Abstract
Sarcoidosis is a complex disease of unknown etiology characterized by the presence of granulomatous inflammation. Though various immune system pathways have been implicated in disease, the relationship between the genetic determinants of sarcoidosis and other inflammatory disorders has not been characterized. Herein, we examined the degree of genetic pleiotropy common to sarcoidosis and other inflammatory disorders to identify shared pathways and disease systems pertinent to sarcoidosis onset. To achieve this, we quantify the association of common variant polygenic risk scores from nine complex inflammatory disorders with sarcoidosis risk. Enrichment analyses of genes implicated in pleiotropic associations were further used to elucidate candidate pathways. In European-Americans, we identify significant pleiotropy between risk of sarcoidosis and risk of asthma (R2=2.03%; p=8.89×10−9), celiac disease (R2=2.03%; p=8.21×10−9), primary biliary cirrhosis (R2=2.43%; p=2.01×10−10), and rheumatoid arthritis (R2=4.32%; p=2.50×10−17). These associations validate in African Americans only after accounting for the proportion of genome-wide European ancestry, where we demonstrate similar effects of polygenic risk for African-Americans with the highest levels of European ancestry. Variants and genes implicated in European-American pleiotropic associations were enriched for pathways involving interleukin-12, interleukin-27, and cell adhesion molecules, corroborating the hypothesized immunopathogenesis of disease.
Keywords: Pleiotropy, sarcoidosis, immunopathogenesis, genetic risk, interleukin-12
Introduction
Genome-wide association (GWA) studies indicate that most variants underlying complex diseases are of modest effect size.1 As these implicated variants may confer risk to more than one clinical phenotype, identifying and characterizing pleiotropy between disorders is necessary to elucidate disease etiology in general,2 especially for inflammatory-related disorders that share pathophysiologic pathways.3 Polygenic risk scores (PRSs) have emerged as a method to assess shared genetic risk between phenotypes. For example, characterizing polygenic risk using thousands of variants in schizophrenia explained ~2% of the variation in a sample of cases with bipolar disorder.4 To better define the genetic architecture of sarcoidosis, a granulomatous inflammatory disease of unknown etiology, we sought to assess the degree of pleiotropic risk between nine well-studied inflammatory diseases and risk of sarcoidosis. This study was conducted in both European-American (EA) and African-American (AA) individuals, the latter being an admixed population with genetic ancestral contributions from both Africa and Europe.5
Sarcoidosis affects people of all race-ethnicities and occurs at all ages.6 Based on a twin study in Danish and Finnish populations, the heritability of sarcoidosis was estimated to be 0.66, confirming a strong genetic component for this disease.7 The pathophysiology of sarcoidosis involves a dysregulated immune reaction to an unknown antigen(s) that leads to granulomatous inflammation and organ dysfunction.6 While sarcoidosis most often affects the lungs in as many as 90% of cases, granulomas can occur in any organ, leading to considerable clinical variance.6, 8 In the United States, a significant disparity is observed in the prevalence and severity of sarcoidosis between AAs and EAs. One study estimated the annual incidence of sarcoidosis in EAs to be 10.9 cases per 100,000, but this value more than triples in AAs (35.5 cases per 100,000).9 In addition, AAs have more extra-thoracic involvement than EA sarcoidosis patients, are less likely to have resolving disease and are more likely to die from sarcoidosis than EA patients.10 The differences in clinical and epidemiological characteristics of these two groups are likely due to both environmental and genetic differences.9, 10 Across ethnicities, assessing the degree of shared polygenic risk of sarcoidosis and other genetically-characterized inflammatory disorders may aid in our understanding of this complex disease.
To date, several putative sarcoidosis susceptibility genes have been revealed by linkage analyses,11 genome-wide association studies,12, 13 admixture mapping,14 Immunochip associations,13, 15 and targeted sequencing studies.16 Recent analyses of organ-specific manifestations of sarcoidosis have also identified genetic risk factors for sarcoidosis phenotypes. For example, novel genetic associations were implicated in sarcoidosis patients with Heerfordt’s syndrome (uveoparotid fever)17 and associated with clinical characteristics similar to Blau Syndrome (an autosomal dominant granulomatous inflammation of skin, eyes and joints).18 Additionally, organ-specific manifestations have been associated with genetic variation in sarcoid-related uveitis,19 cardiac,20 and neurosarcoidosis.16
Though these studies demonstrate specific genetic differences with respect to susceptibility and presence of specific clinical features, no prior study has broadly characterized the genome-wide polygenic risk in any sarcoidosis study sample or compared such profiles to other disorders. While the causal factors of sarcoidosis are largely unknown, the disease shares epidemiological, molecular, and immunological similarities (e.g. racial distribution, a strong effect at the human leukocyte antigen region, and involvement of various T-cell populations) with some autoimmune and inflammatory diseases. If the etiology of sarcoidosis indeed mimics other more extensively studied autoimmune disorders at a genetic level, we hypothesize that investigating known pathways and mechanisms of onset in analogous disease systems may bear new insights into this disorder. Thus, we aimed to identify specific disorders and implicate pathways in related inflammatory disease systems that share genetic determinants with sarcoidosis.
Results
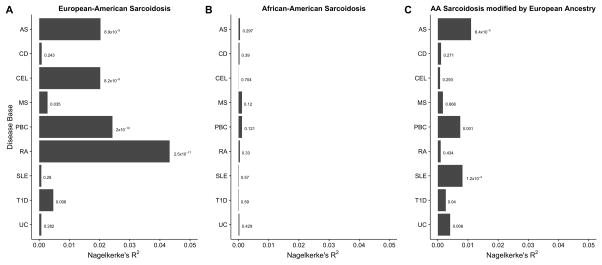
We quantified the degree of pleiotropy between AA and EA sarcoidosis and nine other inflammatory disorders shown in Table 1. Using PRSice,21 we measured the associations of these other disease risk bases and the binary sarcoidosis clinical diagnosis in EA and AA individuals. A summary of these association models is shown in Figure 1. In EAs, sarcoidosis risk was associated with the polygenic risk of asthma, celiac disease, primary biliary cirrhosis, and rheumatoid arthritis disease, each of which explained 2% or more of the observed phenotypic variance (Table S3). While statistically significant, the PRSs for multiple sclerosis and type 1-diabetes models explained less than 1% of variance in sarcoidosis risk. In comparison, none of the complex inflammatory conditions were significantly associated with sarcoidosis disease status in AA under the standard PRS regression model (Table S2). However, when the modifying effect of proportion European genetic ancestry was accounted for, five disease bases (asthma, primary biliary cirrhosis, systemic lupus erythematous, type 1 diabetes, and ulcerative colitis) were associated with disease risk in AAs. The statistical significance of these models could be in part attributed to a multiplicative interaction term of the proportion of European admixture and the PRS (Table S5–S7).
Table 1.
List of summary statistics case/control count and corresponding reference from original association study for nine inflammatory disorders used as prediction bases in this study. Further details of these studies are contained in the corresponding references.
| Disease | # Cases | # Controls | Study Population Ancestry | Reference |
|---|---|---|---|---|
| Asthma | 5,380 | 5,519 | European, African, Hispanic | 40 |
| Celiac Disease | 4,533 | 10,750 | European | 42 |
| Crohn’s Disease | 5,956 | 21,770 | European | 43 |
| Multiple Sclerosis | 9,772 | 17,376 | European | 44 |
| Primary Biliary Cirrhosis | 2,764 | 10,475 | European | 45 |
| Rheumatoid Arthritis | 5,539 | 20,169 | European | 46 |
| Systemic Lupus Erythematous | 7,219 | 15,991 | European | 47 |
| Type 1 Diabetes | 7,514 | 9,045 | European | 48 |
| Ulcerative Colitis | 6,968 | 21,770 | European | 43 |
Figure 1.
Summaries for PRS associations with sarcoidosis risk in the (A) European-Americans, (B) African-Americans samples, and (C) African-Americans accounting for the modifying effect of European ancestry. A variant including threshold of PT = 0.5 was used and the major histocompatibility complex was included where available. The Nagelkerke’s pseudo-R2 term and significance of model were calculated using PRSice21 for panels (A) and (B) and a likelihood ratio test for (C) (see Supplement) using nine inflammatory conditions as a prediction base: asthma (AS), Crohn’s disease (CD), celiac disease (CEL), multiple sclerosis (MS), primary biliary cirrhosis (PBC), rheumatoid arthritis (RA), systemic lupus erythematous (SLE), type 1 diabetes (T1D), and ulcerative colitis (UC).
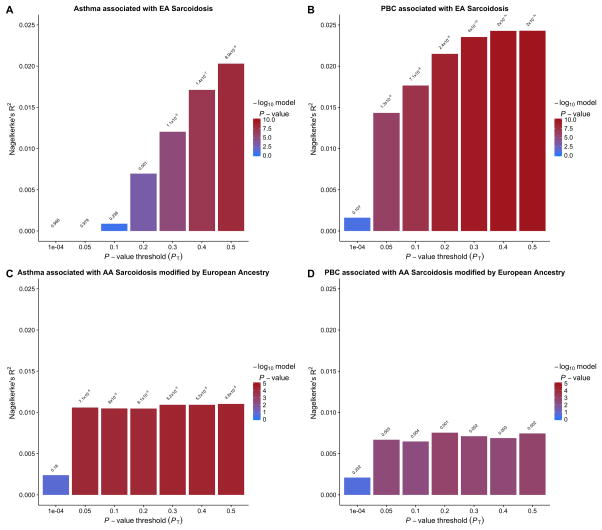
In an effort to better assess polygenic overlap between our strongest associations, we show the variation explained at all variant thresholds for both ethnicities for asthma and primary biliary cirrhosis (Figure 2). In general, as the variant inclusion threshold became less stringent, the degree of variation explained increases before leveling off, an observation consistent with other cross-phenotype polygenic associations.4, 22, 23 In models of sarcoidosis risk using two disease bases, the joint base of asthma and rheumatoid arthritis was the strongest (Table S8, S9), explaining 4.9% of the variance at the variant threshold p ≤ 0.5. For EAs, our four most significant single disease models explain 2.0–4.3% of the variance in sarcoidosis risk. These values are consistent with, and in some cases surpass, the variance explained in previous analyses across phenotypes such as bipolar disorder and schizophrenia.4
Figure 2.
Nagelkerke’s pseudo-R2 value summaries of the association between risk scores derived from the two most significant inflammatory associations using seven significance thresholds and sarcoidosis. Asthma was used as the polygenic risk base associated with (A) EA sarcoidosis and (C) AA sarcoidosis modified by European ancestry. Primary biliary cirrhosis (PBC) was used as the polygenic risk base associated with (B) EA sarcoidosis and (D) AA sarcoidosis modified by European ancestry.
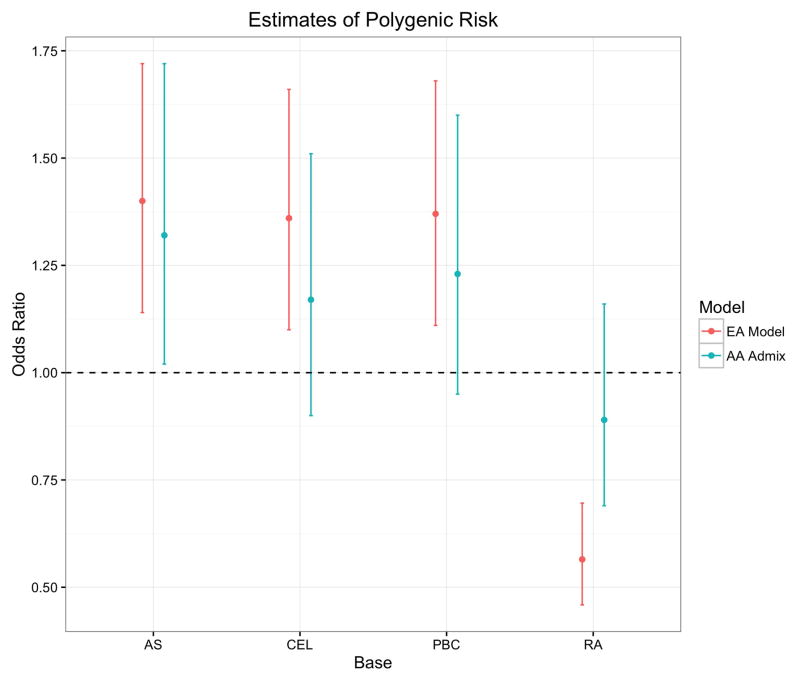
To characterize the direction of the pleiotropic effects, Figure 3 shows the odds ratio (OR) of EA sarcoidosis cases and controls using polygenic risk scores, dichotomized at the median value, as the predictor. To compute ORs for AAs, we restricted the estimation to those in the top tertile of the proportion of European ancestry (individuals with > 0.194 European ancestry), which was comprised of 414 cases and 495 controls. The PRs were again used as the predictor, dichotomized at the median PRS of the high European ancestry subset. While higher PRSs for asthma, celiac disease, and primary biliary cirrhosis were associated with increased risk for sarcoidosis, the higher rheumatoid arthritis risk score group was associated with a protective effect. This result corroborates an epidemiologic finding that rheumatoid arthritis was the only inflammatory condition out of 12 examined that had a lower prevalence in a British cohort of 1,510 sarcoidosis cases compared to the general population.24
Figure 3.
Odds ratios summarizing the magnitude and direction of the association of significant PRSs with sarcoidosis risk by ethnicity. Estimates of the odds ratio with 95% confidence interval are shown comparing EA cases and controls dichotomized at the median polygenic risk score from four conditions: asthma (AS), celiac disease (CEL), primary biliary cirrhosis (PBC), and rheumatoid arthritis (RA). The odds ratios for the same four disorders were computed in the AAs after restricting the sample to the upper tertile of European ancestry, which comprised 414 cases and 495 controls. The PRS in this stratum was again dichotomized at the median to compute the corresponding odds ratios.
After identifying the disorders that share significant genetic risk with sarcoidosis, we sought to examine the genes implicated in the pleiotropic associations. Utilizing the 225,166 variants shared between the five disease sets, we identified 238 common variants significantly associated (FDR < 0.05) with pleiotropy between sarcoidosis, asthma, celiac disease, primary biliary cirrhosis, and rheumatoid arthritis in the EA sample using the GPA framework.25 Using the 238 significant variants from the GPA model, we annotated 85 unique genes (Table S10) using WebGestalt26 before searching for enriched pathway-based sets using ConsensusPathDB.27 Table 2 lists the seven pathways whose members were enriched (FDR < 0.05) in the list of genes implicated in more than one condition (Table S10). These pathways highlight established risk genes characterized in Th1 response in granulomatous tissue, such as interleukin-1228 and interleukin-27,29 while verifying the importance of T-cells and natural-killer cells,30 previously implicated in the pathogenesis of sarcoidosis using other molecular biology techniques.
Table 2.
Significant pathways enriched from the pleiotropic associations between sarcoidosis, asthma, celiac disease, primarily biliary cirrhosis, and rheumatoid arthritis. The statistical significance of 225,116 variants overlapping between these five risk bases was measured using GPA,25 and 238 significant variants (FDR < 0.05) were mapped to 83 unique genes using WebGestalt.26 Enriched pathway-based sets were determined using ConsensusPathDB27 for these 83 genes, leading to the seven statistically significant (FDR < 0.05) pathways shown in the table. Each pathway has a corrected (FDR) and uncorrected (P-value) measure of significance of the overrepresentation of genes in the pathway from the 83 mapped genes. The MHC region was not included in the 225,116 overlapping variants as these were excluded from the celiac disease summary statistics.
| Enriched pathway-based set | Source | P-value | FDR |
|---|---|---|---|
| IL-12 signaling | INOH | 0.00033 | 0.0134 |
| Nef and signal transduction | Reactome | 0.00044 | 0.0134 |
| No2-dependent IL-12 pathway in NK cells | BioCarta | 0.00056 | 0.0134 |
| IL-12 and STAT4 dependent signaling pathway in Th1 Development | BioCarta | 0.0016 | 0.0289 |
| Cell adhesion molecules | KEGG | 0.00247 | 0.0334 |
| T-Cell Receptor (TCR) | NetPath | 0.00278 | 0.0334 |
| IL-27-mediated signaling events | PID | 0.00483 | 0.0497 |
Of the nine inflammatory diseases, our analyses failed to identify significant shared genetic risk associated with AA sarcoidosis when European ancestry was not accounted for. While the most direct way to compare the genetic risk bases between ethnic groups is to compute PRSs within each ethnicity and apply then apply them across ethnicities, the limited sample sizes limits the inferences of these resulting models. Nevertheless, we computed PRSs and applied them cross-ethnically (Table S4). Where the EA PRS was not associated with AA sarcoidosis risk at any variant threshold under the standard PRS model, the AA sarcoidosis base was significantly associated with EA sarcoidosis risk (p=0.020, R2=0.34%). Additionally, the inclusion of the context dependent effect of genome-wide European ancestry in the AA PRS model yielded a statistically significant association with the EA sarcoidosis base (p=0.0011 R2=0.79%) (Table S6). Though relatively few overlapping variants from GWA studies of sarcoidosis have been identified between European and African ethnicities,31, 32 increasing the sample sizes of our sarcoidosis cohorts and refining the population samples based on the proportion of admixture could elucidate the degree of similar polygenic risk between ethnicities of sarcoidosis as well as pleiotropy with other conditions.
Discussion
While our study was able to identify novel evidence of pleiotropy between inflammatory conditions and sarcoidosis in the context of genome-wide European ancestry, our PRS approach could be enhanced by polygenic risk bases derived from studies with more ancestral diversity. Though the asthma meta-analysis results were from three ethnic groups, the other eight inflammatory risk bases were derived from individuals solely of European ancestry. A more exhaustive analytical approach would utilize summary statistics derived from studies of individuals with African ancestry in addition to those derived from Europeans. Our study thus is limited due to the lack of ethnic diversity present in these large genetic association studies. In order to best characterize the etiology of sarcoidosis and other complex traits, future sequencing studies such as the Precision Medicine Initiative must incorporate greater patient ethnic diversity, a “promise yet to be fulfilled” by the National Institutes of Health.33 Our results demonstrate the need and probable utility for performing genetic mapping studies in admixed and minority populations as a means to characterize the genetic risk basis of complex disorders.
An initial interpretation of our findings suggests that genetic risk to sarcoidosis may partially comprise different variants between ethnic populations. However, these differences could also be explained by the design of the genotyping platforms, the variability in LD structure between populations, and the limitations of the PRS algorithm. Due to the short recombination history of the European population relative to the African population, the overrepresentation of subjects of predominantly European ancestry in prior genetic association studies, as well as the historical bias in genotyping arrays designed to capture common genetic variation present in populations of European ancestry,34 the variants included in the summary statistics were more likely to tag causal variants in European populations. While recent methodological advancements such as LDPred have integrated LD structure into PRS models using Bayesian models,35 the behavior of these associations has not been well characterized in AA and other admixed populations due to the absence of genetic information. The lack of methodological development and characterization of behavior of genetic risk in admixed populations can again be partially attributed to the lack of genetic diversity available from publically funded genetic association studies. As such, we used PRSice as a primary association tool to compute our models as this method primarily provides a wrapper for the canonical PRS approach employed in prior studies4 and more extensively reviewed.39
From clinical coincidences,24 previous genome-wide association studies,12, 13 and our present work, evidence of a shared etiology between sarcoidosis and certain autoimmune disorders has been hypothesized. We suggest that pleiotropy between clinical characteristics of sarcoidosis and these associated inflammatory conditions could lead to novel disease categories as has been demonstrated in other inflammatory conditions. For example, pleiotropic analyses of inflammatory bowel disease and primary sclerosing cholangitis suggest that comorbidity between these conditions may be a distinct disorder.3 For our purposes, clinical reports have linked celiac disease to sarcoidosis, including one celiac patient who experienced a reoccurrence of erythema nodosum after being exposed to wheat flour, leading the authors to hypothesize a causal relationship between celiac disease and sarcoidosis in some patients.36, 37 The statistically significant association between polygenic risk of celiac disease and EA sarcoidosis supports this hypothesis of a common causal agent in a subset of patients, which provides a unique avenue to examine the clinical variation of patients in the context of genetic risk.
While the utilization of the PRS algorithm across disorders has been widely implemented in other phenotypes, our study is the first to employ this methodology in the context of sarcoidosis and extend the analyses using GPA to identify candidate pathways shown in Table 2. A prominent genetic effector in our enriched pathways is interleukin-12 (IL-12), which is an established biomarker in active pulmonary sarcoidosis.28 In the hypothesized immunopathogenesis of sarcoidosis, antigen presenting cells are thought to secrete IL-12 in response to interactions with T-cells.6 IL-12 has also been shown to modulate the efficacy of glucocorticoid response in asthma,38 supporting our analyses that suggest a shared genetic risk basis may be active in both sarcoidosis and asthma—the principally pulmonary diseases considered in our analyses. We hypothesize that further analyses characterizing the pleiotropic mechanisms between asthma and sarcoidosis, particularly in the context of IL-12 signaling, may explain some of the variation in clinical features, such as pulmonary severity and steroid response efficacy in these two disorders.
Overall, our discovery of significant pleiotropy between these inflammatory conditions and sarcoidosis provides another perspective on over a half-century of clinical and epidemiological findings into the etiology of this enigmatic condition. In particular, we provide evidence that the inflammatory signaling pathways mediated by IL-12, IL-27, and cell adhesion molecules may be causal in multiple conditions in a subset of patients, providing unique opportunities to measure clinical outcomes from an individual’s genetic risk. Our results suggest that these genes and pathways may also been relevant to the presentation of pulmonary phenotypes in asthma and sarcoidosis. Future studies that measure the pleiotropic risk of sarcoidosis and other phenotypes, including other pulmonary and infectious diseases, may further elucidate candidate pathways for characterizing the pathogenesis of this and other disorders.
Materials and Methods
Our genetic samples comprised 2,738 self-identified AAs (1,273 sarcoidosis cases and 1,465 controls) and 2,726 self-identified EAs (442 sarcoidosis cases and 2,284 controls) genotyped using the Illumina HumanOmni1-Quad array for ~1.1M variants across the genome under standard quality control measures as previously described.12 As the genotyping data used for the sarcoidosis populations were the exact same as those resulting from the quality control described in this prior manuscript,12 details of these steps are excluded from the present manuscript. We note that 1,945 of the 2,284 EA controls were obtained from external studies that do not overlap with the samples used to generate the summary statistics for the other inflammatory disorders. Since the power of PRS increases as the base sample size increases,39 we used disease base sets for nine complex inflammatory disorders from large meta-analyses (Table 1). Each of these meta-analyses exceeded 10,000 combined cases and controls (average > 20,000). Summary statistics for eight of the inflammatory disorders were downloaded from either IBDgenetics.org or Immunobase.org in publically available repositories. The exception was the asthma study, where the EVE asthma meta-analysis consortium provided the results.40 While the asthma meta-analysis summarizes cases of European (39.5%), African (29.1%), and Latino (31.4%) ancestry, the other eight meta-analyses contained samples of only European ancestry.
To compute polygenic risk scores, we used PRSice21 with the default fastscore parameters, which includes p-value informed linkage disequilibrium (LD) clumping with a cutoff of R2 = 0.1, a 250kb distance threshold, and base variant thresholds of p = 10−4, 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5. We emphasize that the LD-clumping step considers the population substructure only in the target genotyping data, and the clumping technique has been successfully applied under similar analysis conditions in associating across disorders.22, 23 PRSice computes a risk score for each individual in the target set (in this case, our AA or EA sarcoidosis sample) using the summary statistics of variants from an independent GWA analysis base (in this case, each of the nine inflammatory diseases). These scores are regressed against the case/control status in the target sample to quantify the phenotypic variation explained by the polygenic risk across disorders. Though previously published PRS variant significance thresholds have ranged from p=0.5 to p=2 ×10−28, less-stringent variant p-value thresholds often produce models that explain a higher proportion of trait variance.4, 39
The summary statistic disease bases consisted of association results for genome-wide autosomal markers including the major histocompatibility complex (MHC) for each of the nine inflammatory disorders except celiac disease, where the MHC was not made available. While the genotyping platforms varied within and between studies, a minimum of 245,000 (average >518,000) variants overlapped between the summary statistics and the sarcoidosis samples before LD-clumping and p-value thresholding (Table S1). Using a threshold of p=0.5, we tested the association between PRSs and case/control status in both the EA and AA sarcoidosis samples. In the AAs, we performed additional regression analyses including a multiplicative interaction term between PRS and genome-wide proportion European ancestry to assess the possibility of a modifying effect of genetic ancestry (see Supplemental PDF). In these models, the Nagelkerke’s adjusted-R2 and corresponding significance correspond to a full model, containing the main effect of PRS and proportion of European ancestry as well as the interaction term. Individual admixture estimates were computed using LAMP41 as we’ve previously described.14
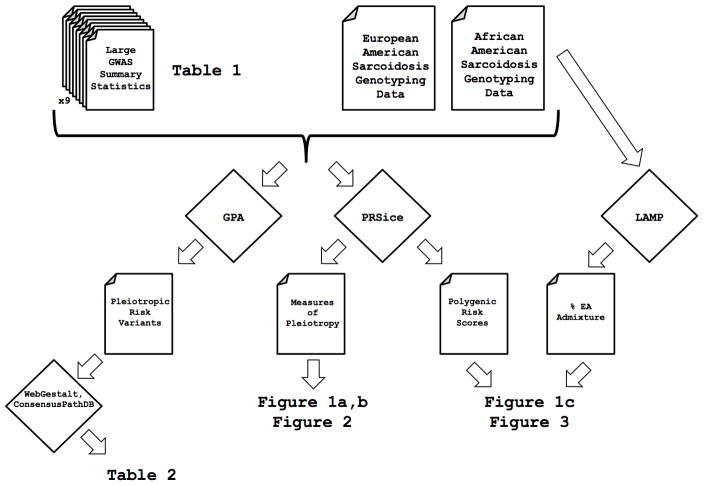
As the polygenic risk methodology provides a measure of the association between disorders (i.e. pleiotropy), we performed additional analyses to target the individual pathways and genetic effectors implicated from the associations between diseases. To achieve this, we used the Genetic analysis incorporating Pleiotropy and Annotation (GPA) methodology, which increases statistical power needed to identify individual risk variants through joint analysis of multiple phenotypes.25 GPA uses an integrated probabilistic model that assigns a new measure of statistical significance for each variant to a phenotype of interest (in this case, sarcoidosis) in the context of associations to other phenotypes (four other disease bases).25 Notably, as the MHC was excluded from the celiac disease summary statistics, variants in this region were excluded from the pleiotropic analysis. As only 3 of the 225,166 were statistically significant (FDR < 0.05) using only the sarcoidosis summary statistics, GPA was able to uncover many associations underpowered in single phenotype associations. After identifying shared genetic variants associated with risk of sarcoidosis and other phenotypes, we annotated statistically significant variants to their corresponding genes using WebGestalt26 before searching for enriched pathway-based sets using ConsensusPathDB.27 Gene sets enriched highlight shared biologic pathways associated with risk of sarcoidosis and other inflammatory disorders. A complete graphical summary of the relationship between the methods, data, figures, tables, and other outputs is shown in Figure 4. Namely, summary statistics from external GWAS studies coupled with the sarcoidosis genotyping data are sufficient inputs to replicate the results using the tools indicated in this figure.
Figure 4.
A graphical overview of the data and methods used to produce data, tables, and figures for this manuscript. Files are indicated by the folded corner whereas methods (such as GPA and PRSice) are indicated in diamonds.
Supplementary Material
Acknowledgments
We thank the OMRF Genomics core for performing the genotyping experiments and the useful discussion. We are also grateful to the EVE asthma analysis consortium (RC2 HL101651) and particularly the efforts of Drs. Carole Ober, Dan Nicolae, and Dara Torgerson to make the asthma summary statistics available to the broader scientific community. Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS), Heart, Lung and Blood Institute (NHLBI), and National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health under award numbers: P20GM103456 and P30GM110766 to IA, R01-HL54306 and U01-HL060263 to MCI, R56-AI072727 and R01-HL092576 to BAR, and P30 GM110766-01, RC2HL101499, and R01HL113326 to CGM. CAL is supported by an NSF Graduate Research Fellowship # DGE1144152. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author Contributions Conceived and designed the study: CAL, CFD, MCI, BAR, AML, CGM. Analyzed data: CAL, CFD, IA, AML. Interpreted the data, drafted/revised the manuscript: CAL, CFD, IA, CJL, PMG, MCI, BAR, AML, CGM.
Conflicts of Interest None declared.
Online Data Supplement This article has an online data supplement, which is accessible from this issue’s table of content online.
Supplemental PDF contains 6 additional figures and 10 tables that provide further details of the results of polygenic scoring under different ancestry conditions and with variable inclusion of the MHC.
References
- 1.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, Manolio T, et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet. 2011;89(5):607–18. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016 doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Schizophrenia C. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63(6):1839–51. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iannuzzi MC, Rybicki BA, Teirstein AS. Sarcoidosis. N Engl J Med. 2007;357(21):2153–65. doi: 10.1056/NEJMra071714. [DOI] [PubMed] [Google Scholar]
- 7.Sverrild A, Backer V, Kyvik KO, Kaprio J, Milman N, Svendsen CB, et al. Heredity in sarcoidosis: a registry-based twin study. Thorax. 2008;63(10):894–6. doi: 10.1136/thx.2007.094060. [DOI] [PubMed] [Google Scholar]
- 8.James DG. Descriptive definition and historic aspects of sarcoidosis. Clin Chest Med. 1997;18(4):663–79. doi: 10.1016/s0272-5231(05)70411-1. [DOI] [PubMed] [Google Scholar]
- 9.Rybicki BA, Major M, Popovich J, Jr, Maliarik MJ, Iannuzzi MC. Racial differences in sarcoidosis incidence: a 5-year study in a health maintenance organization. Am J Epidemiol. 1997;145(3):234–41. doi: 10.1093/oxfordjournals.aje.a009096. [DOI] [PubMed] [Google Scholar]
- 10.Edmondstone WM, Wilson AG. Sarcoidosis in Caucasians, Blacks and Asians in London. Br J Dis Chest. 1985;79(1):27–36. doi: 10.1016/0007-0971(85)90004-x. [DOI] [PubMed] [Google Scholar]
- 11.Rybicki BA, Hirst K, Iyengar SK, Barnard JG, Judson MA, Rose CS, et al. A sarcoidosis genetic linkage consortium: the sarcoidosis genetic analysis (SAGA) study. Sarcoidosis Vasc Diffuse Lung Dis. 2005;22(2):115–22. [PubMed] [Google Scholar]
- 12.Adrianto I, Lin CP, Hale JJ, Levin AM, Datta I, Parker R, et al. Genome-wide association study of African and European Americans implicates multiple shared and ethnic specific loci in sarcoidosis susceptibility. PLoS One. 2012;7(8):e43907. doi: 10.1371/journal.pone.0043907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer A, Ellinghaus D, Nutsua M, Hofmann S, Montgomery CG, Iannuzzi MC, et al. Identification of Immune-Relevant Factors Conferring Sarcoidosis Genetic Risk. Am J Respir Crit Care Med. 2015;192(6):727–36. doi: 10.1164/rccm.201503-0418OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin AM, Iannuzzi MC, Montgomery CG, Trudeau S, Datta I, Adrianto I, et al. Admixture fine-mapping in African Americans implicates XAF1 as a possible sarcoidosis risk gene. PloS one. 2014;9(3):e92646. doi: 10.1371/journal.pone.0092646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivera NV, Ronninger M, Shchetynsky K, Franke A, Nothen MM, Muller-Quernheim J, et al. High-density Genetic Mapping Identifies New Susceptibility Variants in Sarcoidosis Phenotypes and Shows Genomic-driven Phenotypic Differences. Am J Respir Crit Care Med. 2015 doi: 10.1164/rccm.201507-1372OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lareau CA, Adrianto I, Levin AM, Iannuzzi MC, Rybicki BA, Montgomery CG. Fine mapping of chromosome 15q25 implicates ZNF592 in neurosarcoidosis patients. Ann Clin Transl Neurol. 2015;2(10):972–7. doi: 10.1002/acn3.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darlington P, Tallstedt L, Padyukov L, Kockum I, Cederlund K, Eklund A, et al. HLA-DRB1* alleles and symptoms associated with Heerfordt’s syndrome in sarcoidosis. Eur Respir J. 2011;38(5):1151–7. doi: 10.1183/09031936.00025011. [DOI] [PubMed] [Google Scholar]
- 18.Bello GA, Adrianto I, Dumancas GG, Levin AM, Iannuzzi MC, Rybicki BA, et al. Role of NOD2 Pathway Genes in Sarcoidosis Cases with Clinical Characteristics of Blau Syndrome. Am J Respir Crit Care Med. 2015;192(9):1133–5. doi: 10.1164/rccm.201507-1344LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson IA, Liu B, Sen HN, Jiao X, Katamay R, Li Z, et al. Association of complement factor H tyrosine 402 histidine genotype with posterior involvement in sarcoid-related uveitis. Am J Ophthalmol. 2013;155(6):1068–1074e1. doi: 10.1016/j.ajo.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gialafos E, Triposkiadis F, Kouranos V, Rapti A, Kosmas I, Manali E, et al. Relationship between tumor necrosis factor-alpha (TNFA) gene polymorphisms and cardiac sarcoidosis. In Vivo. 2014;28(6):1125–9. [PubMed] [Google Scholar]
- 21.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31(9):1466–8. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Power RA, Steinberg S, Bjornsdottir G, Rietveld CA, Abdellaoui A, Nivard MM, et al. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci. 2015;18(7):953–5. doi: 10.1038/nn.4040. [DOI] [PubMed] [Google Scholar]
- 23.Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajoriya N, Wotton CJ, Yeates DG, Travis SP, Goldacre MJ. Immune-mediated and chronic inflammatory disease in people with sarcoidosis: disease associations in a large UK database. Postgraduate medical journal. 2009;85(1003):233–7. doi: 10.1136/pgmj.2008.067769. [DOI] [PubMed] [Google Scholar]
- 25.Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet. 2014;10(11):e1004787. doi: 10.1371/journal.pgen.1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server issue):W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39(Database issue):D712–7. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moller DR, Forman JD, Liu MC, Noble PW, Greenlee BM, Vyas P, et al. Enhanced expression of IL-12 associated with Th1 cytokine profiles in active pulmonary sarcoidosis. J Immunol. 1996;156(12):4952–60. [PubMed] [Google Scholar]
- 29.Larousserie F, Pflanz S, Coulomb-L’Hermine A, Brousse N, Kastelein R, Devergne O. Expression of IL-27 in human Th1-associated granulomatous diseases. J Pathol. 2004;202(2):164–71. doi: 10.1002/path.1508. [DOI] [PubMed] [Google Scholar]
- 30.Katchar K, Soderstrom K, Wahlstrom J, Eklund A, Grunewald J. Characterisation of natural killer cells and CD56+ T-cells in sarcoidosis patients. Eur Respir J. 2005;26(1):77–85. doi: 10.1183/09031936.05.00030805. [DOI] [PubMed] [Google Scholar]
- 31.Rossman MD, Thompson B, Frederick M, Maliarik M, Iannuzzi MC, Rybicki BA, et al. HLA-DRB1*1101: a significant risk factor for sarcoidosis in blacks and whites. Am J Hum Genet. 2003;73(4):720–35. doi: 10.1086/378097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofmann S, Franke A, Fischer A, Jacobs G, Nothnagel M, Gaede KI, et al. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat Genet. 2008;40(9):1103–6. doi: 10.1038/ng.198. [DOI] [PubMed] [Google Scholar]
- 33.Oh SS, Galanter J, Thakur N, Pino-Yanes M, Barcelo NE, White MJ, et al. Diversity in Clinical and Biomedical Research: A Promise Yet to Be Fulfilled. PLoS Med. 2015;12(12):e1001918. doi: 10.1371/journal.pmed.1001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann TJ, Kvale MN, Hesselson SE, Zhan Y, Aquino C, Cao Y, et al. Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98(2):79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilhjalmsson BJ, Yang J, Finucane HK, Gusev A, Lindstrom S, Ripke S, et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet. 2015;97(4):576–92. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James DG, Sharma OP. Overlap syndromes with sarcoidosis. Postgrad Med J. 1985;61(719):769–71. doi: 10.1136/pgmj.61.719.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowe G, Johnston RN. Sarcoidosis and coeliac disease. Lancet. 1984;2(8403):637. doi: 10.1016/s0140-6736(84)90625-1. [DOI] [PubMed] [Google Scholar]
- 38.Naseer T, Minshall EM, Leung DY, Laberge S, Ernst P, Martin RJ, et al. Expression of IL-12 and IL-13 mRNA in asthma and their modulation in response to steroid therapy. Am J Respir Crit Care Med. 1997;155(3):845–51. doi: 10.1164/ajrccm.155.3.9117015. [DOI] [PubMed] [Google Scholar]
- 39.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9(3):e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74(5):965–78. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47(9):979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.International Multiple Sclerosis Genetics C. Bush WS, Sawcer SJ, de Jager PL, Oksenberg JR, McCauley JL, et al. Evidence for polygenic susceptibility to multiple sclerosis--the shape of things to come. Am J Hum Genet. 2010;86(4):621–5. doi: 10.1016/j.ajhg.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cordell HJ, Han Y, Mells GF, Li Y, Hirschfield GM, Greene CS, et al. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat Commun. 2015;6:8019. doi: 10.1038/ncomms9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bentham J, Morris DL, Cunninghame Graham DS, Pinder CL, Tombleson P, Behrens TW, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47(12):1457–64. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–7. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.