Abstract
Opioids are very effective analgesics, but they are also highly addictive. Methadone is used to treat opioid dependence (OD), acting as a selective agonist at the μ-opioid receptor encoded by the gene OPRM1. Determining the optimal methadone maintenance dose is time-consuming; currently, no biomarkers are available to guide treatment. In methadone-treated OD subjects drawn from a case and control sample, we conducted a genome-wide association study (GWAS) of usual daily methadone dose. In African-American (AA) OD subjects (n = 383), we identified a genome-wide significant association between therapeutic methadone dose (mean = 68.0 mg, standard deviation (SD) = 30.1 mg) and rs73568641 (P = 2.8 × 10−8), the nearest gene (306 kilobases) being OPRM1. Each minor (C) allele corresponded to an additional ~20 mg/day of oral methadone, an effect specific to AAs. In European-Americans (EAs) (n = 1,027), no genome-wide significant associations with methadone dose (mean = 77.8 mg, SD = 33.9 mg) were observed. In an independent set of opioid-naïve AA children being treated for surgical pain, rs73568641-C was associated with a higher required dose of morphine (n = 241, P = 3.9 × 10−2). Similarly, independent genomic loci previously shown to associate with higher opioid analgesic dose were associated with higher methadone dose in the OD sample (AA and EA: n = 1,410, genetic score P = 1.3 × 10−3). The present results in AAs indicate that genetic variants influencing opioid sensitivity across different clinical settings could contribute to precision pharmacotherapy for pain and addiction.
Introduction
Opioids are efficacious analgesics that also have considerable addictive properties. In recent years, the United States has faced an opioid abuse epidemic.1 The rate of fatal overdoses from prescription opioids has quadrupled.2 National prescribing guidelines recently announced by the Centers for Disease Control and Prevention are intended to curb the excessive clinical use of opioids, and to promote evidence-based therapies for patients who develop OD.3
For decades, the mainstay of evidence-based OD treatment has been the pairing of supportive social services with opioid substitution therapy.4, 5 Methadone is an inexpensive and long-acting synthetic opioid, and like the most frequently abused opioids it is a potent μ-opioid receptor agonist.6 Methadone maintenance therapy (MMT) can therefore be used to treat abuse by pharmacologically substituting for other opioids, such as morphine or heroin. MMT reduces craving, withdrawal symptoms, and risk of relapse.6 The initial, or induction, stage of MMT requires considerable care: excessive methadone doses are dangerous,7 while overly conservative dosing is ineffective at preventing relapse to illicit opioid use.8 Determining the clinically optimal dose, one that provides clinical benefit to a particular individual without causing sedation or respiratory depression, is time consuming. Methadone dosing must be adjusted based on clinical signs and symptoms, and patients differ greatly in their dose requirements. Despite the clinical challenges posed by methadone administration, and resistance to MMT for social and cultural reasons,9, 10 MMT remains a vitally important treatment strategy for hundreds of thousands of patients in the United States.11
Opioids such as methadone and morphine are full agonists at the μ-opioid receptor, which is encoded by the gene OPRM1 on chromosome 6.6 OPRM1 has been the subject of intense interest, particularly the common missense single nucleotide polymorphism (SNP) rs1799971, but also non-coding variation, with dozens of candidate gene association studies having examined a wide range of phenotypes.12–14 Many of the initial claims about associations between the candidate missense variant rs1799971 and clinical phenotypes have not proven to be robust,15, 16 although modest effects do appear to be present.17, 18 In addition to OPRM1, studies have also examined the relationship between methadone metabolism and candidate polymorphisms in genes encoding cytochrome P450 enzymes, including CYP3A4, CYP2B6 and CYP2D6.19–21 Neither metabolic enzyme polymorphisms nor serum methadone levels (SMLs) have yet been shown to be reliable predictors of maintenance dose.22, 23 Genes related to both pharmacodynamics and pharmacokinetics may, however, influence each individual’s dosing needs.
Genome-wide association studies (GWASs) survey the entire catalog of common genetic variants in a hypothesis-free manner. OPRM1 is an obvious gene of interest, but prior studies of complex traits have repeatedly demonstrated that unbiased approaches are important for discovering phenotypically relevant SNPs.24–26 We performed a GWAS to search for pharmacogenetic determinants of daily methadone dose in a sample of methadone-treated OD subjects, and followed-up our findings using morphine dose data from an independent clinical sample being treated for acute pain. In this way, we sought to identify and characterize SNPs that associate with therapeutic opioid dose, which could enable personalized treatment of individuals based on their genotype.
Materials and Methods
Recruitment and assessment of subjects with opioid dependence (OD)
Details on the Yale-Penn sample have been published previously24, 27, 28. Briefly, adults with a history of dependence on alcohol, opioids, or cocaine and controls were recruited at five sites in the Eastern United States, primarily via community advertisements and word of mouth, as part of ongoing studies of drug and alcohol dependence genetics. The sample consisted of small nuclear families with affected and unaffected members (originally collected for linkage studies), and unrelated cases and controls. Exclusion criteria included a history of psychotic disorders (schizophrenia, bipolar disorder), serious head injury, or inability to read English at a sixth-grade level. Subjects gave written informed consent as approved by the Institutional Review Board (IRB) at each site, and certificates of confidentiality were obtained from the National Institute on Drug Abuse and the National Institute on Alcohol Abuse and Alcoholism. In-person interviews were conducted by trained interviewers using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA), which is a comprehensive polydiagnostic instrument that yields reliable information on major DSM-IV diagnoses and diagnostic criteria (available at https://nidagenetics.org/filebrowser/download/3765).29, 30 The SSADDA covers psychiatric and substance use disorders, as well as social and medical history and demographic information.
Methadone dose genome-wide association study (GWAS) in methadone-treated OD subjects
DNA from study participants was extracted from blood, saliva, or immortalized cell lines. Subjects were genotyped using either the Illumina HumanOmni1-Quad_v1.0 microarray, or the Illumina Human Core Exome microarray. Subjects were genotyped on the HumanOmni1-Quad_v1.0 at the Yale Center for Genome Analysis or the Center for Inherited Disease Research (CIDR). The HumanOmni1-Quad_v1.0 contains 988,306 autosomal SNPs, and genotypes were called using Illumina Genome Studio software v2011.1, genotyping module v1.8.4. Subjects were initially filtered based on call rate, and SNPs filtered based on call rate and frequency, with identity-by-descent (IBD) estimates used to quantify genetic relatedness between subjects. Extensive details on genotyping and data cleaning procedures have been published previously.24 Subjects genotyped on the HumanOmni1-Quad_v1.0 were included in the present study if they were unrelated and either self-reported African-Americans (AAs) or European-Americans (EAs), with population outliers then removed based on principal component analysis (PCA) of genotype data.31, 32 Subjects not genotyped on the HumanOmni1-Quad_v1.0 were genotyped on the Human Core Exome microarray, which contains both exome-focused SNP content and tagging SNPs for genome-wide imputation. Details about the application of quality control procedures to the Human Core Exome microarray data are provided in Supplemental Methods. Genotype data are being released via the National Institutes of Health (NIH) dbGAP platform (accession number phs000425.v1.p1).
All subjects selected for the GWAS met criteria for DSM-IV OD. Subjects who had been treated with methadone were asked the following question: “When you were taking methadone, what was your usual dosage?” Data on daily methadone dose were available for a total of 383 AAs and 1,027 EAs. Phenotype data were prepared for GWAS using the R statistical computing environment,33 which was also used to generate phenotype data summary statistics (means, standard deviations). Methadone dose data, in milligrams (mg), were transformed to normality with an inverse-normal transformation,34 and used as the dependent variable in the GWAS. As previously described, imputation of genotype data was performed from the 1000 Genomes Project Phase 1 reference panel using Impute2.35–37 The GWAS was carried out with Plink v1.0738, adjusting for age, sex, weight, and 10 principal components (PCs). Within each of the two ancestry groups (EA and AA), separate analyses were run on subjects genotyped on the HumanOmni1-Quadv1.0 and Human Core Exome microarrays. SNPs were filtered out if the minor allele frequency (MAF) was <5%, or if the imputation INFO score was <0.7. Meta-analyses were then performed within ancestry groups using METAL, which was also used to remove SNPs with heterogeneous effect estimates across the two microarrays, and to adjust summary statistics based on the genomic inflation factor (λ).39 LocusZoom was used for regional association plot generation.40 We defined the cutoff for genome-wide significance using the criterion of P = 5.0 × 10−8.
Intravenous morphine dose data from opioid-naïve pediatric surgery patients
Children 4–18 years old who received intravenous morphine during a tonsillectomy and adenoidectomy at the Children’s Hospital of Philadelphia (CHOP) were identified using the Anesthesia Information Management system (CompuRecord, Phillips Medical Systems, Andover, MA). All surgeries were performed between November 1, 2001 and December 1, 2009. Exclusion criteria included obstructive sleep apnea, a combination of tonsillectomy and adenoidectomy with another procedure, or administration of other intraoperative anesthetics. While recovering in the Postanesthesia Care Unit (PACU), children received additional intravenous morphine (in 25–50 μg/kg increments) to control their pain. The CHOP IRB approved collection of these data.
A subset of the patients meeting the above inclusion criteria had previously been consented for genomic study and genotyped (on either the Illumina Human-Hap550 or Illumina Human610-Quad microarray) by the Center for Applied Genomics at CHOP, as approved by the CHOP IRB. Sample details and quality control of phenotype and genotype data have been described previously.41 The 1000 Genomes Project reference panel and Impute2 35–37 were used to impute the top methadone dose GWAS SNP. In this morphine-treated sample, we analyzed total intravenous morphine dose (μg/kg) as a quantitative trait, and used SNPTEST V242 to evaluate the methadone dose GWAS-identified SNP. We used the same statistical model previously developed for this sample, which included age, body mass index, and American Society of Anesthesiologists physical status as covariates.41
Results
Methadone dose genome-wide association study (GWAS) in the Yale-Penn OD sample identifies a significant association upstream of OPRM1 at rs73568641
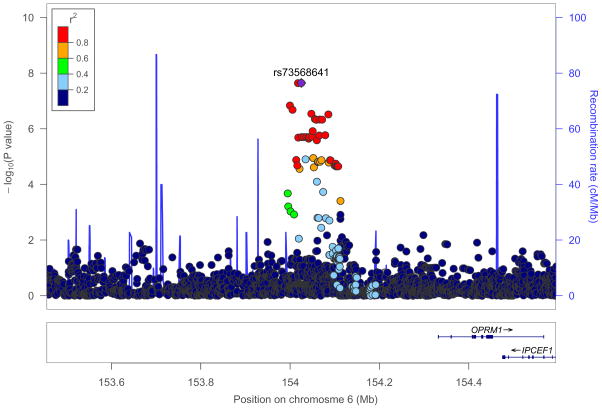
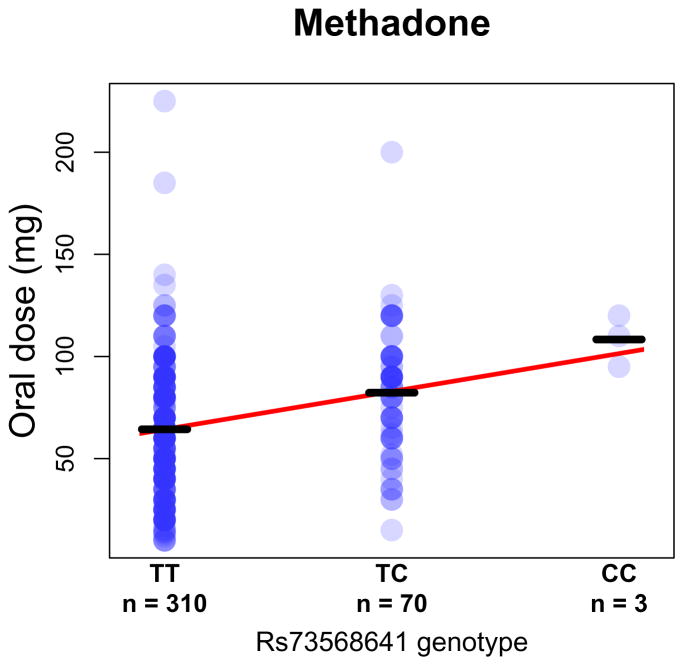
Table 1 provides an overview of the GWAS sample demographics. Dose data for AAs (mean (standard deviation (SD)) = 68.0 mg (30.1 mg)) and EAs (mean (SD) = 77.8 mg (33.9 mg)) are shown in Supplemental Figures 1 and 2, respectively. Summary statistics for all SNPs with P < 5.0 × 10−5 in AAs are provided in Supplemental Table 1, and summary statistics for all SNPs with P < 5.0 × 10−5 in EAs are provided in Supplemental Table 2. The GWAS conducted in AAs identified one genome-wide significant region on chromosome 6 (lead SNP rs73568641, n = 383, P = 2.8 × 10−8, Supplemental Table 3; AA quantile-quantile (QQ) plot is shown in Supplemental Figure 3). Lead SNP rs73568641 tags an association peak approximately ~300 kilobases (kb) upstream of the OPRM1 transcription start site (Figure 1). Rs73568641 genotypes did not deviate from Hardy-Weinberg equilibrium expectiations.43 In AAs, the minor (C) allele of rs73568641 (MAF = 0.1) was associated with a higher daily methadone dose: TT genotype (n = 310), dose mean (SD) = 64.4 mg (29.8 mg); TC genotype (n = 70), dose mean (SD) = 82.3 mg (30.1 mg); CC genotype (n = 3), dose mean (SD) = 108.3 mg (12.6 mg).
Table 1. Overview of methadone dose genome-wide association study (GWAS) sample.
All subjects met criteria for DSM-IV lifetime opioid dependence (OD), had been treated with methadone, and reported their usual daily methadone dose. A maximum of seven DSM-IV OD criteria can be endorsed. Kilograms (kg), milligrams (mg), standard deviation (SD).
| African-Americans | European-Americans | |
|---|---|---|
| Sample size | 383 | 1,027 |
| Men | ||
| Subjects, n (%) | 225 (58.8) | 617 (60.1) |
| DSM-IV OD criteria, mean | 6.5 | 6.7 |
| Age, mean (SD), years | 45.6 (8.4) | 37.2 (10.1) |
| Weight, mean (SD), kg | 87.7 (17.3) | 88.0 (17.6) |
| Methadone dose, mean (SD), mg | 66.1 (29.8) | 77.7 (33.1) |
| Women | ||
| Subjects, n (%) | 158 (41.3) | 410 (39.9) |
| DSM-IV OD criteria, mean | 6.4 | 6.7 |
| Age, mean (SD), years | 43.0 (7.2) | 37.5 (9.8) |
| Weight, mean (SD), kg | 82.4 (21.9) | 71.5 (16.5) |
| Methadone dose, mean (SD), mg | 70.7 (31.8) | 78.1 (35.1) |
Figure 1. Genome-wide significant association with methadone dose in opioid dependent (OD) African-Americans (AAs).
Regional association plot of the implicated locus on chromosome 6, showing a genome-wide significant association between methadone dose and single nucleotide polymorphism (SNP) rs73568641 (purple) (AA n = 383, P = 2.8 × 10−8). The gene nearest to rs73568641 is OPRM1. Each circle corresponds to a SNP, and the vertical position reflects the −log10(P value) (left y-axis). Color coding depicts the degree of linkage disequilibrium (r2) between lead SNP rs73568641 and other SNPs in the region. The blue line indicates the recombination rate (right y-axis). Centimorgan (cM), megabase (Mb).
Figure 2 displays the usual daily methadone dose for each AA subject, stratified by rs73568641 genotype. The association between methadone dose and rs73568641 was specific to AAs (EA n = 1,027, MAF = 0.17, P = 0.32, Supplemental Table 3), and no SNPs were genome-wide significant in the GWAS conducted in the EA sample (EA QQ plot is shown in Supplemental Figure 4). Manhattan plots are shown in Supplemental Figure 5. In an exploratory analysis across samples evaluating previously studied candidate alleles from genes encoding methadone metabolizing enzymes,44 we found suggestive evidence that the CYP2D6 loss of function variant rs3892097 is associated with lower methadone dose (AA and EA n = 1,410, P = 2.6 × 10−3) (Supplemental Table 4). A genome-wide meta-analysis of the AA and EA samples did not reveal additional genome-wide significant variants.
Figure 2. Methadone dose stratified by rs73568641 genotype in opioid dependent (OD) African-Americans (AAs).
Oral methadone dose is shown in milligrams (mg). Bars mark group means. Best fit line is shown in red.
Methadone dose-associated SNP rs73568641 also associates with morphine dose in the CHOP pediatric surgical patients
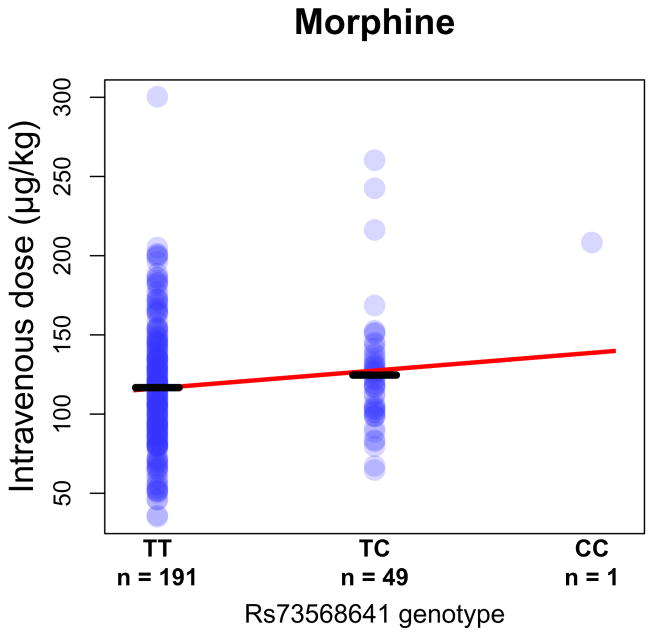
We investigated whether the implicated SNP upstream of OPRM1 also influences sensitivity to the analgesic effects of opioids. Because the observed association between rs73568641 and methadone dose was evident only in AAs, we examined the effect of rs73568641 in an independent AA sample. The only published GWAS of opioid dosage in AAs that we are aware of is our earlier study41, wherein we examined intravenous morphine dose in AA pediatric patients recovering from tonsillectomy and adenoidectomy. In these AA subjects (dose mean (SD) = 118.6 micrograms/kilogram (μg/kg) (39.8 μg/kg)), rs73568641-C was associated with a higher required morphine dose (n = 241, β = 11.6 μg/kg, standard error (SE) = 5.6 μg/kg, two-tailed P = 3.9 × 10−2), the same effect direction as for methadone dose (Figure 3). In EA patients from the CHOP sample (dose mean (SD) = 132.4 μg/kg (40.9 μg/kg)), no association between rs73568641-C and morphine dose was present (n = 277, P = 0.33). These consistent results across independent samples indicate that the effects of the locus are apparent in African-ancestry but not European-ancestry populations (no other populations were tested).
Figure 3. Morphine dose stratified by rs73568641 genotype in pediatric African-American (AA) surgical patients.
Intravenous moprhine dose is shown in micrograms/kilogram (μg/kg). Bars mark group means. Best fit line is shown in red.
Opioid analgesic dose genetic score (GS) associates with higher methadone dose in the Yale-Penn OD sample
Prior GWASs have identified several top SNPs that, while not reaching genome-wide significance, associated replicably to higher opioid analgesic dose, and implicated genes CREB1, TAOK3, and TRPC3.41, 45, 46 We found that a genetic score (GS) calculated using these dose-increasing alleles was associated with higher methadone dose, and that the relationship was more evident in AAs than EAs (AA: n = 383, two-tailed P = 6.6 × 10−4; EA: n = 1,027, two-tailed P = 8.0 × 10−2; meta-analysis P = 1.3 × 10−3, Supplemental Table 5) (an explanation of how the GS was derived is provided in Supplemental Methods).
Discussion
In methadone-treated AA OD subjects, a GWAS identified a genome-wide significant association with methadone dose, with the nearest gene being OPRM1. This same SNP was associated with increased morphine dose in an independent sample of AA surgical patients. We also found evidence that previously identified opioid analgesic dose-associated SNPs (mapping to three different genomic locations, all separate from OPRM1) associated to higher methadone dose in the total sample of methadone-treated OD subjects. These results therefore indicate that the top genetic predictors of opioid dose in the setting of addiction treatment also influence the opioid dose needed to achieve analgesia, and vice-versa.
Despite the current OD epidemic, effective pharmacotherapies are grossly underutilized.47 Buprenorphine has emerged as an office-based treatment for OD, but in a Cochrane review methadone was more effective than buprenorphine when delivered at an adequate dosage.48 Overly conservative dosing undermines clinical effectiveness,49–51 although clinicians must also be careful to minimize the danger of doses that are too high for their patients.52 The initiation of methadone treatment is therefore particularly challenging. The dosage required varies widely, and there are no methods available at the start of treatment to predict the optimal dose for a particular patient.22 Similar patient-to-patient variability is also seen when opioids are used for pain control, and a substantial portion of this variance has been attributed to heritable factors.53
The association signal that we identified is far enough upstream from the OPRM1 coding region – about 300 kb for the lead SNP – to have entirely escaped interrogation in the many previous OPRM1 candidate gene studies.12 The identified locus is non-coding, as is often seen in GWASs, and its molecular function remains unknown. Mechanistic studies in genetically engineered neural cultures might shed light on how the μ-opioid receptor’s response to exogenous opioids differs by genotype.54 We cannot rule out the possibility that, although OPRM1 is the closest gene, the association is partially, or even entirely, attributable to cis- or trans-effects on the expression or function of proteins encoded elsewhere in the genome.
Genetic variants at loci related to methadone metabolism may also be clinically relevant, but their study has been complicated by the presence of differently metabolized optical isomers,55 the use of different experimental paradigms,20, 21, 56–58 and the possible tissue specificity of enzymatic activity.59, 60 Our suggestive finding that a CYP2D6 loss of function allele61, 62 decreases required methadone dose is in the expected direction. Future applications of genetics to dosage prediction could incorporate genetic variation at known pharmacokinetic and pharmacodynamic genes, as well as additional genes such as those implicated by the GS. These additional genes point to relevant biology beyond the interface of opioids with metabolizing enzymes or the μ-opioid receptor, illustrating the importance of unbiased GWASs that are not driven by prior hypotheses.
In the present samples, the association between rs73568641-C and higher opioid dose was observed only in AAs for both methadone and morphine. In our prior OD GWAS,24 which included most of the Yale-Penn subjects in the present study, we similarly reported very different results for AAs and EAs, with the most significant results (which did not include any markers near OPRM1) in AAs. In the present study the GS association signal was stronger in methadone-treated AAs than EAs. There are several possible explanations that could account for associations being preferentially detected in specific populations. GWAS SNPs often tag many common variants, as is the case here (Figure 1), and population-specific GWAS findings63 may be related to linkage disequilibrium between commons SNPs and population-specific rare functional variants.64 Whole genome sequencing approaches and larger samples will be needed to interrogate fully variation across the allele frequency spectrum in multiple ancestry groups. Epistasis provides another possible explanation; some polymorphisms may have phenotypic effects only when population-specific variants in the region or even elsewhere in the genome are present to interact with them.65
Clinicians tend to prescribe lower doses of opioids to minority patients for pain control,66 including minority children.67 While clinical confounds may partly explain this phenomenon, a similar pattern is observed in the setting of substance use disorders: OD treatment programs serving a higher proportion of AA patients are more likely to report under-dosing of methadone.51 We observed lower opioid doses for AAs compared to EAs (methadone: t-test P < 0.001; morphine: t-test P < 0.001). The present data are therefore consistent with the hypothesis that prescriber bias may contribute to differences between population groups in the quantity of opioids dispensed, although we cannot exclude the possibility that the observed differences in dosing may reflect actual differences in medication requirements.68 If EA subjects are dosed more liberally than AA subjects, who receive doses closer to the therapeutic minimum or are undertreated, objective markers to guide dosing could serve to mitigate under-dosing and consequent health disparities.
A limitation of our study is the sample size, which is small compared to case/control mega-GWASs that pool data across many different studies each having only limited phenotype information. In particular, future recruitment of additional methadone- and morphine-treated patients will be needed in order to study large numbers of CC homozygotes. Generally, pharmacogenomics GWASs tend to have many fewer subjects than studies of disease risk, because it is challenging to recruit and clinically characterize informative subjects, although the observed effect sizes are often greater in pharmacogenomics studies.69 Our GWAS is larger than all previously published opioid dose GWASs of which we are aware,41, 45, 46, 70 and our GS finding reinforces that these earlier GWASs were likely successful in identifying real signals despite modest sample sizes. Thus, the larger present sample would seem to be sufficient, especially considering the validation of rs73568641 in an independent sample of morphine-treated patients. Evaluation of rs73568641 using clinically documented morphine dose data also helps to compensate for another limitation of our study, which is that the data on usual daily methadone dose was collected via self-report. The intensive daily nature of methadone treatment, which allows for close monitoring of clinical response and provides frequent reminders to the patient of their dose, further supports the reliability of the reported methadone dose data. One study found that when OD subjects receiving methadone were interviewed in a research setting, the correlation coefficient between self-reported and clinically documented dosage was 0.97.71
In the field of pharmacogenomics, strong positive results have been produced by studying medications that require clinicians to tailor carefully the dose to each individual patient’s needs.69 An initial GWAS of warfarin dose in 181 patients detected a genome-wide significant association signal upstream of the gene (VKORC1) encoding the drug’s target.72 The case of warfarin is instructive for the study of methadone, providing an example of efforts to clinically implement genotype-guided dosing.73 Warfarin differs from methadone, however, in that the International Normalized Ratio (INR) test can provide precise biochemical feedback to guide warfarin dosing, making it difficult for genotype-based algorithms to improve on treatment as usual.74, 75 In contrast, there are currently no biological assays to help clinicians decide which patients will require more aggressive methadone dosing, suggesting that genetics could play a role in improving clinical outcomes. Prospective studies, including randomized controlled trials of genotype guided dosing, are needed to define better the magnitude of the genetic contribution to dose requirements, and to assess the clinical utility of this information.
Conclusions
In methadone-treated OD AAs, we identified a single genome-wide significant association with methadone dosing needs, and found that the closest gene was OPRM1. We validated the genetic marker in an independent sample of AA surgical patients receiving morphine for analgesia. Consistent with the observation that this SNP’s influence is evident across different clinical settings where μ-opioid receptor agonism is employed, top SNPs from prior opioid analgesic dose studies were collectively associated with methadone dose in OD patients. The observed effect of the rs73568641 minor allele on methadone dose requirements could have immediate clinical utility in the therapeutic dosing of methadone, and perhaps other μ-opioid receptor agonists, in AA patients. Prospective replication and further clinical characterization in new samples are needed to realize this potential.
Supplementary Material
Acknowledgments
We wish to thank all of the research participants in this study. Adult subject recruitment and assessment were overseen at the Yale School of Medicine and the APT Foundation by James Poling, Ph.D., and Aryeh Herman, Psy.D.; at McLean Hospital by Roger Weiss, M.D.; at the Medical University of South Carolina by Kathleen Brady, M.D., Ph.D. and Raymond Anton, M.D.; and at the University of Pennsylvania initially by David Oslin, M.D. Genotyping services were provided by the Center for Inherited Disease Research (CIDR) and the Yale University Center for Genome Analysis. Ann Marie Lacobelle, M.S. and Christa Robinson, A.S. provided excellent technical assistance; the SSADDA interviewers devoted substantial time and effort to phenotype the study sample; and Richard Sherva, Ph.D., Ryan Koesterer, M.A., and John Farrell, Ph.D. at Boston University offered valuable assistance with data cleaning and management. Robert T. Malison, M.D., Department of Psychiatry, Yale School of Medicine, provided thoughtful suggestions during preparation of the manuscript.
This study was supported by grants from the National Institutes of Health (NIH) (RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535, MSTP 5T32GM007205-38, CTSA TL1 8UL1TR000142, F30 DA037665, N01-HG-65403, S10 RR19895); a Veterans Affairs VISN1 Career Development Award; the Department of Anesthesiology and Critical Care Medicine at The Children’s Hospital of Philadelphia through Children’s Anesthesia Associates, Ltd., and by The Children’s Hospital of Philadelphia through a grant from the Institutional Development Fund to The Center for Applied Genomics. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Conflict of Interest Disclosures
Dr. Kranzler reports being a consultant, continuing medical education (CME) speaker, or advisory board member for Alkermes, Indivior, Lundbeck, and Otsuka, and a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last three years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, and XenoPort. No other disclosures are reported.
References
- 1.Okie S. A flood of opioids, a rising tide of deaths. N Engl J Med. 2010;363(21):1981–1985. doi: 10.1056/NEJMp1011512. [DOI] [PubMed] [Google Scholar]
- 2.Volkow ND, Frieden TR, Hyde PS, Cha SS. Medication-assisted therapies--tackling the opioid-overdose epidemic. N Engl J Med. 2014;370(22):2063–2066. doi: 10.1056/NEJMp1402780. [DOI] [PubMed] [Google Scholar]
- 3.Dowell D, Haegerich TM, Chou R. CDC Guideline for Prescribing Opioids for Chronic Pain--United States, 2016. JAMA. 2016;315(15):1624–1645. doi: 10.1001/jama.2016.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dole VP. Implications of methadone maintenance for theories of narcotic addiction. JAMA. 1988;260(20):3025–3029. [PubMed] [Google Scholar]
- 5.Dole VP, Nyswander M. A medical treatment for diacetylmorphine (heroin) addiction. A clinical trial with methadone hydrochloride. JAMA. 1965;193:646–650. doi: 10.1001/jama.1965.03090080008002. [DOI] [PubMed] [Google Scholar]
- 6.Kreek MJ, LaForge KS, Butelman E. Pharmacotherapy of addictions. Nat Rev Drug Discov. 2002;1(9):710–726. doi: 10.1038/nrd897. [DOI] [PubMed] [Google Scholar]
- 7.Harding-Pink D. Methadone: one person’s maintenance dose is another’s poison. Lancet. 1993;341(8846):665–666. doi: 10.1016/0140-6736(93)90427-i. [DOI] [PubMed] [Google Scholar]
- 8.Maxwell S, Shinderman MS. Optimizing long-term response to methadone maintenance treatment: a 152-week follow-up using higher-dose methadone. J Addict Dis. 2002;21(3):1–12. doi: 10.1300/J069v21n03_01. [DOI] [PubMed] [Google Scholar]
- 9.Berridge V. Heroin prescription and history. N Engl J Med. 2009;361(8):820–821. doi: 10.1056/NEJMe0904243. [DOI] [PubMed] [Google Scholar]
- 10.Courtwright DT. Preventing and Treating Narcotic Addiction--Century of Federal Drug Control. N Engl J Med. 2015;373(22):2095–2097. doi: 10.1056/NEJMp1508818. [DOI] [PubMed] [Google Scholar]
- 11.Substance Abuse and Mental Health Services Administration CfBHSaQ. The N-SSATS Report: Trends in the Use of Methadone and Buprenorphine at Substance Abuse Treatment Facilities: 2003 to 2011. Rockville, MD: 2013. [PubMed] [Google Scholar]
- 12.Crist RC, Berrettini WH. Pharmacogenetics of OPRM1. Pharmacol Biochem Behav. 2014;123:25–33. doi: 10.1016/j.pbb.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang BZ, Krupitsky E, et al. Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15(6):807–819. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung CC, Chiou MH, Huang BH, Hsieh YW, Hsieh TJ, Huang CL, et al. Impact of genetic polymorphisms in ABCB1, CYP2B6, OPRM1, ANKK1 and DRD2 genes on methadone therapy in Han Chinese patients. Pharmacogenomics. 2011;12(11):1525–1533. doi: 10.2217/pgs.11.96. [DOI] [PubMed] [Google Scholar]
- 15.Oslin DW, Leong SH, Lynch KG, Berrettini W, O’Brien CP, Gordon AJ, et al. Naltrexone vs Placebo for the Treatment of Alcohol Dependence: A Randomized Clinical Trial. JAMA Psychiatry. 2015;72(5):430–437. doi: 10.1001/jamapsychiatry.2014.3053. [DOI] [PubMed] [Google Scholar]
- 16.Gelernter J, Gueorguieva R, Kranzler HR, Zhang H, Cramer J, Rosenheck R, et al. Opioid receptor gene (OPRM1, OPRK1, and OPRD1) variants and response to naltrexone treatment for alcohol dependence: results from the VA Cooperative Study. Alcohol Clin Exp Res. 2007;31(4):555–563. doi: 10.1111/j.1530-0277.2007.00339.x. [DOI] [PubMed] [Google Scholar]
- 17.Schwantes-An TH, Zhang J, Chen LS, Hartz SM, Culverhouse RC, Chen X, et al. Association of the OPRM1 Variant rs1799971 (A118G) with Non-Specific Liability to Substance Dependence in a Collaborative de novo Meta-Analysis of European-Ancestry Cohorts. Behav Genet. 2016;46(2):151–169. doi: 10.1007/s10519-015-9737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock DB, Levy JL, Gaddis NC, Glasheen C, Saccone NL, Page GP, et al. Cis-Expression Quantitative Trait Loci Mapping Reveals Replicable Associations with Heroin Addiction in OPRM1. Biol Psychiatry. 2015;78(7):474–484. doi: 10.1016/j.biopsych.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kharasch ED, Regina KJ, Blood J, Friedel C. Methadone Pharmacogenetics: CYP2B6 Polymorphisms Determine Plasma Concentrations, Clearance, and Metabolism. Anesthesiology. 2015;123(5):1142–1153. doi: 10.1097/ALN.0000000000000867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eap CB, Broly F, Mino A, Hämmig R, Déglon JJ, Uehlinger C, et al. Cytochrome P450 2D6 genotype and methadone steady-state concentrations. J Clin Psychopharmacol. 2001;21(2):229–234. doi: 10.1097/00004714-200104000-00016. [DOI] [PubMed] [Google Scholar]
- 21.Crettol S, Déglon JJ, Besson J, Croquette-Krokar M, Hämmig R, Gothuey I, et al. ABCB1 and cytochrome P450 genotypes and phenotypes: influence on methadone plasma levels and response to treatment. Clin Pharmacol Ther. 2006;80(6):668–681. doi: 10.1016/j.clpt.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 22.Leavitt SB, Shinderman M, Maxwell S, Eap CB, Paris P. When “enough” is not enough: new perspectives on optimal methadone maintenance dose. Mt Sinai J Med. 2000;67(5–6):404–411. [PubMed] [Google Scholar]
- 23.Dennis BB, Bawor M, Thabane L, Sohani Z, Samaan Z. Impact of ABCB1 and CYP2B6 genetic polymorphisms on methadone metabolism, dose and treatment response in patients with opioid addiction: a systematic review and meta-analysis. PLoS One. 2014;9(1):e86114. doi: 10.1371/journal.pone.0086114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, et al. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biological psychiatry. 2014;76(1):66–74. doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson EC, Agrawal A, Heath AC, Bogdan R, Sherva R, Zhang B, et al. Evidence of CNIH3 involvement in opioid dependence. Molecular psychiatry. 2016;21(5):608–614. doi: 10.1038/mp.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, Posthuma D, et al. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry. 2011;16(5):516–532. doi: 10.1038/mp.2010.38. [DOI] [PubMed] [Google Scholar]
- 27.Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. Genome-wide association study of alcohol dependence: significant findings in African- and European-Americans including novel risk loci. Molecular psychiatry. 2014;19(1):41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, et al. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Molecular psychiatry. 2014;19(6):717–723. doi: 10.1038/mp.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug and Alcohol Dependence. 2005;80(3):303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, et al. Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug and alcohol dependence. 2007;91(1):85–90. doi: 10.1016/j.drugalcdep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 32.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- 34.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genetics. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delaneau O, Marchini J, Zagury J. A linear complexity phasing method for thousands of genomes. Nature Methods. 2012;9(2):179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 38.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics (Oxford, England) 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cook-Sather SD, Li J, Goebel TK, Sussman EM, Rehman MA, Hakonarson H. TAOK3, a novel genome-wide association study locus associated with morphine requirement and postoperative pain in a retrospective pediatric day surgery population. Pain. 2014;155(9):1773–1783. doi: 10.1016/j.pain.2014.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genetics. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 43.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76(5):887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Somogyi AA, Barratt DT, Ali RL, Coller JK. Pharmacogenomics of methadone maintenance treatment. Pharmacogenomics. 2014;15(7):1007–1027. doi: 10.2217/pgs.14.56. [DOI] [PubMed] [Google Scholar]
- 45.Nishizawa D, Fukuda K, Kasai S, Hasegawa J, Aoki Y, Nishi A, et al. Genome-wide association study identifies a potent locus associated with human opioid sensitivity. Molecular psychiatry. 2014;19(1):55–62. doi: 10.1038/mp.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aoki Y, Nishizawa D, Hasegawa J, Kasai S, Yoshida K, Koukita Y, et al. Association between the rs1465040 single-nucleotide polymorphism close to the transient receptor potential subfamily C member 3 (TRPC3) gene and postoperative analgesic requirements. J Pharmacol Sci. 2015;127(3):391–393. doi: 10.1016/j.jphs.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Vestal C. In Fighting An Opioid Epidemic, Medication-Assisted Treatment Is Effective But Underused. Health Aff (Millwood) 2016;35(6):1052–1057. doi: 10.1377/hlthaff.2016.0504. [DOI] [PubMed] [Google Scholar]
- 48.Mattick RP, Breen C, Kimber J, Davoli M. Buprenorphine maintenance versus placebo or methadone maintenance for opioid dependence. Cochrane Database Syst Rev. 2014;2:CD002207. doi: 10.1002/14651858.CD002207.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson RE, Chutuape MA, Strain EC, Walsh SL, Stitzer ML, Bigelow GE. A comparison of levomethadyl acetate, buprenorphine, and methadone for opioid dependence. N Engl J Med. 2000;343(18):1290–1297. doi: 10.1056/NEJM200011023431802. [DOI] [PubMed] [Google Scholar]
- 50.Strain EC, Bigelow GE, Liebson IA, Stitzer ML. Moderate- vs high-dose methadone in the treatment of opioid dependence: a randomized trial. JAMA. 1999;281(11):1000–1005. doi: 10.1001/jama.281.11.1000. [DOI] [PubMed] [Google Scholar]
- 51.D’Aunno T, Pollack HA, Frimpong JA, Wuchiett D. Evidence-based treatment for opioid disorders: a 23-year national study of methadone dose levels. J Subst Abuse Treat. 2014;47(4):245–250. doi: 10.1016/j.jsat.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bell JR, Butler B, Lawrance A, Batey R, Salmelainen P. Comparing overdose mortality associated with methadone and buprenorphine treatment. Drug Alcohol Depend. 2009;104(1–2):73–77. doi: 10.1016/j.drugalcdep.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 53.Angst MS, Phillips NG, Drover DR, Tingle M, Ray A, Swan GE, et al. Pain sensitivity and opioid analgesia: a pharmacogenomic twin study. Pain. 2012;153(7):1397–1409. doi: 10.1016/j.pain.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams JT, Ingram SL, Henderson G, Chavkin C, von Zastrow M, Schulz S, et al. Regulation of μ-opioid receptors: desensitization, phosphorylation, internalization, and tolerance. Pharmacol Rev. 2013;65(1):223–254. doi: 10.1124/pr.112.005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang HC, Chu SK, Huang CL, Kuo HW, Wang SC, Liu SW, et al. Genome-Wide Pharmacogenomic Study on Methadone Maintenance Treatment Identifies SNP rs17180299 and Multiple Haplotypes on CYP2B6, SPON1, and GSG1L Associated with Plasma Concentrations of Methadone R- and S-enantiomers in Heroin-Dependent Patients. PLoS Genet. 2016;12(3):e1005910. doi: 10.1371/journal.pgen.1005910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang JS, DeVane CL. Involvement of CYP3A4, CYP2C8, and CYP2D6 in the metabolism of (R)- and (S)-methadone in vitro. Drug Metab Dispos. 2003;31(6):742–747. doi: 10.1124/dmd.31.6.742. [DOI] [PubMed] [Google Scholar]
- 57.Coller JK, Joergensen C, Foster DJ, James H, Gillis D, Christrup L, et al. Lack of influence of CYP2D6 genotype on the clearance of (R)-, (S)- and racemic-methadone. Int J Clin Pharmacol Ther. 2007;45(7):410–417. doi: 10.5414/cpp45410. [DOI] [PubMed] [Google Scholar]
- 58.Fonseca F, de la Torre R, Díaz L, Pastor A, Cuyàs E, Pizarro N, et al. Contribution of cytochrome P450 and ABCB1 genetic variability on methadone pharmacokinetics, dose requirements, and response. PLoS One. 2011;6(5):e19527. doi: 10.1371/journal.pone.0019527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siegle I, Fritz P, Eckhardt K, Zanger UM, Eichelbaum M. Cellular localization and regional distribution of CYP2D6 mRNA and protein expression in human brain. Pharmacogenetics. 2001;11(3):237–245. doi: 10.1097/00008571-200104000-00007. [DOI] [PubMed] [Google Scholar]
- 60.Britto MR, Wedlund PJ. Cytochrome P-450 in the brain. Potential evolutionary and therapeutic relevance of localization of drug-metabolizing enzymes. Drug Metab Dispos. 1992;20(3):446–450. [PubMed] [Google Scholar]
- 61.Levran O, Peles E, Hamon S, Randesi M, Adelson M, Kreek MJ. CYP2B6 SNPs are associated with methadone dose required for effective treatment of opioid addiction. Addict Biol. 2013;18(4):709–716. doi: 10.1111/j.1369-1600.2011.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kagimoto M, Heim M, Kagimoto K, Zeugin T, Meyer UA. Multiple mutations of the human cytochrome P450IID6 gene (CYP2D6) in poor metabolizers of debrisoquine. Study of the functional significance of individual mutations by expression of chimeric genes. J Biol Chem. 1990;265(28):17209–17214. [PubMed] [Google Scholar]
- 63.CONVERGE consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523(7562):588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare Variants Create Synthetic Genome-Wide Associations. PLoS Biology. 2010;8(1):e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Polimanti R, Yang C, Zhao H, Gelernter J. Dissecting ancestry genomic background in substance dependence genome-wide association studies. Pharmacogenomics. 2015;16(13):1487–1498. doi: 10.2217/pgs.15.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pletcher MJ, Kertesz SG, Kohn MA, Gonzales R. Trends in opioid prescribing by race/ethnicity for patients seeking care in US emergency departments. JAMA. 2008;299(1):70–78. doi: 10.1001/jama.2007.64. [DOI] [PubMed] [Google Scholar]
- 67.Goyal MK, Kuppermann N, Cleary SD, Teach SJ, Chamberlain JM. Racial Disparities in Pain Management of Children With Appendicitis in Emergency Departments. JAMA Pediatr. 2015;169(11):996–1002. doi: 10.1001/jamapediatrics.2015.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou HH, Sheller JR, Nu H, Wood M, Wood AJ. Ethnic differences in response to morphine. Clin Pharmacol Ther. 1993;54(5):507–513. doi: 10.1038/clpt.1993.182. [DOI] [PubMed] [Google Scholar]
- 69.Daly AK. Genome-wide association studies in pharmacogenomics. Nat Rev Genet. 2010;11(4):241–246. doi: 10.1038/nrg2751. [DOI] [PubMed] [Google Scholar]
- 70.Mieda T, Nishizawa D, Nakagawa H, Tsujita M, Imanishi H, Terao K, et al. Genome-wide association study identifies candidate loci associated with postoperative fentanyl requirements after laparoscopic-assisted colectomy. Pharmacogenomics. 2016;17(2):133–145. doi: 10.2217/pgs.15.151. [DOI] [PubMed] [Google Scholar]
- 71.Langendam MW, van Haastrecht HJ, van Ameijden EJ. The validity of drug users’ self-reports in a non-treatment setting: prevalence and predictors of incorrect reporting methadone treatment modalities. Int J Epidemiol. 1999;28(3):514–520. doi: 10.1093/ije/28.3.514. [DOI] [PubMed] [Google Scholar]
- 72.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112(4):1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zineh I, Pacanowski M, Woodcock J. Pharmacogenetics and coumarin dosing--recalibrating expectations. N Engl J Med. 2013;369(24):2273–2275. doi: 10.1056/NEJMp1314529. [DOI] [PubMed] [Google Scholar]
- 74.Kimmel SE, French B, Kasner SE, Johnson JA, Anderson JL, Gage BF, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369(24):2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verhoef TI, Ragia G, de Boer A, Barallon R, Kolovou G, Kolovou V, et al. A randomized trial of genotype-guided dosing of acenocoumarol and phenprocoumon. N Engl J Med. 2013;369(24):2304–2312. doi: 10.1056/NEJMoa1311388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.