Abstract
The R-diastereomer of phosphorothioate analogs of cGMP, Rp-cGMPS, is one of few known inhibitors of cGMP-dependent protein kinase I (PKG I); however, its mechanism of inhibition is currently not fully understood. Here, we determined the crystal structure of the PKG Iβ cyclic nucleotide-binding domain (PKG Iβ CNB-B), considered a ‘gatekeeper’ for cGMP activation, bound to Rp-cGMPS at 1.3 Å. Our structural and NMR data show that PKG Iβ CNB-B bound to Rp-cGMPS displays an apo-like structure with its helical domain in an open conformation. Comparison with the cAMP-dependent protein kinase regulatory subunit (PKA RIα) showed that this conformation resembles the catalytic subunit-bound inhibited state of PKA RIα more closely than the apo- or Rp-cAMPS-bound conformations. These results suggest that Rp-cGMPS inhibits PKG I by stabilizing the inactive conformation of CNB-B.
Keywords: cGMP dependent protein kinase, PKG; Second messengers; NO-cGMP signaling; Kinase inhibition
1. Introduction
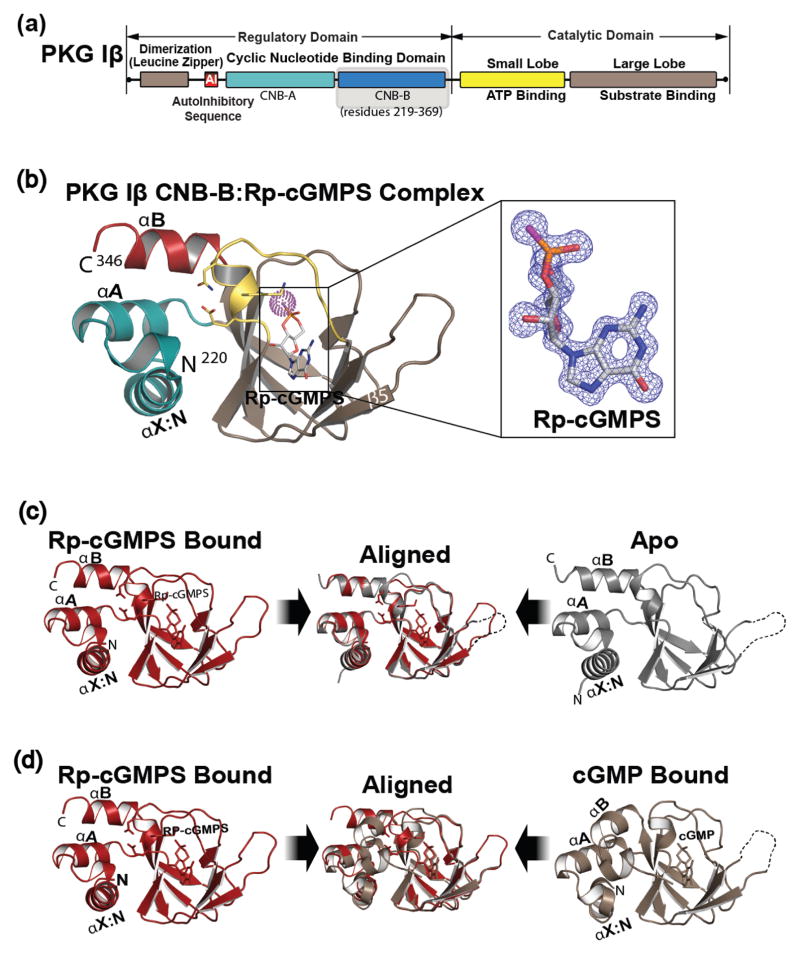
Cyclic GMP is a crucial second messenger that enables translation of extracellular signals into physiological responses[1–3]. As one of the key cGMP receptors, cGMP dependent protein kinase I (PKG) regulates smooth muscle tone, platelet aggregation and memory formation [2,3]. PKG I is a homodimer protein, each monomer of PKG I consists of N-terminal regulatory (R) and C-terminal catalytic (C) domains [4] (Fig. 1a). The R-region contains tandem cyclic nucleotide binding domains (CNB-A and B) with different affinities for cGMP and its analogs[5,6]. While two splice variants, α and β, exist for PKG I, they share the same CNB and C-domains. The CNB domain includes a dynamic α- and a rigid β-subdomains [7,8]. The α subdomain includes the N3A motif and variable numbers of helices, which flank the β subdomain [9,10]. The β subdomain consists of 8 strands that form a barrel and contains a conserved structural motif that specifically binds the cyclic phosphate ribose moiety, referred to as the Phosphate Binding Cassette (PBC) [9,10].
Fig. 1.
Domain organization and overall structure of PKG Iβ CNB-B:Rp-cGMPS complex. (a), domain organization of PKG Iβ with CNB-B domain highlighted. (b), overall structure of PKG Iβ CNB-B (219–351) bound with Rp-cGMPS. The secondary structure elements are labeled. The PBC is colored in yellow, the αB helix is in red, the N-terminal helices are in teal, and the β barrel is in light brown. Rp-cGMPS is colored by atom type. The N and C termini are labeled with their corresponding residue numbers seen in the final model. Rp-cGMPS is shown as sticks, except for sulfur which is highlighted with purple dots. The atoms are colored as follows: carbons in cGMP, white; oxygen, red; nitrogen, blue; sulfur, purple. Inset shows the bound Rp-cGMPS with an |Fo-Fc| omit map (contoured at 1.0 σ level). (c), structural comparison with the apo state. The Rp-cGMPS bound colored in red is shown on left and the apo colored in grey is shown on right. The disordered β4-β5 loop is indicated with a dotted line. Two structures are aligned at the β-barrel region and shown in the middle panel. (d), structural comparison with the cGMP bound state. The Rp-cGMP, colored red and the cGMP bound, colored tan. The middle panel shows their alignment. All structure images were generated using PyMOL (Schrödinger, LLC).
The R-diastereomer of the phosphorothioate analogs of cGMP and cAMP, Rp-cGMPS and Rp-cAMPS, are well known inhibitors of PKG and PKA, respectively [5,11,12]. These Rp-isomers have an equatorial exocyclic sulfur instead of oxygen and due to their antagonistic effects are predicted to bind the cyclic nucleotide pockets without inducing the conformational changes required for kinase activation. Furthermore, structural studies of interactions with PKA and Rp-cAMPS suggest that Rp-cAMPS acts as an inverse agonist to increase the interaction between the R- and C- subunits[13]. Rp-cAMPS selectively binds the CNB domains in a C-subunit bound conformation for its antagonist effect [13]. NMR studies on Rp-cAMPS interactions with PKA and exchange protein activated by cAMP (EPAC) showed that Rp-cAMPS binding alters the allosteric network within the CNB domains and ultimately prevents the propagation of the activation signal out of the domain [7,8,14]. In PKA in particular, Rp-cAMPS binding makes the PBC and adjacent β2–3 more dynamic compared to cAMP or Sp-cAMPS (the S-enantiomer) bound states and, unlike cAMP, stabilizes the inactive state of the CNB-A [7,8]. This similarity between the Apo and Rp-cAMPS bound state in PKA has also been confirmed by hydrogen deuterium exchange mass spectrometry [13].
Recent structural studies highlight the critical role of cGMP binding to CNB-B in activation of PKG I [15,16]. Studies showed that binding of cGMP to CNB-B is required for the large conformational changes associated with the full activation of PKG I [15]. This domain is about 200 fold selective for cGMP compared to cAMP. Mutating key cGMP contact residues in CNB-B significantly increases activation constants for the full length PKG I, demonstrating its critical role in cGMP selective activation [15]. Thus, these findings suggest that Rp-cGMPS binding to CNB-B is critical for the inhibition of PKG I. Rp-cGMPS inhibits activation of native PKG I with a Ki value of 20 μM[17]. However, with no structural information describing the PKG-Rp-cGMPS interaction available, its inhibition mechanism is currently not known.
2. Materials and Methods
2.1. Protein expression, purification, and crystallization
PKG Iβ 219–351 was expressed and purified as previously described for PKG Iβ (219–369) [15,18]. Sequences encoding PKG Iβ 219–369 were cloned into pQTEV using a template containing full-length PKG Iβ (GenBank ID ABQ59040.1, coding for UniProt protein sequence Q13976-2). The purity of the resulting samples was analyzed by SDS-PAGE. For SPR measurement the hepta-histidine tag was not cleaved, prior to the size exclusion chromatography step. The crystallization was performed using the hanging drop method and 400 nL drops. The crystallization solution (0.8 M lithium sulfate monohydrate, 0.1 M sodium acetate trihydrate, pH 4.0, 4% (v/v) polyethylene glycol 200) was mixed 1:1 with freshly purified protein at 50 mg/mL with 10 mM Rp-cGMPS. Crystals grew at 4°C within a week. Crystals were cryoprotected for 5 minutes in the crystallization condition supplemented with 33% ethylene glycol and then flash frozen in liquid nitrogen. Diffraction experiments were performed at the Advanced Light Source (Berkeley, CA). Data were processed using iMOSFLM and resolution limits were set based on CC1/2 [19,20]. Molecular replacement was performed using PHASER and apo PKG Iβ CNB-B structure (PDB ID:4KU8) as a search model[21]. Models were manually built and refined using PHENIX and COOT [22,23]. The experimental structure factors and refined coordinates have been deposited in the Protein Data Bank (PDB) with accession code 5L0N.
2.2. NMR spectroscopy
Samples of apo, Rp-cGMPS bound, and cGMP bound PKG Iβ (219–369) were prepared in 50 mM Tris, pH 7.0, 100 mM NaCl, 1 mM DTT, and 0.02% (w/v) NaN3, and two-dimensional (1H,15N)-HSQC NMR spectra were acquired. A protein concentration of 20 μM was used for all HSQC analyses, and 80 μM of 15N-labelled N-acetylglycine was added to the samples as an internal reference for subsequent alignment of the HSQC spectra with one another. All HSQC overlays were performed using Sparky [University of California, San Francisco], with 15N-labelled N-acetylglycine as an internal reference for spectrum alignment [24]. Sufficient concentrations of Rp-cGMPS and cGMP were used to achieve saturated ligand binding, i.e. > 1 mM, thus minimizing the influence of differing ligand affinities on the final analysis. All NMR spectra were acquired at 306 K with a Bruker Avance 700-MHz NMR spectrometer equipped with a 5 mm TCI cryoprobe. The spectra were processed with NMRPipe, and analyzed using Sparky [24,25]. Peak assignments were obtained from standard three-dimensional triple-resonance NMR spectra (i.e. HNCACB, CBCA(CO)NH, HNCA, HN(CO)CA), using the automated PINE-NMR server to facilitate the assignment process [26].
2.3. Surface plasmon resonance
Surface plasmon resonance (SPR) experiments were performed on a Pioneer FE (Oklahoma City, OK, USA) using OneStep™ injections. OneStep™ injections create a continuous concentration gradient of analyte during the association phase and are a reliable alternative to fixed concentration injections [27,28]. Instead of a full dilution series as with other SPR instrumentation, OneStep™ injections generate analyte concentrations gradients which fully characterize kinetic and affinity interactions. A his-tagged PKG Iβ CNB-B (219–351) surface was prepared using standard Ni-NTA capture-coupling protocols and amine coupling with HisCap chips (Oklahoma City, OK, USA) [29]. A surface of ~3400 RU was generated to minimize mass transport and rebinding artifacts. OneStep™ injections were performed in triplicate from 0–10 μM cGMP and 0–100 μM Rp-cGMPS using HBS-T buffer (10 mM HEPES, pH 7.5, 150 mM NaCl, and 0.005% Tween-20) at a flow rate of 60 uL min−1 using 50% of the dispersion loop volume. 3% (w/v) sucrose was also prepared in running buffer as a diffusion standard. Double referenced, experimental data were fit to a mass transport limited model using Qdat software (Oklahoma City, OK, USA). No mass transport was observed in the Rp-cGMPS curves.
2.4 Kinase activity assays
Kinase activity was determined using a microfluidic mobility-shift assay on a Caliper DeskTop Profiler (PerkinElmer, Waltham, MA, USA), as previously described [18]. Activation of PKG Iβ in dependence of cGMP or Rp-cGMPS, respectively, was determined. cGMP activates PKG with an activation constant (Ka) of 537 ± 10 nM (n=2), while PKG Iβ remains inactive in the presence of Rp-cGMPS. 5 nM FLAG-hPKG Iβ was incubated with varying concentrations of cGMP or Rp-cGMPS, respectively, in assay buffer (20 mM MOPS, 150 mM NaCl, 1 mM ATP, 10 mM MgCl2, 0.1 mg/ml BSA, 0.05% L-31, and 1 mM DTT, pH 7.0) with 990 μM Kemptide and 10 μM FITC-Kemptide in a 384-well assay plate (Corning, low volume, non-binding surface) for 2 h at room temperature. Substrate and product peaks were separated using a LabChip EZ reader 4-sipper ProfilerPro™ chip (PerkinElmer, Waltham, MA, USA) in off-chip mode with downstream and upstream voltages of −150 V and −1800 V, respectively, and a screening pressure of −1.7 psi. Rp-cGMPS competitively inhibits cGMP-dependent activation of PKG Iβ. Partially cGMP-activated PKG Iβ can be inactivated in the presence of micromolar concentrations of Rp-cGMPS (Ki= 49 ± 10 μM (n=2)). 1 U (enzymatic unit) is equal to 1 μmol substrate conversion per min. Each data point depicts the mean of duplicate measurements, while error bars show the standard error of the mean (SEM). 5 nM FLAG-hPKG Iβ was incubated with 1 μM cGMP and various concentrations of Rp-cGMPS in assay buffer in a 384-well assay plate for 2 h and substrate conversion was determined as described above. Substrate conversion was plotted against the logarithmic cyclic nucleotide concentration and activation constants (Ka) and inhibition constants (Ki) were determined from sigmoidal dose-response curves employing GraphPad Prism 6.01.
3. Results and Discussion
3.1 Structure determination
To understand how Rp-cGMPS binds CNB-B of PKG Iβ, we solved a crystal structure of their complex at 1.3 Å resolution (Table 1 and Fig. 1B). The PKG Iβ CNB-B : Rp-cGMPS complex was crystallized in the P212121 space group with one molecule per asymmetric unit. The final model includes the entire CNB-B domain used for crystallization (residues 219–351) with the exception of the αC helix (residues 344–351), due to missing electron density. Within its cGMP pocket there is clear electron density for Rp-cGMPS and a neighboring sulfate molecule, due to the high sulfate in the crystallization solution. The structure shows the CNB-B domain in an apo like conformation when bound to Rp-cGMPS [15] (Fig. 1c). It shows an open PBC interacting with the N3A motif (in) while the αB helix at the C-terminus positions away (up) from the PBC and the αC helix is disordered as seen in the apo structure (PDB ID: 4KU8). This contrasts the cGMP bound conformation where the PBC becomes more compact (closed), the αB/C helices come down toward the PBC (down) as the αC helix becomes ordered and N3A moves away from the PBC (out) [15] (Fig. 1d).
Table 1.
Data collection and refinement statistics
| PKG Iβ CNB-B: Rp-cGMPS | |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.97741 |
| Space group | P 21 21 21 |
| Cell dimensions | |
| a,b,c (Å) | 39.9, 54.3, 56.4 |
| α,β,δ (°) | 90.00 90.00 90.00 |
| Resolution (Å) | 39.8-1.29 |
| Rsym or Rmerge | 0.050 (0.470) |
| I/σI | 18.4 (2.9) |
| CC1/2a,b | 0.999 (0.915) |
| Completeness (%) | 99.8 (99.9) |
| Redundancy | 8.0 (7.0) |
| Refinement | |
| Resolution (Å) | 32.577-1.285 |
| No. reflections | 31845 |
| Rwork/Rfreec | 0.182/0.167 |
| No. atoms | |
| Proteins | 1937 |
| Ligand/ion | 70 |
| Water | 141 |
| B-factors | |
| Protein | 19.7 |
| Ligand/ion | 32.2 |
| Water | 30.2 |
| r.m.s.d deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.0 |
| PDB ID | 5L0N |
Highest resolution shell is shown in parenthesis.
CC1/2, Pearson correlation coefficient.
5.0% of the observed intensities was excluded from refinement for cross validation purposes.
r.m.s., root mean square.
3.2 Detailed interactions of the PBC residues
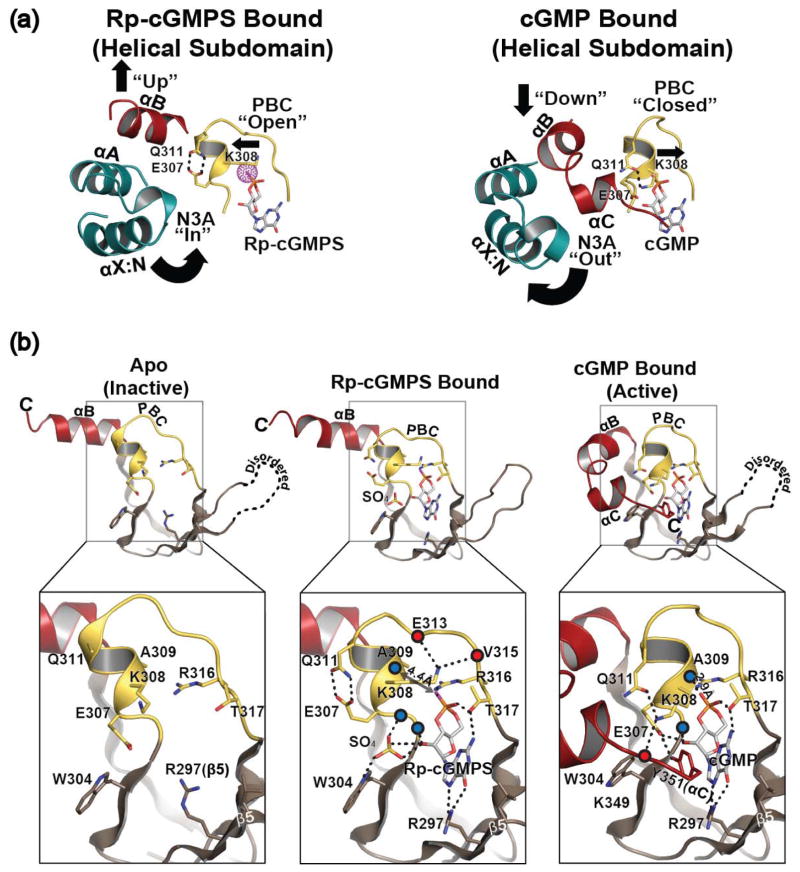
Although interactions with Rp-cGMPS, especially between the guanine moiety and base binding region, are almost identical to those seen when cGMP bound, The Rp-cGMPS bound structure shows significant rearrangement of the hydrogen bonding pattern for PBC residues compared to those seen in the apo and cGMP bound conformations (Fig. 2). Firstly, the helical region of the PBC (αP) no longer interacts with Rp-cGMPS and the loop region of PBC, allowing the open conformation (Fig. 2a). In both cGMP and cAMP bound structures, the exposed backbone atoms of αP provide strong hydrogen bonds with Arg316 and cyclic nucleotides at the loop of the PBC, stabilizing the closed conformation [15] (Fig. 2b). These include a hydrogen bond between the backbone amide of Ala309 and the equatorial exocyclic oxygen. Additionally, the backbone carbonyl of Gly307 forms a hydrogen bond with the side chain of Arg316, which also binds the equatorial exocyclic oxygen of cyclic nucleotides. In the Rp bound conformation, replacing the equatorial exocyclic oxygen with sulfur disrupts these hydrogen bonds, leading to an opening of the PBC, similar to what is seen in the apo structure [15](Fig. 2b, middle panel). The Rp-cGMPS bound structure shows αP stretched away from the loop region of the PBC with the backbone atoms of Gly306 and Ala309 located 4 Å away from the side chain of Arg316 and the equatorial exocyclic sulfur respectively. Secondly, the structure shows that Glu307 no longer interacts with the ribose, but alternately with Gln311 through hydrogen bonds (Fig. 2). In the cGMP bound conformation, the side chain of Glu307 forms a hydrogen bond with 2′OH of the ribose, while the side chain of Gln311 interacts with the side chain of Lys308. Also, a sulfate anion occupies the same site that Glu307 in the cGMP bound structure used to occupy and interacts with the ribose and Trp304 through hydrogen bonds (Fig. 2b). In the cGMP bound conformation, the side chain of Lys308 points downward interacting with the side chains of Gln311 (PBC) and the backbone carbonyl of Lys349 (the C-terminal loop) through hydrogen bonds stabilizing the closed PBC [15]. In the apo structure, the side chain of Lys308 points toward solvent without any interacting partner while the side chain of Gln311 is disordered [15](Fig. 2b, left panel). In major contrast, the side chain of Lys308 flips towards the helical tip of the PBC, forming hydrogen bonds with the backbone carbonyl atoms of Glu313 and Val315 further stabilizing the open PBC in the Rp-cGMPS bound state.
Fig. 2.
Rp-cGMPS stabilizes the open PBC. (a), conformational changes upon cGMP binding. Only helical subdomain and PBC are shown, same color scheme as Fig. 1. PBC of PKG Iβ CNB-B bound to Rp-cGMPS, left and cGMP, right. Interactions of key residues shown as dotted lines. (b) comparison with apo and cGMP bound states shown with detailed interactions at PBC. For clarity only β4 through the C-terminus are shown. Same color scheme is used as Fig. 1, with the addition of blue and red spheres to represent the backbone amides and carbonyls respectively. Interactions of key residues are shown as dotted lines.
3.3 NMR studies
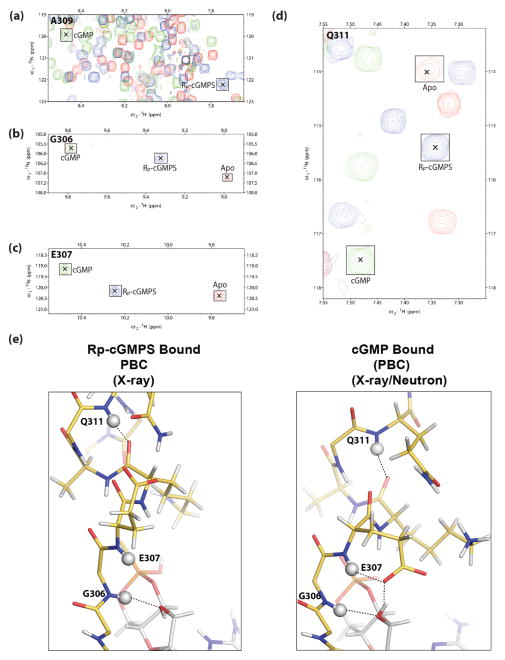
To further investigate if this apo like conformation exists in the presence of Rp-cGMPS in solution, we turned to NMR. Backbone assignments were obtained for selected, representative PBC residues (Fig. 3). Although complete assignment of the PBC in all three forms (i.e. apo, cGMP- and Rp-cGMPS-bound) was not possible for all PBC amino acids due to line-broadening and/or peak overlap, the assigned NMR cross-peaks indicate that in solution the Rp bound PBC exists in a distinct conformation from that of the cGMP bound state [30]. For example, Ala309 showed a proton shift from ~8.5ppm to ~7.7ppm when bound to Rp-cGMPS, consistent with the loss of the interaction between its backbone amide and the equatorial oxygen of cGMP and in agreement with our previous X-ray and joint X-ray/neutron crystal structures [15,18] (Fig. 3a). Gly306, Glu307, and Gln311 also showed clear cross peaks at intermediate positions between the apo (inactive) and cGMP (active) cross-peaks (Fig. 3b–e). Considering that exchange between the inactive and active states is fast on the chemical shift NMR time scale, the intermediate position of the Rp cross-peak between apo and cGMP points to a net shift in the Rp bound towards the inactive apo state [30]. These results strongly support that in solution Rp-cGMPS stabilizes the inactive apo-like conformation better than cGMP. However, it also shows that the conformational ensemble of the Rp-cGMPS bound CNB-B in solution might include a subset of active state conformers. These data agree with previous cyclic phosphorothioate NMR experiments, which found that Rp-cAMPS partially shifts the conformational equilibrium of the cAMP-binding domain-A of PKA towards an inhibitory C-bound like structure [7,31].
Fig. 3.
NMR confirms the stabilization of a unique PBC conformation by Rp-cGMPS. (a–d), expansions of the overlaid (1H,15N)-HSQC spectra of apo (red contours), Rp-cGMPS-bound (blue contours) and cGMP-bound (green contours) PKG Iβ CNB-B (219–369), illustrating the Rp-GMPS-associated perturbations of key PBC residues: (a), A309, (b), G306; (c), E307; (d), Q311. [24]. For clarity, the peaks are marked with “×” and outlined with labelled squares. Deviations from linearity in the apo vs. Rp-cGMPS vs. cGMP peak positions are likely to arise due to nearest neighbor effects. (e), structures of the PBC of PKG Iβ CNB-B bound to Rp-cGMPS (left) and cGMP (right) (PDB ID: 4KU7). PBC residues (residues 306–313) are shown as sticks complete with protons using the same color scheme as Fig. 1. Protons are colored in white. Backbone amide protons of G305, E307 and Q311 are shown as white spheres. Hydrogen bonding interactions involving these protons are shown as dotted lines.
3.4 Affinity and inhibition constant
To investigate how these changes in interactions with Rp-cGMPS affect the affinity, we determined the equilibrium dissociation constant (KD) of Rp-cGMPS and cGMP for PKG Iβ CNB-B using Surface Plasmon Resonance (SPR) (Supplementary Table 1). In agreement with previously reported competition based SPR and fluorescence polarization assays, PKG Iβ CNB-B showed a KD of 108 nM for cGMP[15] (Supplementary Fig. 1a–b). As for Rp-cGMPS, a KD value of 8.8 μM was determined, representing ~81-fold reduction in its affinity compared to cGMP (Supplementary Fig. 1b). This is consistent with the loss of key hydrogen bonds as seen in the structure and NMR. Our data also showed that Rp-cGMPS does not activate the full-length PKG Iβ (residues 5-686) even at as high as 3 mM confirming its role as an inhibitor [17] (Supplementary Fig. 1c–d). Finally, we determined its Ki value to be 49 μM using microfluidic mobility-shift assay (Supplementary Fig. 1e). This value is similar to what previous reported using a radiolabeled ATP assay [17].
3.5 Conclusions about inhibition mechanism
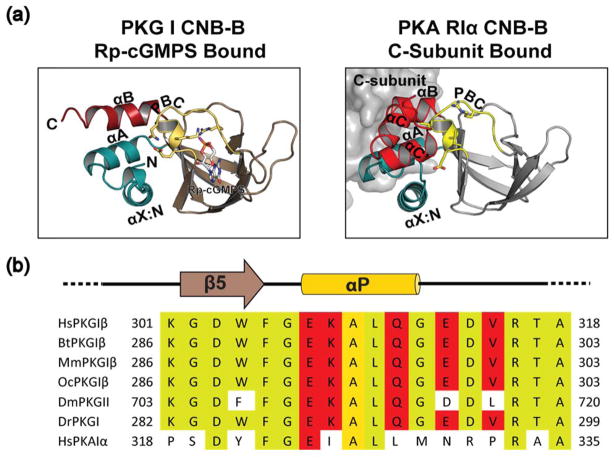
The co-crystal structure of PKG Iβ:Rp-cGMPS reveals that Rp-cGMPS stabilizes an inactive conformation with a distinctly rearranged hydrogen bonding pattern between PBC residues, which is unseen in either apo or cGMP bound conformations [15]. Its comparison with PKA RIα CNB domain suggests that the stabilization of the open PBC upon Rp-cGMPS binding is a unique feature in PKG Iβ (Fig. 4a). Due to the lack of structural information for the R-C complex which represents an inhibited state of PKG I, we compared PKG Iβ CNB-B:Rp-cGMPS complex with structures of PKA RIα CNB-B [32]. Interestingly, this comparison revealed that the PKG Iβ CNB-B:Rp-cGMPS complex assumes a conformation that resembles the C-subunit bound conformation of PKA RIα CNB-B domain, and not the apo or Rp-cAMPS bound. In fact, the apo and Rp-cAMPS bound structures showed an active conformation similar to the cAMP bound structure. Sequence comparison at the PBC shows indeed that PKA RIα lacks key PBC residues that stabilize the inactive state, suggesting that this is a unique feature for PKG Iβ (Fig. 4b). In summary, our structure combined with NMR data suggests that Rp-cGMPS may inhibit PKG by locking CNB-B in the inactive conformation keeping the kinase inhibited.
Fig. 4.
Comparison of PKA RIα:C complex with PKG Iβ CNB-B Rp-cGMPS structure. (a) structures of PKG Iβ CNB-B:Rp-cGMPS, left and the CNB-B of PKA RIα:C holoenzyme (PDB ID:2QCS), right. Same color scheme as Fig. 1, C-subunit shown as a grey surface. (b) sequence alignments of PKG Iβ in various species and human PKA RIα. Yellow shading indicates conserved residues and red shading those residues which stabilizes the apo like state when bound to Rp-cGMPS.
Supplementary Material
Affinity and inhibition of PKG Iβ by Rp-cGMPS. (a–b), surface plasmon resonance experiments with hepta-his tagged PKG Iβ CNB-B (219–351) interacting with cGMP and Rp-cGMPS. Overlaid experimental data for three replicates are shown in black and fitted model in orange. OneStep injections were performed with a SensiQ Pioneer FE, which titrates the analyte over 3–4 orders of magnitude in concentration. This creates gradients of 0–10 and 0–100 μM for cGMP and Rp-cGMPS, respectively. cGMP and Rp-cGMPS shows KD values of 108.2±0.7 nM and 8.8±0.3 μM, respectively. (c), activation of PKG Iβ by cGMP. cGMP activates PKG Iβ with an activation constant (Ka) of 537 ± 10 nM (n=2) (d), no activation in the presence of Rp-cGMPS. (e), inhibition by Rp-cGMPS. Rp-cGMPS inhibits PKG Iβ with a Ki value of 49 ± 1.
Acknowledgments
We thank Darren E. Casteel and Kim lab members for critical reading of the manuscript and Jason Davenport and Aaron Martin (SensiQ Technologies) for the technical support with SPR. G.M. is supported by Canadian Institutes of Health Research (grant number: MOP-68897 http://www.cihr-irsc.gc.ca/e/193.html) and Natural Sciences and Engineering Research Council of Canada (grant number: RGPIN-2014–04514 http://www.nserc-crsng.gc.ca/index_eng.asp). F.W.H is supported by the Federal Ministry of Education and Research, fund number 0316177F No Pain and the German Research Foundation grant He 1818/10. J.C.C is supported by the Training Program in Pharmacological Sciences fellowship, National Institute of General Medical Science grant no. 32GM089657-04. C.K. is funded by National Institutes of Health (NIH) grant R01 GM090161. The Berkeley Center for Structural Biology is supported in part by the NIH, the National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. The SPR experiments were performed in the Drug Discovery Core in the Center for Drug Discovery at Baylor College of Medicine.
References
- 1.Goldberg ND, Haddox MK, Nicol SE, Glass DB, Sanford CH, Kuehl FA, Jr, Estensen R. Biologic regulation through opposing influences of cyclic GMP and cyclic AMP: the Yin Yang hypothesis. Adv Cyclic Nucleotide Res. 1975;5:307–330. [PubMed] [Google Scholar]
- 2.Francis SH, Busch JL, Corbin JD, Sibley D. cGMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol Rev. 2010;62:525–563. doi: 10.1124/pr.110.002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3ª.Hofmann F, Bernhard D, Lukowski R, Weinmeister P. cGMP regulated protein kinases (cGK) Handb Exp Pharmacol. 2009:137–162. doi: 10.1007/978-3-540-68964-5_8. [DOI] [PubMed] [Google Scholar]; 228 FEBS Letters. 2017;591:221–230. doi: 10.1002/1873-3468.12505. 2016 Federation of European Biochemical Societies Crystal structure of PKG Ib : Rp-cGMPS complex J. C. Campbell et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francis SH, Corbin JD. Cyclic nucleotidedependent protein kinases: intracellular receptors for cAMP and cGMP action. Crit Rev Clin Lab Sci. 1999;36:275–328. doi: 10.1080/10408369991239213. [DOI] [PubMed] [Google Scholar]
- 5.Schwede F, Maronde E, Genieser H, Jastorff B. Cyclic nucleotide analogs as biochemical tools and prospective drugs. Pharmacol Ther. 2000;87:199–226. doi: 10.1016/s0163-7258(00)00051-6. [DOI] [PubMed] [Google Scholar]
- 6.Corbin JD, Ogreid D, Miller JP, Suva RH, Jastorff B, Doskeland SO. Studies of cGMP analog specificity and function of the two intrasubunit binding sites of cGMP-dependent protein kinase. J Biol Chem. 1986;261:1208–1214. [PubMed] [Google Scholar]
- 7.Das R, Chowdhury S, Mazhab-Jafari MT, Sildas S, Selvaratnam R, Melacini G. Dynamically driven ligand selectivity in cyclic nucleotide binding domains. J Biol Chem. 2009;284:23682–23696. doi: 10.1074/jbc.M109.011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das R, Melacini G. A model for agonism and antagonism in an ancient and ubiquitous cAMP-binding domain. J Biol Chem. 2007;282:581–593. doi: 10.1074/jbc.M607706200. [DOI] [PubMed] [Google Scholar]
- 9.Kornev AP, Taylor SS, Ten Eyck LF. A generalized allosteric mechanism for cis-regulated cyclic nucleotide binding domains. PLoS Comput Biol. 2008;4:e1000056. doi: 10.1371/journal.pcbi.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehmann H, Wittinghofer A, Bos JL. Capturing cyclic nucleotides in action: snapshots from crystallographic studies. Nat Rev Mol Cell Biol. 2007;8:63–73. doi: 10.1038/nrm2082. [DOI] [PubMed] [Google Scholar]
- 11.Rothermel JD, Perillo NL, Marks JS, Botelho LH. Effects of the specific cAMP antagonist, (Rp)- adenosine cyclic 30 ,50 -phosphorothioate, on the cAMPdependent protein kinase-induced activity of hepatic glycogen phosphorylase and glycogen synthase. J Biol Chem. 1984;259:15294–15300. [PubMed] [Google Scholar]
- 12.Poppe H, Rybalkin SD, Rehmann H, Hinds TR, Tang X-B, Christensen AE, Schwede F, Genieser H-G, Bos JL, Doskeland SO, et al. Cyclic nucleotide analogs as probes of signaling pathways. Nat Meth. 2008;5:277–278. doi: 10.1038/nmeth0408-277. [DOI] [PubMed] [Google Scholar]
- 13.Badireddy S, Yunfeng G, Ritchie M, Akamine P, Wu J, Kim CW, Taylor SS, Qingsong L, Swaminathan K, Anand GS. Cyclic AMP analog blocks kinase activation by stabilizing inactive conformation: conformational selection highlights a new concept in allosteric inhibitor design. Mol Cell Proteomics. 2011;10:M110.004390. doi: 10.1074/mcp.M110.004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc Natl Acad Sci U S A. 2011;108:6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang GY, Kim JJ, Reger AS, Lorenz R, Moon EW, Zhao C, Casteel DE, Bertinetti D, Vanschouwen B, Selvaratnam R, et al. Structural basis for cyclicnucleotide selectivity and cGMP-selective activation of PKG I. Structure. 2014;22:116–124. doi: 10.1016/j.str.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JJ, Casteel DE, Huang G, Kwon TH, Ren RK, Zwart P, Headd JJ, Brown NG, Chow DC, Palzkill T, et al. Co-crystal structures of PKG Ibeta (92- 227) with cGMP and cAMP reveal the molecular details of cyclic-nucleotide binding. PLoS One. 2011;6:e18413. doi: 10.1371/journal.pone.0018413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butt E, van Bemmelen M, Fischer L, Walter U, Jastorff B. Inhibition of cGMP-dependent protein kinase by (Rp)-guanosine 30 ,50 - monophosphorothioates. FEBS Lett. 1990;263:47–50. doi: 10.1016/0014-5793(90)80702-k. [DOI] [PubMed] [Google Scholar]
- 18.Huang GY, Gerlits OO, Blakeley MP, Sankaran B, Kovalevsky AY, Kim C. Neutron diffraction reveals hydrogen bonds critical for cGMP-selective activation: insights for cGMP-dependent protein kinase agonist design. Biochemistry. 2014;53:6725–6727. doi: 10.1021/bi501012v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diederichs K, Karplus PA. Better models by discarding data? Acta Crystallogr D Biol Crystallogr. 2013;69:1215–1222. doi: 10.1107/S0907444913001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 24.Goddard TD, Sparky DGK. NMR Assignment and Integration Software. University of California; San Francisco, CA: [Google Scholar]
- 25.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 26.Bahrami A, Assadi AH, Markley JL, Eghbalnia HR. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput Biol. 2009;5:e1000307. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27ª.Quinn JG. Modeling Taylor dispersion injections: determination of kinetic/affinity interaction constants and diffusion coefficients in label-free biosensing. Anal Biochem. 2012;421:391–400. doi: 10.1016/j.ab.2011.11.024. [DOI] [PubMed] [Google Scholar]; FEBS Letters. 2017;591:221–230. doi: 10.1002/1873-3468.12505. 2016 Federation of European Biochemical Societies 229 J. C. Campbell et al. Crystal structure of PKG Ib : Rp-cGMPS complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinn JG. Evaluation of Taylor dispersion injections: determining kinetic/affinity interaction constants and diffusion coefficients in label-free biosensing. Anal Biochem. 2012;421:401–410. doi: 10.1016/j.ab.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 29.Kimple AJ, Muller RE, Siderovski DP, Willard FS. A capture coupling method for the covalent immobilization of hexahistidine tagged proteins for surface plasmon resonance. Methods Mol Biol. 2010;627:91–100. doi: 10.1007/978-1-60761-670-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.VanSchouwen B, Selvaratnam R, Giri R, Lorenz R, Herberg FW, Kim C, Melacini G. Mechanism of cAMP Partial Agonism in Protein Kinase G (PKG) J Biol Chem. 2015;290:28631–28641. doi: 10.1074/jbc.M115.685305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akimoto M, Selvaratnam R, McNicholl ET, Verma G, Taylor SS, Melacini G. Signaling through dynamic linkers as revealed by PKA. Proc Natl Acad Sci U S A. 2013;110:14231–14236. doi: 10.1073/pnas.1312644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim C, Cheng CY, Saldanha SA, Taylor SS. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130:1032–1043. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Affinity and inhibition of PKG Iβ by Rp-cGMPS. (a–b), surface plasmon resonance experiments with hepta-his tagged PKG Iβ CNB-B (219–351) interacting with cGMP and Rp-cGMPS. Overlaid experimental data for three replicates are shown in black and fitted model in orange. OneStep injections were performed with a SensiQ Pioneer FE, which titrates the analyte over 3–4 orders of magnitude in concentration. This creates gradients of 0–10 and 0–100 μM for cGMP and Rp-cGMPS, respectively. cGMP and Rp-cGMPS shows KD values of 108.2±0.7 nM and 8.8±0.3 μM, respectively. (c), activation of PKG Iβ by cGMP. cGMP activates PKG Iβ with an activation constant (Ka) of 537 ± 10 nM (n=2) (d), no activation in the presence of Rp-cGMPS. (e), inhibition by Rp-cGMPS. Rp-cGMPS inhibits PKG Iβ with a Ki value of 49 ± 1.