization/Loss of Slit Diaphragms and Labyrinthine Channels
In humans, mutations of COQ2 have been identified as a treatable cause of SRNS2,27,28 although the pathogenesis remains unclear.29 Hence we explored the role of Coq2 in the Drosophila nephrocyte. Staining Sns and Kirre upon Coq2 silencing we noticed a partial displacement from the membrane for both proteins (Figure 4, A–A’’). Confocal sections through the surface showed areas lacking Sns/Kirre, misspacing of the lines of the fingerprint-like pattern, and clusters of slit diaphragm proteins (Figure 4, B–B’’). Accordingly, although slit diaphragms densely populate the surface of control GCN (Figure 4C), Coq2 silencing causes large areas without slit diaphragms (Figure 4D). When labyrinthine channels were still present, they were elongated and thinner. Frequently multiple consecutive slit diaphragms were present inside these channels (Figure 4, D and E). To quantify loss of slit diaphragms, we evaluated the cell membrane of a full diameter of six cells each for control-RNAi and Coq2-RNAi. We categorized slit diaphragm frequency ≥2/µm as normal (i.e., distance between slits ≤500 nm). On average, nephrocytes expressing control-RNAi exhibited normal slit diaphragm frequency on 87% of the analyzed cell surface. In contrast, only 4% of the surface of nephrocytes expressing Coq2-RNAi showed the regular frequency whereas the remainder displayed a reduced frequency (0.5–2 slits/µm, 24%) or only sporadic slit diaphragms (≤0.5 slits/µm, 72%, Figure 4F). This amounts to a strong loss of slit diaphragms upon Coq2 silencing.
Figure 4.
Coq2-RNAi results in mislocalization and loss of slit diaphragm proteins and can be partially rescued by feeding vanillic acid. (A–A”) Garland nephrocytes expressing Coq2-RNAi costained for Sns and Kirre reveal partial protein mislocalization from the slit diaphragm on the cell surface toward the interior of the cell (for comparison see Figure1, A–A’’). The peripheral line of slit diaphragm proteins seems replaced by clustered signals (white arrow) or lost (white arrow head). (B–B’’) Surface section shows a mixture of clustering in puncta, loss, or increased distance of ridges formed by slit diaphragm proteins (for comparison see Figure1, B–B’’). (C) EM image of control-RNAi (GFP) shows slit diaphragms at regular distances (black arrow heads) and labyrinthine channels (white asterisks) that rarely exceed a depth of 500 nm. (D and E) EM images of nephrocytes expressing Coq2-RNAi show reduced number of slit diaphragms (arrow heads) and labyrinthine channels (asterisks). Where present, channels appear elongated and narrower compared with control (C). Slit diaphragms are often mislocalized into the labyrinthine channels and multiplied (red arrow heads). (F) Quantitation of frequency of slit diaphragms measured along one complete GCN diameter for control-RNAi compared with Coq2-RNAi as mean of six cells from three different animals. Frequency of slit diaphragms was classified into three groups: normal (>2 slits/µm), reduced (0.5–2 slits/µm), and sporadic (<0.5 slits/µm). Note the strong reduction of slit diaphragms in Coq2-RNAi. (G and H) Supplementing vanillic acid with the food increases FITC-albumin uptake compared with control treatment with H2O, suggesting a partially restored nephrocyte function. Feeding vanillic acid to flies expressing control-RNAi has no relevant influence on FITC-albumin uptake. (H) Quantitation of experiments from (G). Rescue with vanillic acid blunts the phenotype of Coq2 silencing. n=4 per genotype and intervention. (I–J’’) Nephrocytes from flies treated with vehicle (H2O) show a phenotype similar to untreated cells (A–A’’). (K–L’’) Upon treatment with vanillic acid slit diaphragm proteins are more adherent to cell surface and the fingerprint pattern of Sns/Kirre is restored (L–L’’). (M) EM image of a nephrocyte expressing Coq2-RNAi after treatment with vehicle (H2O) shows a similar phenotype to untreated cells (see D–E). (N) Treating flies expressing Coq2-RNAi with vanillic acid improves frequency of slit diaphragms formed on the surface (black arrow heads). Slit diaphragms inside labyrinthine channels (red arrow head) and elongated labyrinthine channels are less frequently observed than with vehicle alone. (O) Quantitation of slit diaphragm frequency of one complete diameter for Coq2-RNAi with vehicle (H2O) compared with Coq2-RNAi treated with vanillic acid as mean of six cells from three different animals. Frequency of slit diaphragms was classified into three groups: normal (>2 slits/µm), reduced (0.5–2 slits/µm), and sporadic (<0.5 slits/µm). Note the increase of slit diaphragms on surface upon application of vanillic acid. All scale bars represent 500 nm in electron microscopy images and 5 µm in confocal images.
Sns/Kirre-staining and TEM using a second Coq2-RNAi matched the findings with the first Coq2-RNAi (Supplemental Figure 7, A–E’’).
To examine the specificity for CoQ10 synthesis we used vanillic acid which had previously been shown to bypass a block in the CoQ10 biosynthesis pathway downstream of COQ6 and COQ2 in yeast.30 Feeding flies with this compound partially restored GCN function in Coq2 silencing whereas the function of control nephrocytes remained unchanged (Figure 4, G–H). Vanillic acid treatment also largely restored the staining pattern of Sns/Kirre (Figure 4, K–L’’) compared with treatment with vehicle (H2O) alone (Figure 4, I–J’’). In the ultrastructural analysis vanillic acid increased areas of normal slit diaphragm frequency from about 15% to 60% compared with vehicle control (Figure 4, M–O). Elongated channels and duplications of slit diaphragms were still observed occasionally (Figure 4N). The Coq2-RNAi phenotype thus was partially rescued by substitution of vanillic acid.
Mitochondrial ROS Formation Occurs in Coq2 Silencing
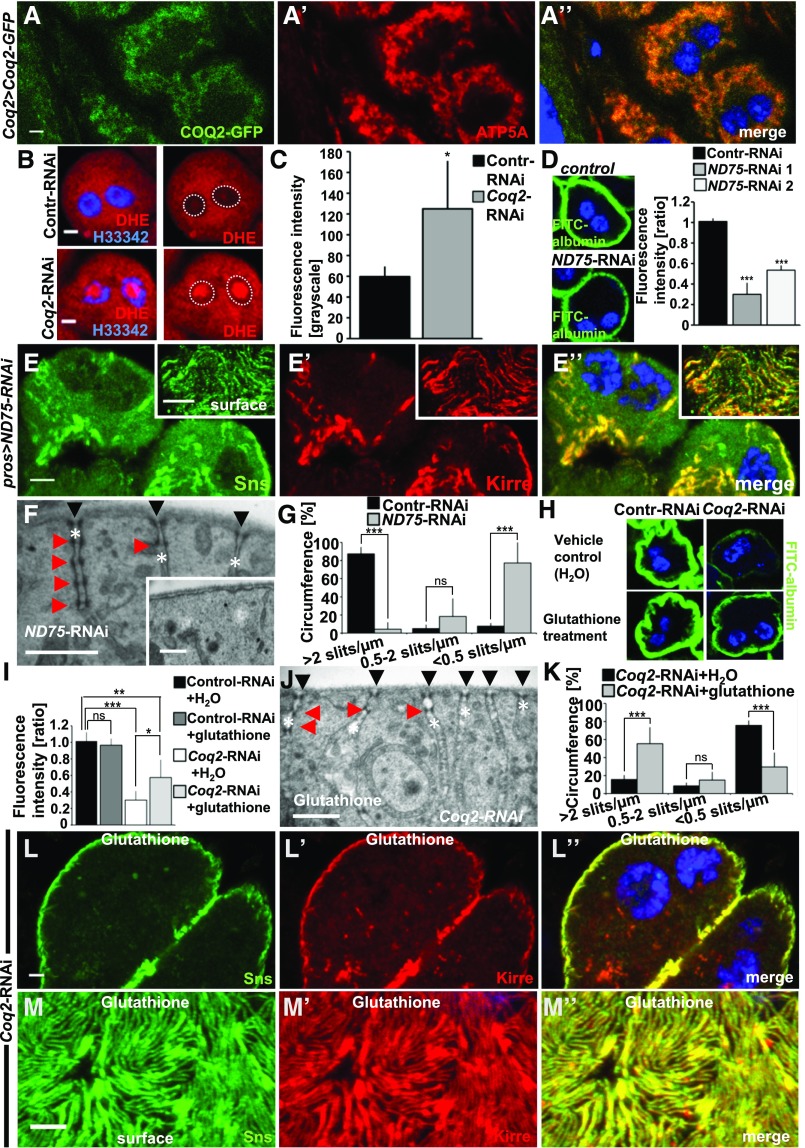
The pathogenesis of COQ2-nephropathy could be conveyed through defects directly affecting the slit diaphragm via lack of CoQ10, e.g., by affecting lipid rafts or lipid oxidation. An indirect mechanism of COQ2 deficiency via increased ROS-formation and mitochondrial dysfunction has also been suggested.31 To analyze this in GCN, we first studied localization of Coq2. As no anti-Coq2 antibodies are available we employed a GFP-reporter allele that drives Coq2 under endogenous promoter.32 Costaining cells expressing this reporter with the mitochondrial marker ATP5A showed colocalization, indicating that Coq2 most likely resides in mitochondria (Figure 5A). Lack of Coq2-dependent CoQ10 synthesis is known to interfere with electron transfer at the mitochondrial respiratory chain, which in turn increases ROS formation.33 Therefore we analyzed the redox state of these cells with the ROS indicator dihydroethidium (DHE). This compound is blue-fluorescent in the reduced state but shifts toward the red spectrum upon oxidation by superoxide anions. The oxidized form of DHE intercalates with DNA. We incubated GCN expressing control-RNAi or Coq2-RNAi with DHE and recorded red fluorescence (Figure 5B). Quantitation of the nuclear signal revealed a significant increase demonstrating intensified ROS formation (Figure 5C). Then we tested if interference with respiratory chain function independent of CoQ10 synthesis results in a phenotype similar to Coq2 knockdown. This would suggest an indirect pathogenesis related to mitochondrial energy metabolism and not directly to lack of CoQ10 itself. We silenced ND75, a subunit of the respiratory chain complex, which is known to increase ROS formation.34,35 Uptake of FITC-albumin was reduced upon ND75 silencing suggesting impairment of nephrocyte function (Figure 5D). Testing localization of slit diaphragm proteins upon ND75 silencing we found that Sns/Kirre were displaced from the cell membrane and mislocalized intracellularly (Figure 5, E–E’’). When slit diaphragm proteins were detectable on the surface, the fingerprint pattern was discontinuous and irregular (Figure 5, E–E’’, insets). This phenotype was highly reminiscent of the phenotype of Coq2 silencing. We performed TEM analysis of ND75 knockdown and observed widespread loss of labyrinthine channels and slit diaphragms with only 4% of the cell surface displaying normal slit diaphragm frequency (Figure 5, F–G). The few remaining labyrinthine channels were elongated and narrowed with frequently multiple slit diaphragms inside the channels (Figure 5F). Knockdown of ND75 thus results in a phenocopy of Coq2-RNAi. To investigate the role of ROS formation further we fed the ROS scavenger glutathione to Drosophila larvae. Supplementing glutathione resulted in an increased FITC uptake when Coq2 was silenced, whereas there was no relevant effect on control nephrocytes (Figure 5, H and I). In TEM we observed a partial rescue of the slit diaphragm frequency (55%, Figure 5, J and K) and the fingerprint-like staining pattern of Sns/Kirre was partially restored (Figure 5, L and M). ROS scavenging thus partially rescues the phenotype of Coq2 silencing.
Figure 5.
Increased ROS formation in Coq2 loss-of-function. (A–A”) GCNs expressing a third-copy reporter allele of Coq2 with an insertion of GFP in frame costained for the mitochondrial marker ATP5A indicates localization of Coq2 to the mitochondria. (B) GCN expressing control-RNAi or Coq2-RNAi after 5 minutes of incubation with ROS indicator DHE. Stronger intranuclear signal of DHE upon knockdown of Coq2 indicates increased ROS (dotted circles in right panel without DAPI mark nuclei). (C) Quantitation of nuclear DHE signal indicates significant increase of ROS formation upon knockdown of Coq2. (D) GCN show a decrease of FITC-albumin uptake upon knockdown of ND75, a subunit of the complex I of the respiratory chain, as shown by representative images for one RNAi line and quantitation for two independent RNAis. (E–E’’) Silencing of ND75 results in displacement of Sns/Kirre from the membrane toward the interior of the cell. Areas denuded from labyrinthine channels appear on the cell surface. Knockdown of ND75-RNAi thus results in a phenocopy of Coq2 silencing. (F) TEM analysis of GCN expressing ND75-RNAi reveals loss of slits/labyrinthine channels or elongated channels with multiple slit diaphragms (red arrow heads). (G) Quantitation of slit diaphragm frequency on the full circumference of cells expressing ND75-RNAi. One complete diameter of six cells from three different animals was analyzed. Frequency of slit diaphragms was classified into three groups: normal (>2 slits/µm), reduced (0.5–2 slits/µm), and sporadic (<0.5 slits/µm). Values are presented as mean percentage±SD for each category. Note strong reduction of slit diaphragms upon knockdown of ND75 similar to knockdown of Coq2. Control is identical to control from Figure 4F. (H) Treatment of control or Coq2-RNAi with glutathione for 5 days partially restores uptake of FITC-albumin in Coq2-RNAi nephrocytes but has no effect on control-RNAi–expressing cells. (I) Quantitation of experiments in (H), n=3–5 per genotype and intervention. (J) TEM imaging after treatment of flies expressing Coq2-RNAi with the ROS scavenger glutathione for 5 days partially restores slit diaphragm frequency on the cell surface in ultrastructural analysis. Slit diaphragm localized in labyrinthine channels and elongated labyrinthine channels may still be found. (K) Quantitation of experiments in (J) for six cells from three animals. Control is identical to control from Figure 4O. (L–M’’) Treatment of Coq2-RNAi with glutathione partially restores typical localization including fingerprint pattern of Sns/Kirre (M–M’’). Compare with vehicle-only controls in Figure 4, I–J’’. All scale bars represent 500 nm in electron microscopy images and 5 µm in confocal images unless otherwise stated.
In summary, our data suggest that loss of Coq2 exerts its pathologic effects by ROS formation induced by CoQ10 deficiency.
Discussion
Here, we systematically screened the orthologs of genes that if mutated cause SRNS in humans using tracer endocytosis in Drosophila GCN. We detected a loss-of-function phenotype in this system for 16 out of 29 genes, thereby demonstrating its relevance as a model system for human SRNS. The genes we found to be required for GCN function pertained to the functional categories of slit diaphragm complex, interaction with the ECM, actin regulation, regulation of gene expression, endocytosis, and CoQ10 biosynthesis.
Mutations in CoQ10 biosynthesis genes represent the only monogenic form of human SRNS, in whom gene identification led to definition of a treatment approach. We employed GCN to model human COQ2 nephropathy and found Coq2 to be required for slit diaphragm morphology and function. Vanillic acid supplementation of Drosophila rescued the Coq2 deficiency in GCN. This compound was shown to restore yeast CoQ10 biosynthesis30,36 but also is a mild antioxidant37 and regulates gene expression in certain bacteria.38 Vanillic acid has been suggested as a treatment for deficient human CoQ10 biosynthesis.30 Our findings imply that the pathogenesis of Coq2 is linked to mitochondrial ROS formation but not to CoQ10 function outside of mitochondria, such as a role as an antioxidant at lipid rafts. In podocytes, excess ROS may cause podocyte apoptosis39,40 or cytoskeletal rearrangements.41,42
The use of GCN to model podocyte disease is an attractive alternative to pericardial nephrocytes as GCN are discernible anatomically without use of a marker. Both nephrocyte models are limited by their evolutionary distance and the morphologic and functional disparities between fly and humans. However, no mammalian in vitro model that forms functional slit diaphragms is available. The rapid, versatile, and inexpensive analysis this model offers facilitates screening approaches and dissection of pathways of human SRNS.
Our findings lay the groundwork for using GCN to study pathogenesis of SRNS, and we employ this model to identify ROS formation as a potential mechanism of COQ2 nephropathy.
Concise Methods
Fly Husbandry and Genetics
Overexpression and transgenic RNAi studies were performed using the UAS/GAL4 system (RNAi crosses grown at 25 or 29°C). Supplementation experiments were performed by adding 200 µl H2O ± 0.25 mM glutathione or 10 mM vanillic acid consecutively 0, 1, 3, and 4 days before dissection.
The RNAi stocks used throughout the study where obtained from the Vienna Drosophila Resource Center or Bloomington Drosophila Stock Center at Indiana University (BDSC). RNAi-lines and the CG7277 allele and deficiency are specified in Supplemental Table 2. Wild type flies were obtained from Bloomington (BDSC # 8522). Prospero-GAL46 or Hand-GAL4 (kindly provided by A. Paululat via H. Jasper) were used to drive expression in nephrocytes.
The CRISPR gRNA construct targeting sns was generated using a described protocol43 using pCFD4 (#49411; Addgene) and injected into flies expressing phiC31 integrase under vasa promoter with an attP landing site in 51C (#24482; BDSC) by Bestgene. Tandem guide RNA sequences are as follows: AGTGCCAGGTGGGACCGGCT and CTACGGAGCTTATGAGTGCG. Restricted Cas9 expression was achieved by genetic combination of Hand-GAL4 and UAS-Cas9.P (#54594; BDSC) by standard crosses.
Immunohistochemistry and TUNEL Detection
GCN were dissected, fixed for 15 minutes in PBS containing 4% paraformaldehyde, and stained according to the standard procedure. The following primary antibodies were used: rabbit anti-sns17 (1:500, gift from S. Abmayr), guinea pig anti-Kirre18 (1:200, gift from S. Abmayr), mouse anti-ATP5A (1:200, ab14748; Abcam), rat anti-Crumbs44 (1:500, gift from U. Tepass), Alexa Fluor 647 anti-HRP (1:1000, 323–605–021; Jackson Immuno Research), mouse anti-mys (1:50, CF.6G11; DSHB), and mouse anti-mew (1:40, DK.1A4; DSHB). Hoechst 33342 (1:1000; Thermofisher) was used to visualize nuclei. Apoptotic cells were visualized using the In Situ Cell Death Detection Kit (Thermofisher) according to the manufacturer’s instructions.
For imaging, a Leica SP5x confocal microscope was used. Image processing was done by ImageJ and Adobe Photoshop CS4 software.
Fluorescent Tracer Uptake and ROS Measurement
Nephrocytes were dissected in PBS and incubated with DHE (30 μM, 5 min), FITC-albumin (Sigma) or Texas-Red–dextran (10 kD), Texas-Red–avidin, Texas-Red–transferrin (80 kD), HRP-avidin (154 kD), and FITC-dextran (500 kD) (all from Thermofisher) for 30 seconds or 5 minutes as indicated. After a fixation of 5 minutes in 8% paraformaldehyde cells were rinsed in PBS and exposed to Hoechst 33342 (1:1000) for 20 seconds and mounted in Antifade Diamond (Thermofisher). Cells were imaged using a Leica SP5x confocal microscope. Quantification of fluorescent tracer uptake was performed with ImageJ software. Average fluorescence of the three brightest cells was measured and intensity of the background subtracted. If applicable, the results are expressed as a ratio to a control experiment with EGFP-RNAi that was done in parallel.
Electron Microscopy
Dissected GCN were fixed in 4% formaldehyde and 0.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, and TEM was carried out using standard techniques. One complete diameter of six cells from three different animals was analyzed. Frequency of slit diaphragms was classified into three groups: normal (>2 slits/µm), reduced (0.5–2 slits/µm), and sporadic (<0.5 slits/µm). Areas where the labyrinthine channels are cut obliquely are recognizable by elongated stretches of higher electrodensity along the cell surface. These areas were excluded from the quantitation.
Statistical Analyses
Paired t test was used to determine the statistical significance between two interventions. ANOVA followed by Sidak correction (unless otherwise indicated) was used for multiple comparisons (GraphPad Prism software). Asterisks indicate significance as follows: *P<0.05, **P<0.01, ***P<0.001. A statistically significant difference was defined as P<0.05.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank Susan Abmayr, Alvaro Glavic, Renjie Jiao, David Bilder, Zhe Han, Ulrich Tepass, Karl-Friedrich Fischbach, Paul Hartley, and Heinrich Jasper for sharing reagents, the Developmental Studies Hybridoma Bank for antibodies, Bloomington Drosophila Stock Center and Vienna Drosophila RNAi Center for fly stocks. We thank Maria Ericsson (Harvard Medical School Electron Microscopy Facility) for technical assistance.
This research was supported by grants from the National Institutes of Health (DK076683 and DK086542) to F.H. and by a Research Fellowship from the Deutsche Forschungsgemeinschaft (DFG) to T.H. (HE 7456/1-1). T.B.H. was supported by the DFG and European Research Council.
F.H. is the Warren E. Grupe Professor. T.H. designed and performed the experiments, D.A.B. critically reviewed the paper. M.H., and T.B.H. were involved in adult nephrocyte analysis. T.H. wrote the paper with help from F.H., F.H. conceived of and directed the project.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016050517/-/DCSupplemental.
References
- 1.Vivante A, Hildebrandt F: Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol 12: 133–146, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadowski CE, Lovric S, Ashraf S, Pabst WL, Gee HY, Kohl S, Engelmann S, Vega-Warner V, Fang H, Halbritter J, Somers MJ, Tan W, Shril S, Fessi I, Lifton RP, Bockenhauer D, El-Desoky S, Kari JA, Zenker M, Kemper MJ, Mueller D, Fathy HM, Soliman NA, Hildebrandt F; SRNS Study Group : A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 26: 1279–1289, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trautmann A, Bodria M, Ozaltin F, Gheisari A, Melk A, Azocar M, Anarat A, Caliskan S, Emma F, Gellermann J, Oh J, Baskin E, Ksiazek J, Remuzzi G, Erdogan O, Akman S, Dusek J, Davitaia T, Özkaya O, Papachristou F, Firszt-Adamczyk A, Urasinski T, Testa S, Krmar RT, Hyla-Klekot L, Pasini A, Özcakar ZB, Sallay P, Cakar N, Galanti M, Terzic J, Aoun B, Caldas Afonso A, Szymanik-Grzelak H, Lipska BS, Schnaidt S, Schaefer F; PodoNet Consortium : Spectrum of steroid-resistant and congenital nephrotic syndrome in children: The PodoNet registry cohort. Clin J Am Soc Nephrol 10: 592–600, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giglio S, Provenzano A, Mazzinghi B, Becherucci F, Giunti L, Sansavini G, Ravaglia F, Roperto RM, Farsetti S, Benetti E, Rotondi M, Murer L, Lazzeri E, Lasagni L, Materassi M, Romagnani P: Heterogeneous genetic alterations in sporadic nephrotic syndrome associate with resistance to immunosuppression. J Am Soc Nephrol 26: 230–236, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovric S, Ashraf S, Tan W, Hildebrandt F: Genetic testing in steroid-resistant nephrotic syndrome: When and how? Nephrol Dial Transplant 31: 1802–1813, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weavers H, Prieto-Sánchez S, Grawe F, Garcia-López A, Artero R, Wilsch-Bräuninger M, Ruiz-Gómez M, Skaer H, Denholm B: The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature 457: 322–326, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhuang S, Shao H, Guo F, Trimble R, Pearce E, Abmayr SM: Sns and Kirre, the Drosophila orthologs of Nephrin and Neph1, direct adhesion, fusion and formation of a slit diaphragm-like structure in insect nephrocytes. Development 136: 2335–2344, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denholm B, Skaer H: Bringing together components of the fly renal system. Curr Opin Genet Dev 19: 526–532, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang F, Zhao Y, Han Z: An in vivo functional analysis system for renal gene discovery in Drosophila pericardial nephrocytes. J Am Soc Nephrol 24: 191–197, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang F, Zhao Y, Chao Y, Muir K, Han Z: Cubilin and amnionless mediate protein reabsorption in Drosophila nephrocytes. J Am Soc Nephrol 24: 209–216, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmstädter M, Lüthy K, Gödel M, Simons M, Ashish, Nihalani D, Rensing SA, Fischbach KF, Huber TB: Functional study of mammalian Neph proteins in Drosophila melanogaster. PLoS One 7: e40300, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Na J, Sweetwyne MT, Park AS, Susztak K, Cagan RL: Diet-Induced podocyte dysfunction in Drosophila and mammals. Cell Rep 12: 636–647, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gee HY, Zhang F, Ashraf S, Kohl S, Sadowski CE, Vega-Warner V, Zhou W, Lovric S, Fang H, Nettleton M, Zhu JY, Hoefele J, Weber LT, Podracka L, Boor A, Fehrenbach H, Innis JW, Washburn J, Levy S, Lifton RP, Otto EA, Han Z, Hildebrandt F: KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest 125: 2375–2384, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashraf S, Gee HY, Woerner S, Xie LX, Vega-Warner V, Lovric S, Fang H, Song X, Cattran DC, Avila-Casado C, Paterson AD, Nitschké P, Bole-Feysot C, Cochat P, Esteve-Rudd J, Haberberger B, Allen SJ, Zhou W, Airik R, Otto EA, Barua M, Al-Hamed MH, Kari JA, Evans J, Bierzynska A, Saleem MA, Böckenhauer D, Kleta R, El Desoky S, Hacihamdioglu DO, Gok F, Washburn J, Wiggins RC, Choi M, Lifton RP, Levy S, Han Z, Salviati L, Prokisch H, Williams DS, Pollak M, Clarke CF, Pei Y, Antignac C, Hildebrandt F: ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest 123: 5179–5189, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gee HY, Saisawat P, Ashraf S, Hurd TW, Vega-Warner V, Fang H, Beck BB, Gribouval O, Zhou W, Diaz KA, Natarajan S, Wiggins RC, Lovric S, Chernin G, Schoeb DS, Ovunc B, Frishberg Y, Soliman NA, Fathy HM, Goebel H, Hoefele J, Weber LT, Innis JW, Faul C, Han Z, Washburn J, Antignac C, Levy S, Otto EA, Hildebrandt F: ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest 123: 3243–3253, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tutor AS, Prieto-Sánchez S, Ruiz-Gómez M: Src64B phosphorylates Dumbfounded and regulates slit diaphragm dynamics: Drosophila as a model to study nephropathies. Development 141: 367–376, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Bour BA, Chakravarti M, West JM, Abmayr SM: Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev 14: 1498–1511, 2000 [PMC free article] [PubMed] [Google Scholar]
- 18.Galletta BJ, Chakravarti M, Banerjee R, Abmayr SM: SNS: Adhesive properties, localization requirements and ectodomain dependence in S2 cells and embryonic myoblasts. Mech Dev 121: 1455–1468, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Kosaka T, Ikeda K: Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J Cell Biol 97: 499–507, 1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreenkova OV, Karpova EK, Menshanov PN, Rauschenbach IY: [Knockdown of InR gene in ventral nephrocytes promotes resistance to toxic stress in Drosophila melanogaster females]. Genetika 51: 263–265, 2015 [PubMed] [Google Scholar]
- 21.Gleixner EM, Canaud G, Hermle T, Guida MC, Kretz O, Helmstädter M, Huber TB, Eimer S, Terzi F, Simons M: V-ATPase/mTOR signaling regulates megalin-mediated apical endocytosis. Cell Rep 8: 10–19, 2014 [DOI] [PubMed] [Google Scholar]
- 22.Seiler MW, Venkatachalam MA, Cotran RS: Glomerular epithelium: Structural alterations induced by polycations. Science 189: 390–393, 1975 [DOI] [PubMed] [Google Scholar]
- 23.Hamano Y, Grunkemeyer JA, Sudhakar A, Zeisberg M, Cosgrove D, Morello R, Lee B, Sugimoto H, Kalluri R: Determinants of vascular permeability in the kidney glomerulus. J Biol Chem 277: 31154–31162, 2002 [DOI] [PubMed] [Google Scholar]
- 24.Scott RP, Quaggin SE: Review series: The cell biology of renal filtration. J Cell Biol 209: 199–210, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozyraki R, Fyfe J, Verroust PJ, Jacobsen C, Dautry-Varsat A, Gburek J, Willnow TE, Christensen EI, Moestrup SK: Megalin-dependent cubilin-mediated endocytosis is a major pathway for the apical uptake of transferrin in polarized epithelia. Proc Natl Acad Sci USA 98: 12491–12496, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE: An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12: 357, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diomedi-Camassei F, Di Giandomenico S, Santorelli FM, Caridi G, Piemonte F, Montini G, Ghiggeri GM, Murer L, Barisoni L, Pastore A, Muda AO, Valente ML, Bertini E, Emma F: COQ2 nephropathy: A newly described inherited mitochondriopathy with primary renal involvement. J Am Soc Nephrol 18: 2773–2780, 2007 [DOI] [PubMed] [Google Scholar]
- 28.McCarthy HJ, Bierzynska A, Wherlock M, Ognjanovic M, Kerecuk L, Hegde S, Feather S, Gilbert RD, Krischock L, Jones C, Sinha MD, Webb NJ, Christian M, Williams MM, Marks S, Koziell A, Welsh GI, Saleem MA; RADAR the UK SRNS Study Group : Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 8: 637–648, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desbats MA, Lunardi G, Doimo M, Trevisson E, Salviati L: Genetic bases and clinical manifestations of coenzyme Q10 (CoQ 10) deficiency. J Inherit Metab Dis 38: 145–156, 2015 [DOI] [PubMed] [Google Scholar]
- 30.Doimo M, Trevisson E, Airik R, Bergdoll M, Santos-Ocaña C, Hildebrandt F, Navas P, Pierrel F, Salviati L: Effect of vanillic acid on COQ6 mutants identified in patients with coenzyme Q10 deficiency. Biochim Biophys Acta 1842: 1–6, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinzii CM, Garone C, Emmanuele V, Tadesse S, Krishna S, Dorado B, Hirano M: Tissue-specific oxidative stress and loss of mitochondria in CoQ-deficient Pdss2 mutant mice. FASEB J 27: 612–621, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarov M, Barz C, Jambor H, Hein MY, Schmied C, Suchold D, Stender B, Janosch S, Kj VV, Krishnan RT, Krishnamoorthy A, Ferreira IR, Ejsmont RK, Finkl K, Hasse S, Kampfer P, Plewka N, Vinis E, Schloissnig S, Knust E, Hartenstein V, Mann M, Ramaswami M, VijayRaghavan K, Tomancak P, Schnorrer F: A genome-wide resource for the analysis of protein localisation in. eLife 5: e12068, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turrens JF: Mitochondrial formation of reactive oxygen species. J Physiol 552: 335–344, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owusu-Ansah E, Banerjee U: Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 461: 537–541, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hochmuth CE, Biteau B, Bohmann D, Jasper H: Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell 8: 188–199, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozeir M, Mühlenhoff U, Webert H, Lill R, Fontecave M, Pierrel F: Coenzyme Q biosynthesis: Coq6 is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem Biol 18: 1134–1142, 2011 [DOI] [PubMed] [Google Scholar]
- 37.Kumar S, Prahalathan P, Raja B: Antihypertensive and antioxidant potential of vanillic acid, a phenolic compound in L-NAME-induced hypertensive rats: A dose-dependence study. Redox Rep 16: 208–215, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gitzinger M, Kemmer C, Fluri DA, El-Baba MD, Weber W, Fussenegger M: The food additive vanillic acid controls transgene expression in mammalian cells and mice. Nucleic Acids Res 40: e37, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Susztak K, Raff AC, Schiffer M, Böttinger EP: Glucose-induced reactive oxygen species cause apoptosis of podocytes and podocyte depletion at the onset of diabetic nephropathy. Diabetes 55: 225–233, 2006 [PubMed] [Google Scholar]
- 40.Das R, Xu S, Quan X, Nguyen TT, Kong ID, Chung CH, Lee EY, Cha SK, Park KS: Upregulation of mitochondrial Nox4 mediates TGF-β-induced apoptosis in cultured mouse podocytes. Am J Physiol Renal Physiol 306: F155–F167, 2014 [DOI] [PubMed] [Google Scholar]
- 41.Stanley A, Thompson K, Hynes A, Brakebusch C, Quondamatteo F: NADPH oxidase complex-derived reactive oxygen species, the actin cytoskeleton, and Rho GTPases in cell migration. Antioxid Redox Signal 20: 2026–2042, 2014 [DOI] [PubMed] [Google Scholar]
- 42.Nimnual AS, Taylor LJ, Bar-Sagi D: Redox-dependent downregulation of Rho by Rac. Nat Cell Biol 5: 236–241, 2003 [DOI] [PubMed] [Google Scholar]
- 43.Port F, Chen HM, Lee T, Bullock SL: Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA 111: E2967–E2976, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pellikka M, Tanentzapf G, Pinto M, Smith C, McGlade CJ, Ready DF, Tepass U: Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature 416: 143–149, 2002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.