Abstract
Idiopathic membranous nephropathy (MN) is associated with HLA; however, the HLA allele involved remains unknown. To identify the HLA risk alleles associated with phospholipase A2 receptor (PLA2R)-related MN in the Chinese population, we sequenced the entire MHC region in DNA samples from 99 patients with PLA2R-related MN, 50 patients with PLA2R-unrelated MN, and 100 healthy subjects. Two HLA risk alleles, HLA-DRB1*15:01 and HLA-DRB3*02:02, independently and strongly associated with an increased risk of PLA2R-related MN. After adjusting for HLA-DRB1*15:01 and HLA-DRB3*02:02, no other alleles showed significant association with PLA2R-related MN. A replication study in an independent cohort of 293 participants with PLA2R-related MN and 285 healthy controls validated these findings. In a joint analysis, a multivariate logistic regression model confirmed that HLA-DRB1*15:01 (odds ratio [OR], 24.9; 95% confidence interval [95% CI], 15.3 to 42.6; P=2.3×10−35) and HLA-DRB3*02:02 (OR, 17.7; 95% CI, 11.0 to 30.3; P=8.0×10−29) independently and strongly associated with PLA2R-related MN. As many as 98.7% of patients with PLA2R-related MN, compared with 43.9% of control subjects, carried at least one HLA risk allele. Subjects with either risk allele had higher odds of developing PLA2R-related MN than those without a risk allele (OR, 98.9; 95% CI, 44.4 to 281.7; P=2.5×10−23). These HLA risk alleles also associated with the age at disease onset in patients with PLA2R-related MN. In conclusion, our findings provide clear evidence that the HLA-DRB1*15:01 and HLA-DRB3*02:02 alleles independently and strongly associate with PLA2R-related MN in the Chinese population.
Keywords: membranous nephropathy, human leukocyte antigen, autoimmune disease, HLA Typing, Target sequencing
Idiopathic membranous nephropathy (IMN) is an organ-specific autoimmune disease and a major cause of adult nephrotic syndrome. Over the past few years, great advances have been made in understanding the pathogenesis of membranous nephropathy (MN) due to the identification of podocyte autoantigens. The phospholipase A2 receptor (PLA2R) has been identified as the main podocyte antigen in IMN,1 and approximately 70% of affected individuals have circulating antibodies against PLA2R.2,3 An understanding of how particular HLA alleles at high resolution confer susceptibility is essential for a comprehensive understanding of the pathogenesis of autoimmune diseases.4 Identifying HLA risk alleles is also important to develop antigen-specific therapies for patients with autoimmune diseases. Using the candidate gene approach, previous studies have shown that IMN is associated with HLA class 2 genes.5–10 Recently, a genome-wide association study (GWAS) has shown strong associations of the 6p21 HLA-DQA1 and 2q24 PLA2R1 loci with IMN in patients of European ancestry.11 However, this GWAS had an insufficient variant density to define the associations between IMN and high-resolution HLA alleles.12 More importantly, there are various podocyte autoantigens in IMN, including PLA2R, neutral endopeptidase, THSD7A, and other unknown antigens.3,13–15 Each HLA molecule binds to sets of peptides with particular sequences or physical features and presents them on the cell surface for recognition by T cells via T cell receptor engagement. Thus, each autoantigen in IMN could have its own particular HLA risk alleles. Because previous studies did not exclude patients with PLA2R-unrelated MN,11,16 the enrollment of patients with different autoantibodies may greatly reduce the statistical power and decrease the estimated effect size.17
The identification of PLA2R as the main podocyte autoantigen in IMN makes it possible to precisely define a homogeneous subgroup of patients with IMN in whom the pathogenesis of the disease is associated with PLA2R antibodies. In this study, patients who were positive for both serum anti-PLA2R antibodies and glomerular PLA2R antigen staining were classified as having PLA2R-related MN, whereas patients who were negative for both were classified as having PLA2R-unrelated MN. Previously, we performed a microarray analysis of microdissected glomeruli from 44 biopsy renal tissues from patients with MN (23 with PLA2R-related MN and 20 with PLA2R-unrelated MN) and 21 paracancerous renal tissues as normal controls from patients who underwent a nephrectomy for renal cancer. We found that the expression of HLA-DRB5 showed the most significant change of all of the differentially expressed mRNAs among the three groups (Supplemental Figure 1, Supplemental Table 1). Because not all people within the population carry the three additional distinct HLA-DRB genes (HLA-DRB3/4/5), the DRB5 signal in the microarray analysis manifested as an all or none expression pattern. The patients with PLA2R-related MN had a higher frequency of HLA-DRB5 (82.6%) than the patients with PLA2R-unrelated MN (10%) and normal controls (33.3%), indicating that some particular HLA-DRB5 alleles or HLA-DRB5–linked HLA alleles are strongly associated with the pathogenesis of PLA2R-related MN. These results also suggest that there were different HLA risk alleles between PLA2R-related MN and PLA2R-unrelated MN. In this study, to precisely identify the particular HLA risk alleles associated with PLA2R-related MN, we conducted a full human HLA gene capture sequencing study and validated the associations in an independent cohort of patients.
Results
Association Testing for Whole HLA Alleles in the Discovery Samples
To initially explore which particular HLA alleles confer susceptibility to PLA2R-related MN, 249 DNA samples (99 patients with PLA2R-related MN, 50 patients with PLA2R-unrelated MN, and 100 healthy controls) were captured for sequencing of the entire HLA region of the human genome (Supplemental Tables 2 and 3). The patients and controls were genetically well matched (Supplemental Figures 2 and 3). All HLA genes from the 249 samples were successfully genotyped at a four-digit resolution. In total, 290 four-digit resolution HLA alleles were detected, and no novel HLA alleles were discovered. We first performed the associations for HLA alleles in patients with PLA2R-related MN. Of the detected HLA alleles, six of them (DRB1*15:01, DRB5*01:01, DQA1*01:02, DQB1*06:02, DRB4*01:03, and DRB3*02:02) were significantly associated with PLA2R-related MN (Figures 1A and 2A, Supplemental Table 4) (P<1×10−5). All significant HLA alleles associated with PLA2R-related MN were aggregated in the HLA class 2 gene region, and no significant signals were found in the regions for HLA class 1 or other genes. HLA-DRB1*15:01 (odds ratio [OR], 16.9; 95% confidence interval [95% CI], 8.4 to 34.2; P=2.7×10−15) had the most significant associations on the basis of the univariate analysis. It is not surprising that three alleles highly linked to HLA-DRB1*15:01 (DRB5*01:01, DQA1*01:02, and DQB1*06:02) were also significantly associated with the disease.18,19 Because the HLA-DRB1*15:01 was the most significant signal, we defined the HLA-DRB1*15:01-DRB5*01:01-DQA1*01:02-DQB1*06:02 haplotype as the HLA-DRB1*15:01 haplotype. HLA-DRB4*01:03 (OR, 0.22; 95% CI, 0.12 to 0.41; P=6.5×10−7) was associated with a decreased risk of the disease.
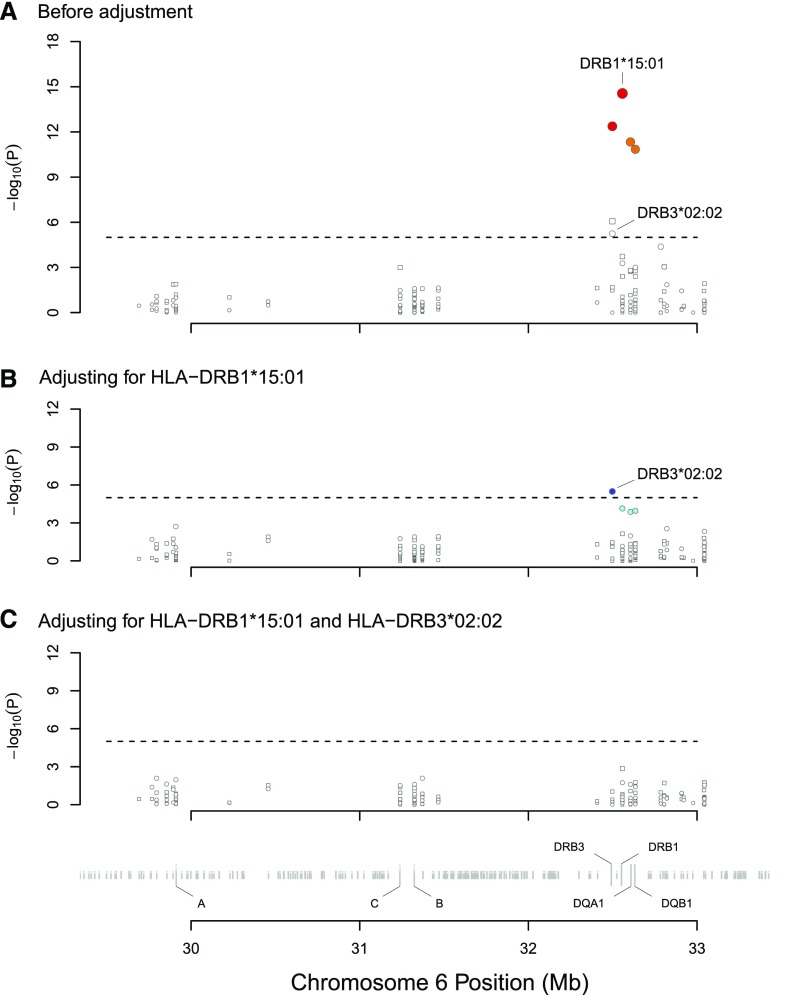
Figure 1.
Association tests between all HLA alleles and PLA2R-related MN in the discovery dataset. Circles denote alleles with an OR>1, whereas squares denote alleles with an OR≤1. (A) shows that the significant signals (P<1×10−5) were aggregated in the HLA class 2 region; no significant signals were found in the HLA class 1 region. HLA-DRB1*15:01 (OR, 16.9; 95% CI, 8.4 to 34.2; P=2.7×10−15) showed the strongest signal. Colors denote the strength of the LD of the alleles compared with HLA-DRB1*15:01. Red denotes an R2 value of 0.8 or more, orange denotes an R2 value of 0.5 to <0.8, and white denotes an R2 value of <0.25. Three alleles that were tightly linked to HLA-DRB1*15:01 (DRB5*01:01, DQA*01:02, and DQB1*06:02) also showed significant associations. The HLA-DRB3*02:02 (OR, 4.0; 95% CI, 2.2 to 7.2; P=5.7×10−6) was also significantly associated with PLA2R-related MN. (B) shows the association plot after adjusting for HLA-DRB1*15:01. The allele HLA-DRB3*02:02 maintained its independent significant association. Colors denote the strength of the LD of the alleles compared with HLA-DRB3*02:02. Blue denotes an R2 value of 0.8 or more, cyan denotes an R2 value of 0.25 to <0.5, and white denotes an R2 value of <0.25. (C) shows that none of the other alleles were independently associated with the disease after adjusting for HLA-DRB1*15:01 and HLA-DRB3*02:02.
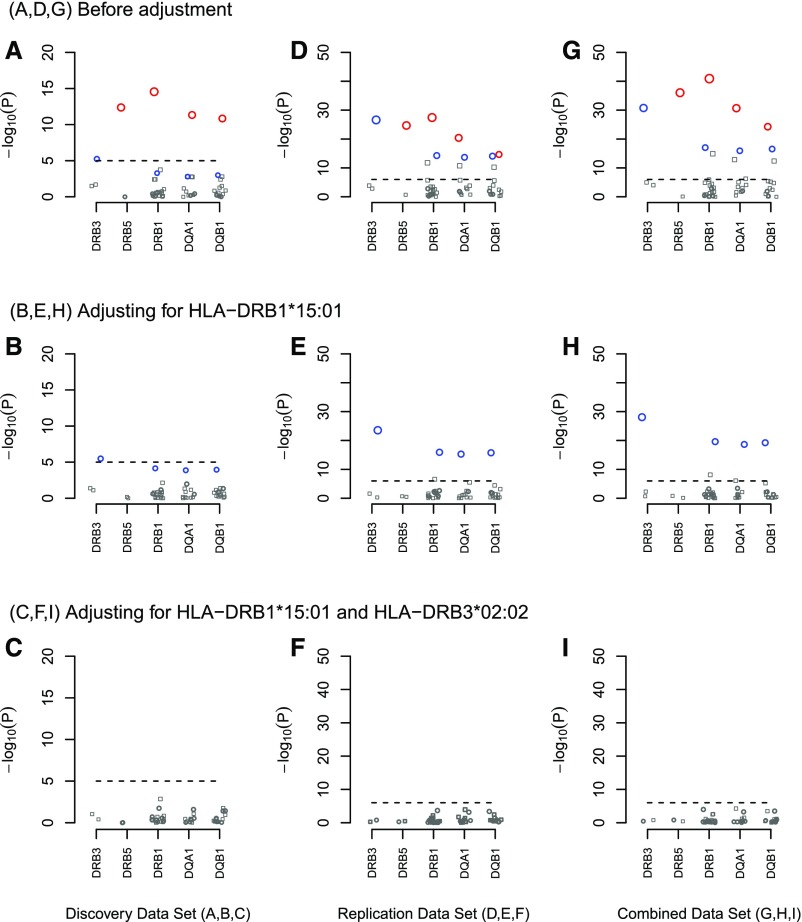
Figure 2.
Association tests for the five most significant HLA genes in PLA2R-related MN. Circles denote alleles with an OR>1, whereas squares denote alleles with an OR≤1. Red circles denote the HLA-DRB1*15:01 allele and its linked alleles, and blue circles denote the HLA-DRB3*02:02 allele and its linked alleles. (A–C) highlight the five most significant HLA genes in the discovery study as shown in Figure 1. (D–F) show the independent replication analysis of the five selected HLA genes. (G–I) show the combined analysis. (B, E, and H) After adjusting for HLA-DRB1*15:01, the effects of the HLA-DRB1*15:01–linked HLA alleles were eliminated, whereas the effects of the HLA-DRB3*02:02 allele and its linked HLA alleles remained significant. (C, F, and I) After adjusting for both HLA-DRB1*15:01 and HLA-DRB3*02:02, we found that none of the alleles were associated with PLA2R-related MN.
After adjusting for HLA-DRB1*15:01, the effect of HLA-DRB3*02:02 remained significant (OR, 18.3; 95% CI, 5.4 to 62.3; P=3.2×10−6), whereas the effects of the other alleles were eliminated (Figure 1B). Three alleles closely linked to HLA-DRB3*02:02 (DRB1*03:01, DQB1*02:01, and DQA1*05:01) were marginally associated with the disease (Figures 1B and 2B).18,19 Because the HLA-DRB3*02:02 was the most significant signal, we defined the HLA-DRB3*02:02-DRB1*03:01-DQB1*02:01-DQA1*05:01 haplotype as the HLA-DRB3*02:02 haplotype. Moreover, we found that the OR of HLA-DRB3*02:02 was obviously increased from 4.0 (95% CI, 2.2 to 7.2) to 18.3 (95% CI, 5.4 to 62.3) after the adjustment analysis. The effect of HLA-DRB1*15:01 remained significant in all models. The NMDP database19 showed that the frequency of the HLA-DRB3*02:02-DRB1*15:01 haplotype was <0.001% in various populations, indicating that HLA-DRB3*02:02 and HLA-DRB1*15:01 are not of the same haplotype in worldwide populations. After further adjusting for HLA-DRB1*15:01 and HLA-DRB3*02:02, none of the other HLA alleles retained their independent associations with PLA2R-related MN (Figures 1C and 2C).
HLA-DRB1*15:01 was present in 81.8% of patients with PLA2R-related MN and 21.0% of healthy controls, whereas HLA-DRB3*02:02 was present in 60.6% of the patients and 28.0% of the controls. The frequencies of HLA-DRB1*15:01 and HLA-DRB3*02:02 in healthy controls in this study were similar to those in Chinese volunteers from the bone marrow donor program.19,20 Remarkably, of the 18 patients who were negative for the HLA-DRB1*15:01 allele, all were positive for HLA-DRB3*02:02. Thus, all of the patients (100%) with PLA2R-related MN in the discovery cohort carried at least one of the two HLA risk alleles (HLA-DRB1*15:01 and/or HLA-DRB3*02:02). We also analyzed the associations for single-nucleotide polymorphisms (SNPs) and amino acid variants with PLA2R-related MN in the discovery samples. We found that SNPs and amino acid variants linked with the two HLA risk alleles were also independently associated with PLA2R-related MN (Supplemental Figure 4). We also used a multiplicative genetic model in the logistic regression and obtained similar results for the HLA alleles and SNP/amino variants (Supplemental Figure 5). Taken together, we preliminarily identified two risk HLA alleles, HLA-DRB1*15:01 and HLA-DRB3*02:02, that were strongly and independently associated with an increased risk of PLA2R-related MN.
To assess the allele frequency of these HLA alleles in patients with PLA2R-unrelated MN, 50 patients with PLA2R-unrelated MN were also enrolled in the discovery study (Supplemental Figures 6 and 7). We found that HLA-DRB3*02:02 was associated with PLA2R-unrelated MN (OR, 4.6; 95% CI, 2.2 to 9.4; P=3.9×10−5), and 64.0% of patients with PLA2R-unrelated MN carried HLA-DRB3*02:02 compared with 26.0% of healthy controls. Interestingly, only 16% of patients with PLA2R-unrelated MN carried HLA-DRB1*15:01 compared with 81.8% of patients with PLA2R-related MN (81.8%; P=4.1×10−14) and 21.0% of the healthy controls (P=0.61), suggesting that HLA-DRB1*15:01 is not associated with PLA2R-unrelated MN.
Independent Replication Study and Combined Analyses
To validate the HLA risk alleles of PLA2R-related MN that we identified in the discovery study, we selected and genotyped the five most strongly associated HLA genes (HLA-DRB3, HLA-DRB5, HLA-DRB1, HLA-DQA1, and HLA-DQB1) in an independent sample of Han Chinese individuals (293 patients with PLA2R-related MN and 285 controls). The patients with PLA2R-unrelated MN were not enrolled in the validation study, because the main goal of this study was to identify the HLA risk alleles associated with PLA2R-related MN. All patients were successfully genotyped at a four-digit resolution, except for two patients in the HLA-DQB1 loci. There were 80 total HLA alleles detected in the replication study. Seven novel HLA alleles were identified, four of which were in the HLA-DQB1 loci and three of which were in the HLA-DQA1 loci. As with the discovery samples, HLA-DRB1*15:01 presented the strongest signal, and HLA-DRB3*02:02 showed the second strongest signal (Figure 2, D–F, Supplemental Table 5). The three HLA-DRB1*15:01–linked alleles and the three HLA-DRB3*02:02–linked alleles also showed significant but weaker signals.
The combined analyses of the 392 patients with PLA2R-related MN and 385 healthy controls were consistent with separate analyses of the discovery and replication samples (Figure 2, G–I, Supplemental Table 6). The most significant independent signals were observed for HLA-DRB1*15:01 (OR, 9.7; 95% CI, 7.0 to 13.6; P=1.1×10−41) and HLA-DRB3*02:02 (OR, 6.4; 95% CI, 4.7 to 8.8; P=1.7×10−31) in the univariate models. After adjusting for HLA-DRB1*15:01 and HLA-DRB3*02:02, none of the other alleles were associated with the disease. When the two independent HLA risk alleles were both included in a multivariate model, HLA-DRB1*15:01 (OR, 24.9; 95% CI, 15.3 to 42.6; P=2.3×10−35) and HLA-DRB3*02:02 (OR, 17.7; 95% CI, 11.0 to 30.3; P=8.0×10−29) showed stronger associations than their univariate counterparts.
We evaluated the effects of both independent HLA risk alleles in combination on the risk of developing PLA2R-related MN (Figure 3). Of the 392 patients with PLA2R-related MN, 283 patients (72.2%) carried HLA-DRB1*15:01, 274 patients (69.9%) carried HLA-DRB3*02:02, 387 patients (98.7%) carried at least one of the two HLA risk alleles, and only five patients (1.7%) carried neither of the HLA risk alleles. Of the 385 healthy controls, 81 individuals (21.0%) carried HLA-DRB1*15:01, 102 individuals (26.5%) carried HLA-DRB3*02:02, 169 individuals (43.8%) carried at least one of the two HLA risk alleles, and 216 individuals (56.1%) carried neither of the HLA risk alleles. According to these results, individuals carrying either of the HLA risk alleles have a 98.9-fold increased odds of developing PLA2R-related MN compared with those who do not carry either risk allele (95% CI, 44.4 to 281.7; P=2.5×10−23).
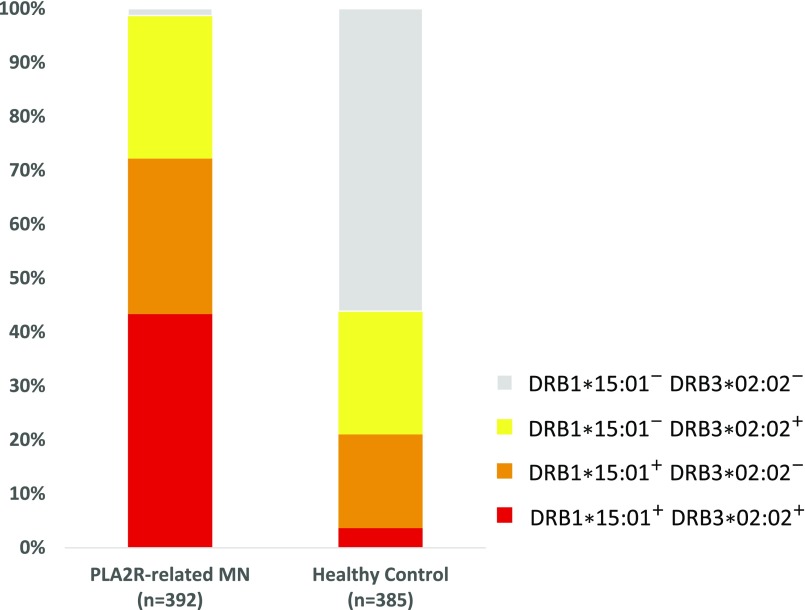
Figure 3.
Distribution of the two HLA risk alleles in patients with PLA2R-related MN and healthy controls. As many as 98.7% of patients with PLA2R-related MN carried at least one HLA risk allele compared with 43.9% of healthy controls. Of the 392 patients with PLA2R-related MN, 283 patients (72.2%) carried the HLA-DRB1*15:01 allele, 274 patients (69.9%) carried the HLA-DRB3*02:02 allele, 387 patients (98.7%) carried at least one of the two HLA risk alleles, and only five patients (1.7%) carried neither of the HLA risk alleles. Of the 385 healthy controls, 81 individuals (21.0%) carried HLA-DRB1*15:01, 102 individuals (26.5%) carried HLA-DRB3*02:02, 169 individuals (43.8%) carried at least one of the two HLA risk alleles, and 216 individuals (56.1%) carried neither of the HLA risk alleles.
Associations between the HLA Risk Alleles and the Baseline Demographic and Clinical Characteristics of Patients with PLA2R-Related MN
We performed an association analysis of the patient’s demographic and clinical data (Table 1). There were 104 patients identified as HLA-DRB1*15:01+DRB3*02:02−, 113 patients identified as HLA-DRB1*15:01−DRB3*02:02+, and 170 patients identified as HLA-DRB1*15:01+DRB3*02:02+. We found that the HLA risk alleles are associated with patient age at disease onset. The median age at the disease onset was 35 years old in patients with HLA-DRB1*15:01+DRB3*02:02−, which was remarkably younger than those with HLA-DRB1*15:01−DRB3*02:02+ (50 years old; P<0.001). The probability distribution of the age at disease onset was apparently different between patients with HLA-DRB1*15:01+DRB3*02:02− and those with HLA-DRB1*15:01−DRB3*02:02+ (Supplemental Figures 8 and 9). In addition, patients with HLA-DRB1*15:01−DRB3*02:02+ had higher levels of circulating anti-PLA2R antibodies, lower eGFR, more global glomeruli sclerosis, and more severe interstitial fibrosis than patients with HLA-DRB1*15:01+DRB3*02:02−. Other parameters, including sex, serum albumin, and proteinuria, were not statistically significantly associated with these two HLA risk alleles. The demographic and clinical data on the basis of the HLA-DRB3*02:02 in patients with PLA2R-unrelated MN were also analyzed, but no significant associations were observed (Supplemental Tables 7 and 8).
Table 1.
Demographic and clinical data stratified by the HLA risk allele status in patients with PLA2R-related MN
| Characteristics | DRB1*15:01+RB3*02:02− | DRB1*15:01−DRB3*02:02+ | DRB1*15:01+DRB3*02:02+ | P Valuea |
|---|---|---|---|---|
| Patients | 104 | 113 | 170 | |
| Men, % | 62 | 69 | 56 | 0.33 |
| Age at disease onset, yr | 35 (22–58) | 50 (42–58) | 44 (30–56) | <0.001 |
| Serum anti-PLA2R level, RU/ml | 65.1 (38.1–142.7) | 92.8 (48.2–208.7) | 82.5 (38.5–167.7) | 0.03 |
| eGFR | 110.9±24.3 | 93.9±25.7 | 100.8±25.5 | <0.001 |
| Serum uric acid, μmol/L | 371±90 | 381±91 | 362±99 | 0.45 |
| Serum albumin, g/L | 28.1±5.4 | 28.4±5.3 | 28.3±5.2 | 0.76 |
| 24-h Proteinuria, g/24 h | 3.9 (2.2–5.9) | 4.0 (2.5–6.6) | 3.9 (2.3–5.5) | 0.75 |
| Global glomeruli sclerosis, % | 0 (0–5.0) | 4.9 (0–10.7) | 2.9 (0–6.8) | <0.001 |
| Interstitial fibrosis, % | 0.002 | |||
| None or trace: <10% | 85.6 | 66.7 | 68.1 | |
| Mild: 10%–25% | 13.5 | 22.2 | 26.3 | |
| Moderate: 26%–50% | 0.96 | 9.1 | 5.6 | |
| Severe: >50% | 0 | 2.0 | 0 |
HLA-DRB1*15:01−RB3*02:02+ group versus HLA-DRB1*15:01+DRB3*02:02− group.
Discussion
In this study, we sequenced the whole HLA region and identified two independent risk HLA alleles: the HLA-DRB1*15:01 and HLA-DRB3*02:02, both of which are strongly and independently associated with PLA2R-related MN in the Chinese population. Individuals carrying HLA-DRB1*15:01 and/or HLA-DRB3*02:02 had a 99-fold increased odds of developing PLA2R-related MN compared with those who do not carry either of these HLA risk alleles. More importantly, we found that almost all patients with PLA2R-related MN (98.7%) carried at least one of these two HLA risk alleles. The largest differences between our study and previously reported association studies in MN were due to the homogeneity of the patients enrolled in this study. Phenotype resolution is a major determinant of the success or failure of large-scale genetic studies.21 An imprecise phenotype could reduce the power to localize susceptibility variants and greatly decrease the estimated risks attributed to generic variation.17,22 There are various podocyte autoantigens in patients with IMN.13–15 To identify HLA risk alleles that are precisely associated with PLA2R-related MN, the qualifying patients enrolled in this study were positive for both serum anti-PLA2R antibodies and PLA2R antigen staining in glomeruli.
Another difference between our study and previously reported HLA associations in MN is that we captured and sequenced the full HLA region, which allowed us to analyze the associations for all high-resolution HLA alleles.23 There is no evidence that novel rare HLA alleles are associated with the development of PLA2R-related MN, because we did not detect any novel HLA alleles in patients during the discovery study and only identified several novel HLA alleles in the replication study. Notably, although all copies of chromosome 6 in the population contain the DRB1 locus, most (but not all) DRB loci have an expressed second DRB locus, which can include DRB3, DRB4, or DRB5. The loci of HLA-DRB3/4/5 are in close proximity to those of HLA-DRB1; thus, the alleles between HLA-DRB3/4/5 and DRB1 are tightly linked. For example, HLA-DRB1*15:01 is almost exclusively linked with HLA-DRB5*01:01 in most ethnic populations, which explains the most significant signal observed in the gene expression profile of microdissected glomeruli in our previous study. The full sequencing of the entire HLA genome was important for identifying the independent risk allele HLA-DRB3*02:02. Recently, two other studies used the candidate gene approach and showed that HLA-DRB1*1501 is an HLA risk allele in Chinese and Japanese populations.9,10 Because the typing of the HLA-DRB3 gene was not performed, the risk of HLA-DRB3*02:02 was not identified in these two studies.
Recently, a GWAS of MN showed that the HLA-DRB1*03:01-DQA1*05:01 haplotype, which is linked with HLA-DRB3*02:02, was associated with MN in European whites; however, HLA-DRB1*15:01 was not identified in that study.16 Several reasons might explain the different findings between this study and our results. First, there might be different risk HLA alleles between Asian and European white patients, because previous studies also showed that HLA-DRB1*15:01 is associated with IMN in both Chinese and Japanese patients.9,10 Second, the study of European whites did not exclude patients with PLA2R-unrelated MN. Our data show that the HLA risk alleles are different between patients with PLA2R-related MN and those with PLA2R-unrelated MN. Also, our data show that PLA2R-unrelated MN is associated with HLA-DRB3*02:02 but not HLA-DRB1*15:01. Third, another possible reason is that the age of patients enrolled is different between the two studies. Patients were 52.5±13.3 years old at the time of diagnosis in the study of European whites, which was obviously older than age of patients enrolled in this study (43.4±16.3 year old).16 We found that patients carrying HLA-DRB1*15:01−DRB3*02:02+ are most likely to develop the disease at approximately 55 years of age, whereas patients carrying HLA-DRB1*15:01+DRB3*02:02− are more likely to develop the disease at a younger age.
As is the case in other autoimmune diseases, most individuals who carry high-risk alleles do not develop PLA2R-related MN. Therefore, other environmental factors or precipitating events must converge to trigger the disease. Precipitating events might include incidental infection and/or purely stochastic processes that lead to the emergence and expansion of T and B cells with their respective disease-causing T and B cell receptor repertoires.24 The structure of HLA molecules is important for understanding of the pathogenesis of autoimmune diseases, because HLA molecules determine which fragments of an autoantigen will be presented on the surface of antigen-presenting cells for subsequent recognition by T cells. SNPs in PLA2R1 on chromosome 2 have been previously reported to be associated with the occurrence of IMN, and a strong interaction between HLA and PLA2R1 was observed in this disease.11 In this study, we also genotyped the SNP rs35771982 within PLA2R1, which encodes an amino acid substitution (His300Asp) in the first C-type lectin-like domain of PLA2R. We found that as many as 99.5% of patients with PLA2R-related MN carried genotype GC or GG at rs35771982 compared with 91.4% of healthy controls (Supplemental Tables 9–11). Recently, studies showed no difference in the binding of anti-PLA2R antibodies to wild-type PLA2R and His300Asp-PLA2R.25,26 Meanwhile, rare genetic variants within PLA2R1 are unlikely to contribute to the pathogenesis of MN.27 These findings indicate that polymorphisms of PLA2R1 are not related to the B cell epitope; instead, these polymorphisms could be related to the T cell epitope in the development of the anti-PLA2R autoantibodies. Future studies are needed to determine the T cell epitope of PLA2R when HLA-DRB1*15:01, HLA-DRB3*02:02, or both risk alleles are present.
It is important to notice that our findings are on the basis of the population genetics of Han Chinese patients and need validation in various populations. It remains unknown whether HLA-DRB1*15:01 is also an HLA risk allele for young-onset PLA2R-related MN in European whites. Moreover, the relationship between the two HLA risk alleles and the development of the disease is complex due to the high degree of LD between alleles in the HLA region. In most ethnic populations, HLA-DRB1*15:01 is strongly linked with HLA-DRB5*01:01-DQA1*0102-DQB1*0602, whereas HLA-DRB3*02:02 is strongly linked with HLA-DRB1*03:01-DQA1*05:01-DQB1*02:01. Although all HLA loci were sequenced in the discovery cohort and HLA-DRB1*15:01 and HLA-DRB3*02:02 had stronger effects than the other alleles in both discovery and validation cohorts, direct biologic evidence of the causal HLA alleles is necessary to further elucidate this phenomenon. In addition, we also found that HLA-DRB3*02:02 is the most significant signal for PLA2R-unrelated MN. However, this association requires replication in larger cohorts of various ethnic populations, because there were various autoantigens in PLA2R-unrelated MN, and each autoantigen might have its own particular HLA risk alleles. Thus, the HLA risk alleles in PLA2R-unrelated MN need additional investigation, especially if the unknown podocyte autoantigens were identified in the future.
In conclusion, we found that the HLA-DRB1*15:01 and HLA-DRB3*02:02 alleles are independently and strongly associated with PLA2R-related MN in the Chinese population. Virtually all patients with PLA2R-related MN carry either or both of the HLA-DRB1*15:01 and HLA-DRB3*02:02 alleles, suggesting that having either the HLA-DRB1*15:01 or HLA-DRB3*02:02 allele may be necessary but not sufficient for PLA2R-related MN development in the Chinese population. Additional direct biologic evidence of the causal HLA alleles should be obtained in the future.
Concise Methods
Participants and Samples
In total, 573 patients who were diagnosed with biopsy-proven IMN between December of 2011 and December of 2013 were screened for this study from the Nanjing Glomerulonephritis Registry Study (Supplemental Figure 2). All of these patients were clinically ruled out for secondary MN, including positive antinuclear antibodies, hepatitis B virus infection, and cancer. Patients who were positive for both serum anti-PLA2R antibodies (>20 RU/ml) and glomerular PLA2R antigen staining were defined as PLA2R-related MN in this study, whereas patients who were negative for both markers were defined as PLA2R-unrelated MN. Serum anti-PLA2R antibody levels were detected by ELISA, and glomerular PLA2R antigen staining was detected by immunofluorescence as described previously.28 To further reduce the heterogeneity, 131 patients negative for serum anti-PLA2R antibodies but positive for glomerular PLA2R antigen staining and five patients positive for serum anti-PLA2R antibodies but negative for glomerular PLA2R antigen staining were also excluded in this study. Five patients were excluded due to lack of a DNA sample, and two patients were excluded because of the low DNA quality. Finally, a total of 392 patients with PLA2R-related MN (99 patients in the discovery analysis and 293 patients in the replication analysis) and 50 patients with PLA2R-unrelated MN were enrolled in this study. A total of 385 ethnically matched healthy control samples were also include from the biobank (100 individuals in the discovery analysis and 285 in the replication analysis) (Supplemental Table 2). The research protocol was approved by the Institutional Review Board of the Jinling Hospital, Nanjing University and conducted according to the principles of the Declaration of Helsinki. Written informed consent was obtained from all participants. All human renal and blood specimens were obtained from the Biobank of the National Clinical Research Center of Kidney Diseases at Jinling Hospital.
HLA Target Sequence, HLA Typing, Variant Calling, and Quality Control
All samples in the discovery cohort were captured by using an HLA array.18,23 The HLA region located at 6p21.3 contains >200 genes, including the HLA class 1 genes (HLA-A, HLA-B, and HLA-C), class 2 genes (HLA-DRB1 and HLA-DQB1), and other nonclassic HLA genes.29,30 We sequenced the target to 99-fold with a coverage of 97% of the HLA region in 199 samples (Supplemental Table 3). Target sequencing was performed on the HiSEquation 2000 platform to generate 100-bp paired end reads.
HLA typing in the discovery cohort was performed by SOAPHLA.18,23 The following HLA genes were typed: HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, HLA-G, HLA-H, HLA-J, HLA-K, HLA-L, HLA-P, HLA-V, HLA-DRA, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1, HLA-DPB1, HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-MICA, HLA-MICB, HLA-TAP1, and HLA-TAP2; those are the most polymorphic genes in the HLA region. The amino acid sequence of each HLA allele was determined according to the IMGT/HLA database (Release 3.22.0). Additionally, HLA-DRB3, DRB4, and DRB5 were typed by using an algorithm in SOAPHLA.18 Notably, only one or none of the HLA-DRB3, HLA-DRB4, and HLA-DRB5 haplotypes could exist on the same chromosome, indicating the presence of copy number variations in the HLA-DRB3/4/5 loci. The determination of copy number variations for HLA-DRB3/4/5 was on the basis of a ratio that was calculated by using the mean depth of one gene in contrast to the mean depth of the entire HLA region. According to the MHC haplotypes project, HLA-DRB3 and HLA-DRB4 do not exist in PGF haplotypes; thus, we located the variation of the positions of HLA-DRB3 and HLA-DRB4 relative to the distance to HLA-DRB1. In the replication study, five selected HLA genes (HLA-DRB1, HLA-DQB1, HLA-DQA1, HLA-DRB3, and HLA-DRB5) were typed using the Sanger platform in BGI-Shenzhen.
The samples were aligned to the NCBI human genome reference assembly (GRCh37) by using BWA (version 0.5.9). The parameters were “-o 1 -e 63 -i 15 -L -k 2 -l 31 -t 2 -q 10 –I.” The BAM file was produced using SAMtools (version 0.1.17), and GATK (version 1.4) was used to precisely call SNPs and InDels to perform a realignment around known InDels identified by the 1000 Genomes Project. After the initial variant calls were generated, we enacted a strict filtering to identify credible SNPs and InDels using the following criteria: (1) a call rate of 90% (sites with a depth four or more times were treated as high quality, whereas those not meeting this criterion were considered to be missing); (2) a major allele frequency >0.01; and (3) a Hardy–Einberg test P value > 1×10−4 in the controls. Annovar31 was used to annotate the SNPs and InDels according to their genetic locations and expected effects on encoded gene products on the basis of information from the RefSeq database.
Statistical Analyses
Logistic regression with dominant, and multiplicative models were used to test the association of the HLA alleles, SNPs, and amino acid variants in the discovery study. For each HLA allele and amino acid variant in the dominant model, the association was analyzed with the use of a group of biallelic tests for every HLA allele and amino acid, which were marked as present or absent.32 To assess candidate independent effects outside of the HLA alleles identified in the univariate logistic analysis, we included the most significant HLA alleles as covariates to test all of the other markers in a multivariate logistic analysis. If other independently associated HLA alleles were identified, we further included them as additional covariates in subsequent multivariate analyses to identify additional independent effects. The Bonferroni correction was used to control for multiple testing; the association tests applied for the HLA alleles were adjusted at P< 1×10−5 and the association tests applied for the SNPs and amino acid variants were adjusted at P<1×10−7. All association analyses were performed by means of PLINK software (version 1.07). The association analysis for the baseline demographic and clinical data from the time of renal biopsy included eGFR, albumin levels, 24-hour protein levels, and the serum anti-PLA2R levels. Nonparametric variables are expressed as median (range or interquartile range) and were compared using either the Mann–Whitney or the Kruskal–Wallis test. Qualitative data are described as percentages and were analyzed using either the chi-squared or the Fisher exact test.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank the physicians, research nurses, patients, and healthy volunteers who contributed to this study.
The authors acknowledge the support from National Basic Research Program of China 973 Program 2012CB517600 grant 2012CB517606, Natural Science Foundation of China grants 81500547 and 81500556, Natural Science Foundation of Jiangsu Province grant BK20150560, Major International (Regional) Joint Research Project grant 81320108007, Major Research Plan of the National Natural Science Foundation grant 91442104, and Natural Science Foundation of Jinling Hospital grant 2013041.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Genetic Complexities of the HLA Region and Idiopathic Membranous Nephropathy,” on pages 1331–1334.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016060644/-/DCSupplemental.
References
- 1.Debiec H, Ronco P: PLA2R autoantibodies and PLA2R glomerular deposits in membranous nephropathy. N Engl J Med 364: 689–690, 2011 [DOI] [PubMed] [Google Scholar]
- 2.Qin W, Beck LH Jr., Zeng C, Chen Z, Li S, Zuo K, Salant DJ, Liu Z: Anti-phospholipase A2 receptor antibody in membranous nephropathy. J Am Soc Nephrol 22: 1137–1143, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronco P, Debiec H: Pathogenesis of membranous nephropathy: Recent advances and future challenges. Nat Rev Nephrol 8: 203–213, 2012 [DOI] [PubMed] [Google Scholar]
- 4.Sollid LM, Pos W, Wucherpfennig KW: Molecular mechanisms for contribution of MHC molecules to autoimmune diseases. Curr Opin Immunol 31: 24–30, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salant DJ: Genetic variants in membranous nephropathy: Perhaps a perfect storm rather than a straightforward conformeropathy? J Am Soc Nephrol 24: 525–528, 2013 [DOI] [PubMed] [Google Scholar]
- 6.Vaughan RW, Demaine AG, Welsh KI: A DQA1 allele is strongly associated with idiopathic membranous nephropathy. Tissue Antigens 34: 261–269, 1989 [DOI] [PubMed] [Google Scholar]
- 7.Klouda PT, Manos J, Acheson EJ, Dyer PA, Goldby FS, Harris R, Lawler W, Mallick NP, Williams G: Strong association between idiopathic membranous nephropathy and HLA-DRW3. Lancet 2: 770–771, 1979 [DOI] [PubMed] [Google Scholar]
- 8.Ogahara S, Naito S, Abe K, Michinaga I, Arakawa K: Analysis of HLA class II genes in Japanese patients with idiopathic membranous glomerulonephritis. Kidney Int 41: 175–182, 1992 [DOI] [PubMed] [Google Scholar]
- 9.Xie L-j, Qu Z, Cui Z, Liu G, Liao Y-h, Zhao M-h: The susceptible human leukocyte antigen class II genes and the encoding amino acid residues on major histocompatibility complex molecules to primary membranous nephropathy [Abstract]. J Am Soc Nephrol 26: 392A:FR-PO159, 2015 [Google Scholar]
- 10.Honda K, Thir M, Okamoto K, Doi K, Hodaka, Suzuki, Watanabe T, Nangaku M, Tokunaga K, Noiri E: Interaction of risk alleles in Japanese idiopathic membranous nephropathy [Abstract]. J Am Soc Nephrol 26: 739A26:SA-PO502, 2015 [Google Scholar]
- 11.Stanescu HC, Arcos-Burgos M, Medlar A, Bockenhauer D, Kottgen A, Dragomirescu L, Voinescu C, Patel N, Pearce K, Hubank M, Stephens HA, Laundy V, Padmanabhan S, Zawadzka A, Hofstra JM, Coenen MJ, den Heijer M, Kiemeney LA, Bacq-Daian D, Stengel B, Powis SH, Brenchley P, Feehally J, Rees AJ, Debiec H, Wetzels JF, Ronco P, Mathieson PW, Kleta R: Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. N Engl J Med 364: 616–626, 2011 [DOI] [PubMed] [Google Scholar]
- 12.Kiryluk K: Risk alleles in idiopathic membranous nephropathy. N Engl J Med 364: 2072–2073, 2011 [DOI] [PubMed] [Google Scholar]
- 13.Ronco P, Debiec H: Pathophysiological advances in membranous nephropathy: Time for a shift in patient’s care. Lancet 385: 1983–1992, 2015 [DOI] [PubMed] [Google Scholar]
- 14.Debiec H, Guigonis V, Mougenot B, Decobert F, Haymann JP, Bensman A, Deschênes G, Ronco PM: Antenatal membranous glomerulonephritis due to anti-neutral endopeptidase antibodies. N Engl J Med 346: 2053–2060, 2002 [DOI] [PubMed] [Google Scholar]
- 15.Tomas NM, Beck LH Jr., Meyer-Schwesinger C, Seitz-Polski B, Ma H, Zahner G, Dolla G, Hoxha E, Helmchen U, Dabert-Gay AS, Debayle D, Merchant M, Klein J, Salant DJ, Stahl RA, Lambeau G: Thrombospondin type-1 domain-containing 7A in idiopathic membranous nephropathy. N Engl J Med 371: 2277–2287, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekula P, Li Y, Stanescu HC, Wuttke M, Ekici AB, Bockenhauer D, Walz G, Powis SH, Kielstein JT, Brenchley P, Eckardt KU, Kronenberg F, Kleta R, Köttgen A, Köttgen A; GCKD Investigators : Genetic risk variants for membranous nephropathy: Extension of and association with other chronic kidney disease aetiologies [published online ahead of print February 4, 2016]. Nephrol Dial Transplant 10.1093/ndt/gfw1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sham PC, Purcell SM: Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet 15: 335–346, 2014 [DOI] [PubMed] [Google Scholar]
- 18.Zhou F, Cao H, Zuo X, Zhang T, Zhang X, Liu X, Xu R, Chen G, Zhang Y, Zheng X, Jin X, Gao J, Mei J, Sheng Y, Li Q, Liang B, Shen J, Shen C, Jiang H, Zhu C, Fan X, Xu F, Yue M, Yin X, Ye C, Zhang C, Liu X, Yu L, Wu J, Chen M, Zhuang X, Tang L, Shao H, Wu L, Li J, Xu Y, Zhang Y, Zhao S, Wang Y, Li G, Xu H, Zeng L, Wang J, Bai M, Chen Y, Chen W, Kang T, Wu Y, Xu X, Zhu Z, Cui Y, Wang Z, Yang C, Wang P, Xiang L, Chen X, Zhang A, Gao X, Zhang F, Xu J, Zheng M, Zheng J, Zhang J, Yu X, Li Y, Yang S, Yang H, Wang J, Liu J, Hammarström L, Sun L, Wang J, Zhang X: Deep sequencing of the MHC region in the Chinese population contributes to studies of complex disease. Nat Genet 48: 740–746, 2016 [DOI] [PubMed] [Google Scholar]
- 19.Gragert L, Madbouly A, Freeman J, Maiers M: Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum Immunol 74: 1313–1320, 2013 [DOI] [PubMed] [Google Scholar]
- 20.Zhou XY, Zhu FM, Li JP, Mao W, Zhang DM, Liu ML, Hei AL, Dai DP, Jiang P, Shan XY, Zhang BW, Zhu CF, Shen J, Deng ZH, Wang ZL, Yu WJ, Chen Q, Qiao YH, Zhu XM, Lv R, Li GY, Li GL, Li HC, Zhang X, Pei B, Jiao LX, Shen G, Liu Y, Feng ZH, Su YP, Xu ZX, Di WY, Jiang YQ, Fu HL, Liu XJ, Liu X, Zhou MZ, Du D, Liu Q, Han Y, Zhang ZX, Cai JP: High-resolution analyses of human leukocyte antigens allele and haplotype frequencies based on 169,995 volunteers from the China Bone Marrow Donor Registry Program. PLoS One 10: e0139485, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacRae CA, Vasan RS: Next-generation genome-wide association studies: Time to focus on phenotype? Circ Cardiovasc Genet 4: 334–336, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manchia M, Cullis J, Turecki G, Rouleau GA, Uher R, Alda M: The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS One 8: e76295, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao H, Wu J, Wang Y, Jiang H, Zhang T, Liu X, Xu Y, Liang D, Gao P, Sun Y, Gifford B, D’Ascenzo M, Liu X, Tellier LC, Yang F, Tong X, Chen D, Zheng J, Li W, Richmond T, Xu X, Wang J, Li Y: An integrated tool to study MHC region: Accurate SNV detection and HLA genes typing in human MHC region using targeted high-throughput sequencing. PLoS One 8: e69388, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koning F, Thomas R, Rossjohn J, Toes RE: Coeliac disease and rheumatoid arthritis: Similar mechanisms, different antigens. Nat Rev Rheumatol 11: 450–461, 2015 [DOI] [PubMed] [Google Scholar]
- 25.Kao L, Lam V, Waldman M, Glassock RJ, Zhu Q: Identification of the immunodominant epitope region in phospholipase A2 receptor-mediating autoantibody binding in idiopathic membranous nephropathy. J Am Soc Nephrol 26: 291–301, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seitz-Polski B, Dolla G, Payré C, Girard CA, Polidori J, Zorzi K, Birgy-Barelli E, Jullien P, Courivaud C, Krummel T, Benzaken S, Bernard G, Burtey S, Mariat C, Esnault VL, Lambeau G: Epitope spreading of autoantibody response to PLA2R associates with poor prognosis in membranous nephropathy. J Am Soc Nephrol 27: 1517–1533, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coenen MJ, Hofstra JM, Debiec H, Stanescu HC, Medlar AJ, Stengel B, Boland-Augé A, Groothuismink JM, Bockenhauer D, Powis SH, Mathieson PW, Brenchley PE, Kleta R, Wetzels JF, Ronco P: Phospholipase A2 receptor (PLA2R1) sequence variants in idiopathic membranous nephropathy. J Am Soc Nephrol 24: 677–683, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin HZ, Zhang MC, Le WB, Ren Q, Chen DC, Zeng CH, Liu L, Zuo K, Xu F, Liu ZH: Combined assessment of phospholipase A2 receptor autoantibodies and glomerular deposits in membranous nephropathy. J Am Soc Nephrol 27: 3195–3203, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiina T, Hosomichi K, Inoko H, Kulski JK: The HLA genomic loci map: Expression, interaction, diversity and disease. J Hum Genet 54: 15–39, 2009 [DOI] [PubMed] [Google Scholar]
- 30.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC Jr., Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S: Gene map of the extended human MHC. Nat Rev Genet 5: 889–899, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Wang K, Li M, Hakonarson H: ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang FR, Liu H, Irwanto A, Fu XA, Li Y, Yu GQ, Yu YX, Chen MF, Low HQ, Li JH, Bao FF, Foo JN, Bei JX, Jia XM, Liu J, Liany H, Wang N, Niu GY, Wang ZZ, Shi BQ, Tian HQ, Liu HX, Ma SS, Zhou Y, You JB, Yang Q, Wang C, Chu TS, Liu DC, Yu XL, Sun YH, Ning Y, Wei ZH, Chen SL, Chen XC, Zhang ZX, Liu YX, Pulit SL, Wu WB, Zheng ZY, Yang RD, Long H, Liu ZS, Wang JQ, Li M, Zhang LH, Wang H, Wang LM, Xiao P, Li JL, Huang ZM, Huang JX, Li Z, Liu J, Xiong L, Yang J, Wang XD, Yu DB, Lu XM, Zhou GZ, Yan LB, Shen JP, Zhang GC, Zeng YX, de Bakker PI, Chen SM, Liu JJ: HLA-B*13:01 and the dapsone hypersensitivity syndrome. N Engl J Med 369: 1620–1628, 2013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.