Abstract
Effective and rapid dereplication is a hallmark of present-day drug discovery from natural sources. This project strove to both decrease the time and expand the structural diversity associated with dereplication methodologies. A 5 min liquid chromatographic run time employing heated electrospray ionization (HESI) was evaluated to determine whether it could be used as a faster alternative over the 10 min ESI method we reported previously. Results revealed that the 5 min method was as sensitive as the 10 min method and, obviously, was twice as fast. To facilitate dereplication, the retention times, UV absorption maxima, full-scan HRMS and MS/MS were cross-referenced with an in-house database of over 300 fungal secondary metabolites. However, this strategy was dependent upon the makeup of the screening in-house database. Thus, mass defect filtering (MDF) was explored as an additional targeted screening strategy to permit identification of structurally related components. The use of a dereplication platform incorporating the 5 min chromatographic method together with MDF facilitated rapid and effective identification of known compounds and detection of structurally related analogs in extracts of fungal cultures.
Introduction
As Ōmura and colleagues1 showed the world via their research on avermectins1, 2, 3 (and many other compounds), nature is a prolific source of chemical diversity, providing humanity with many drugs and drug leads for a suite of diseases.4, 5, 6, 7 A common characteristic of natural product extracts is that they consist of a mixture of compounds, often structurally related, although present at varying abundance. As such, it is desirable to have a means to prioritize samples, such that valuable time and financial resources can be focused on those extracts most likely to yield new structural diversity. An important part of this discovery process involves dereplication, a rapid identification of known compounds in extracts in order to focus on the isolation of new chemical entities. Several recent advancements in dereplication strategies, most involving hyphenated spectroscopic techniques, have been pivotal to the increased efficiency in natural product-based drug discovery.8, 9, 10 In particular, MS coupled with electrospray ionization (ESI) has become increasingly more important in natural product drug discovery, especially for chemical profiling of complex extracts.11, 12, 13 Hyphenation of liquid chromatography with ESI-MS has increased the pace of dereplication by providing an efficient means of identifying the multiple components of complex extracts before further purification and isolation work.10, 14
Our research team has been working on the dereplication of fungal cultures for about a decade, particularly within a drug discovery program that aims to identify new anticancer lead compounds.15, 16 This started with a generalized dereplication methodology utilizing UV and 1H NMR spectra and targeting distinct classes of mycotoxins,17 as these nuisance compounds yield positive results in cytotoxicity-based assays. This protocol evolved significantly to target a wider range of secondary metabolites in fungal extracts using a complementary suite of hyphenated techniques, specifically ultraperformance liquid chromatography-photodiode array-high resolution tandem mass spectrometry (UPLC-PDA-HRMS-MS/MS). Finally, the latter was augmented with a droplet liquid microjunction surface sampling probe so as to analyze the secondary metabolite profile of fungal cultures in situ.18
Mass defect filtering (MDF) was developed recently and is finding growing use in drug metabolite profiling studies.19, 20, 21 This approach utilizes the full-scan HRMS data, and detection of drug metabolites is accomplished via data mining after acquisition. MDF is based on the fact that each isotope of every element has a defined mass defect (that is, the nonintegral portion of an m/z value). This is why each molecule of a defined elemental composition will have a unique exact mass, typically observed in the 1000ths to 10 000ths place.
The useful application of MDF in drug metabolism studies is centered on the observation that the core structure of a drug does not change significantly during biotransformation.19, 22, 23 Thus, the mass defect between the parent drug and its metabolites will fall within a narrow range (typically within 50 mDa). If a mass defect filter window is centered on the mass defect of the parent drug, then ions with a mass defect outside of this window will be excluded, resulting in the removal of interference signals in the total ion chromatogram. Importantly, this then facilitates the selective identification of drug metabolite peaks from the noise in complex biological matrices.
Aside from drug metabolite identification, MDF has been adapted for use in the qualitative examination of natural products, particularly those based on herbals.24, 25, 26 However, this data-mining tool has not been used in the analysis of fungal secondary metabolites. Theoretically, compounds produced by fungi can be classified into distinct classes, each class sharing the same core structure. Thus, if a filter reference and substituents around the core structure are carefully defined, then mass defect filters can be constructed to exclude the majority of unrelated ions. This should, in turn, facilitate the identification of structural analogs in the extract.
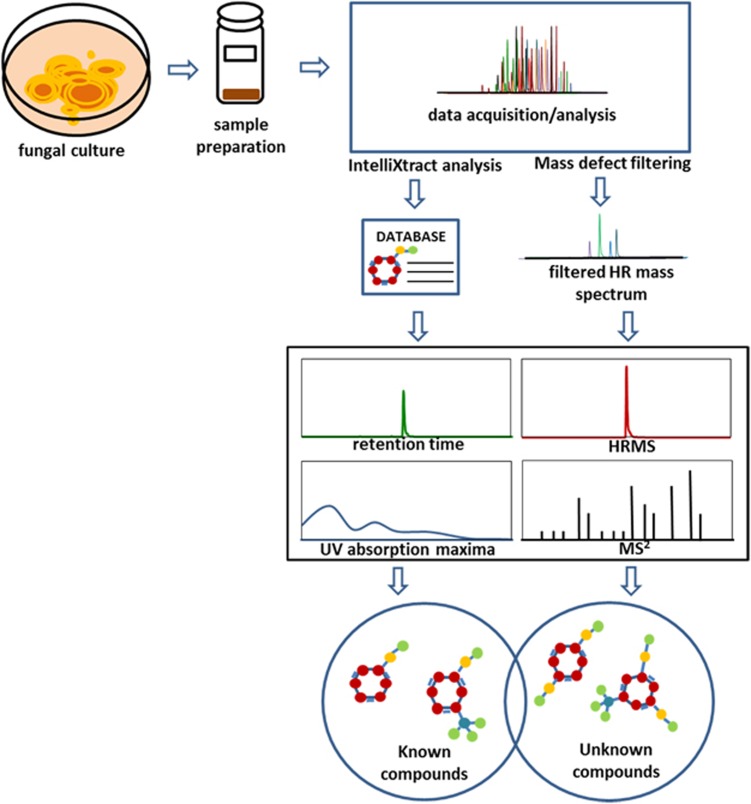
In this study, MDF is presented as an additional approach for the targeted screening of secondary metabolites in fungal extracts. The merger of UPLC-PDA-HRMS-MS/MS dereplication with MDF permits additional profiling for structurally related components in the sample based on their mass defects (Scheme 1). This workflow enhances the dereplication methods so as to screen for a wider range of secondary metabolites produced by fungal cultures.
Results and discussion
The 5 min HESI method
Previously, we developed a dereplication method for diverse classes of fungal secondary metabolites16 in extracts on an LTQ Orbitrap.27 That protocol utilized a 10 min liquid chromatographic run time to acquire full-scan HRMS and MS/MS (using a normalized collision energy of 30) spectra in both positive and negative ESI modes, with UV absorption maxima and retention times used as orthogonal data. To facilitate dereplication, these data were uploaded into ACD MS Manager with add-in Intelliextract software that cross-references the molecular ion peaks and retention times with an in-house database of over 300 fungal secondary metabolites. Molecular ions identified by the software that matched the database were confirmed by comparison of the HRMS, MS/MS, retention time and UV data to that of the standard in the database. With this method, nearly all of the fungal metabolites (>98%) in the in-house database ionized efficiently in the ESI+ mode, whereas ∼90% of the samples ionized in the ESI− mode. In total, this suggested the ESI+ mode as the ionization of choice for sample dereplication, unless knowledge of the samples or extracts indicated ionization in ESI− mode.
Although valuable for routine analysis of a large number of samples (batches of >50 samples), the 10 min liquid chromatographic method was not ideal, particularly for shared instruments. Thus, a dereplication method utilizing a 5 min run time at a 0.6 ml min−1 flow rate via heated electrospray ionization (HESI) was evaluated to determine whether it could be used as a faster alternative. The use of HESI was important,28, 29 as it would allow better desolvation, given that a higher flow rate (twice as fast) was used for the 5 min method compared with the 0.3 ml min−1 flow rate utilized in the 10 min method.
As proof of concept, about a third of the fungal library (>100 compounds) was analyzed in the HESI+ using the 5 min method to obtain the retention times, UV absorption maxima, full-scan HRMS and MS/MS spectra. In general, data obtained for all compounds matched with data in the database built using the 10 min dereplication method,27 except for the retention times.
To compare the performance characteristics of the 10 min ESI dereplication method with the 5 min HESI dereplication, the two related parameters, limit of detection (LOD) and limit of quantitation (LOQ), were measured for each method, both in the positive ionization mode. The LOD and LOQ values were obtained from the calibration data and regression line directly (Supplementary Figures S1 and S2). Five representative compounds (1–5) were tested that had molecular weights ranging from 373 to 1963 amu, and this included compounds that were based on polyketide building blocks, amino acid building blocks and those with both in their scaffolds (Supplementary Table S1). In general, the 5 min HESI method had comparable sensitivity to the 10 min ESI method based on the LOD and LOQ values obtained for the five analytes tested (Supplementary Table S1).
Dereplication of fungal extracts
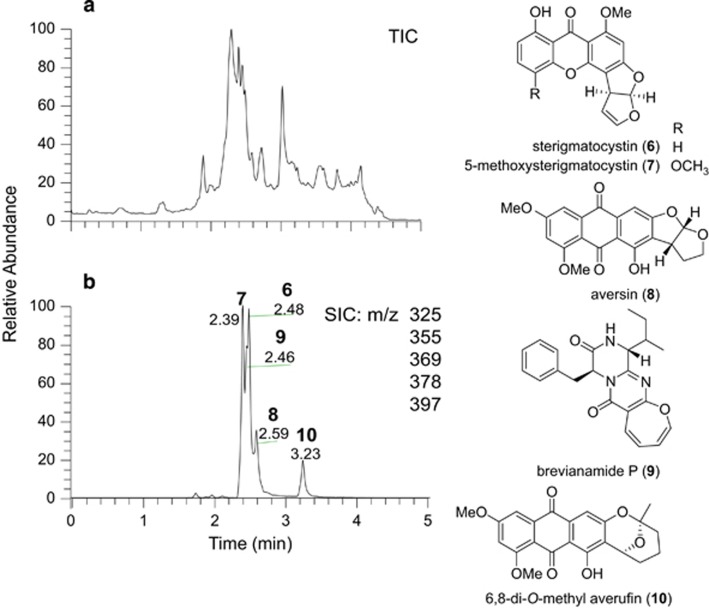
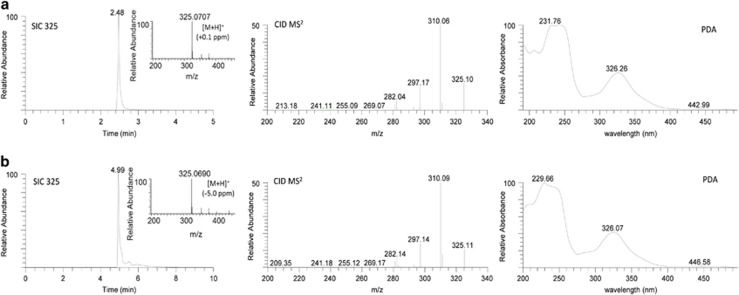
An extract of MSX40080 was dereplicated and determined to contain aflatoxins, thus explaining its potent cytotoxicity.27 To test whether the 5 min method would be able to identify the same aflatoxins, the extract was reanalyzed (Figure 1). Five aflatoxins, including sterigmatocystin (6), 5-methoxysterigmatocystin (7), aversin (8), brevianamide P (9) and 6,8-di-O-methyl averufin (10), were identified, consistent with previous results.27 Comparison of data obtained from both methods indicated that only the retention times were different for the detected compounds, as shown for 6 (Figure 2) and 7–10 (Supplementary Figure S3). In the context of discovering leads for an anticancer drug discovery program, aflatoxins are an obvious nuisance, making their rapid dereplication critical.
Figure 1.
(a) The (+)-ESI-TIC of MSX40080. (b) The selected ion chromatogram (SIC) of dereplicated aflatoxins (6–10) in MSX40080. ESI, electrospray ionization; TIC, total ion chromatogram.
Figure 2.
Comparison of SIC (m/z 325), (+)-HRMS, CID MS/MS (30 eV) and UPLC-PDA data of sterigmatocystin (6) detected in MSX40080 using (a) the 5 min and (b) the 10 min dereplication protocols. CID, collision-induced dissociation; PDA, photodiode array; SIC, selected ion chromatogram; UPLC, ultraperformance liquid chromatography.
Dereplication using UPLC-PDA-HRMS-MS/MS with MDF
The first step of the dereplication strategy was focused on the targeted screening of secondary metabolites in our library using the 5 min UPLC-HRMS-MS/MS methodology. To take this dereplication strategy a step further, identified hits were expanded upon by screening for potential analogs with the aid of Compound Discoverer data-processing software utilizing MDF. MDF was essential to remove the majority of interfering ions and facilitate detection of the potential structural class of related compounds via post acquisition processing of HRMS data. Theoretically, each structural analog possessed relatively minor and well-defined changes in the mass defect from filter reference compounds. Therefore, the mass defects between the reference skeletons and related analogs should fall within a defined mass defect window.
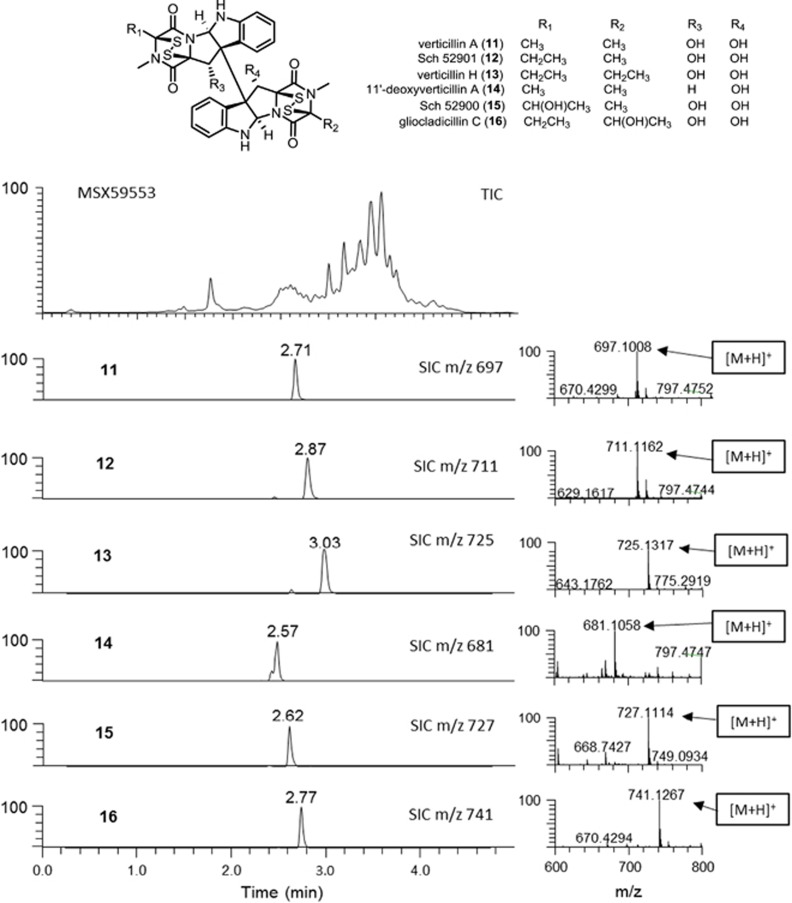
The case of identifying epipolythiodioxopiperazine (ETP) alkaloid analogs in different fungal cultures illustrates the use of the UPLC-PDA-HRMS-MS/MS dereplication strategy and the benefits of MDF. Recent studies have identified verticillin A (11) as an effective tumor suppressor, inducing tumor cell apoptosis in metastatic human colorectal cancer cells at nanomolar concentrations,30, 31 and structurally related analogs have been shown to have potent biological activities.32, 33, 34 Therefore, it was valuable to identify these compounds in extracts, and at the same time determine the presence of structurally related analogs that were not part of our library. In this case we were using dereplication as a means to identify other fungi that biosynthesize these compounds, with the aim of enhancing scale-up and production, as will be reported in the future.
A common characteristic of the 15 analogs, all being dimeric ETPs, is a polysulfide bridge in the molecule (Dictionary of Natural Products, Taylor and Francis Group, Oxford, UK). Supplementary Figure S4 is a graphical representation of the accurate masses and the corresponding mass defects of these compounds. As summarized in Supplementary Table S2, the compounds fall into a narrow range of nominal masses and mass defects. To identify these analogs, a filtering template was set using the structure of Sch 52901 (12) having a nominal mass that is the midpoint of the target compounds, and setting a mass window of ±150 Da. The analogs show a narrow range of mass shifts as a result of modifications in the structures, yielding only a small difference in mass defects relative to the template core structure 12 (Supplementary Table S2). Using a slightly wider mass defect range covering all the potential target compounds (Supplementary Table S2), a mass defect window of ±50 mDa was implemented.
The applicability of MDF for detecting these analogs was illustrated with the analysis of an extract obtained from a culture of Clonostachys rogersoniana (MSX59553) that was previously identified by our research group as a Bionectriaceae sp. (Hypocreales) based on partial 28S rDNA sequence data (JQ749725) and was known to biosynthesize ETP alkaloids.34 Dereplication of this strain using the 5 min HESI+ method identified six known ETPs, including verticillin A (11), Sch 52901 (12), verticillin H (13), 11′-deoxyverticillin (14), Sch 52900 (15), and gliocladicillin C (16) based on retention times, HRMS and MS/MS data (Figure 3 and Supplementary Figure S5). The total ion chromatogram of MSX59553 was analyzed further by MDF with the aim of revealing additional structurally related analogs. MDF significantly reduced the amount of interfering ions (Supplementary Figure S6), facilitating identification of related analogs that were present in the extract. The characteristic peaks of ETPs captured by MDF was consistent with the verticillins identified in the previous analysis.
Figure 3.
The (+)-ESI-TIC of MSX59553 and epipolythiodioxopiperazine analogs (11–16) dereplicated via the 5 min UPLC-PDA-HRMS-MS/MS (CID 30 eV) method. CID, collision-induced dissociation; ESI, electrospray ionization; PDA, photodiode array; SIC, selected ion chromatogram; TIC, total ion chromatogram; UPLC, ultraperformance liquid chromatography.
Identification of the fragmentation pattern for ETP alkaloids
The benefit of using MDF in conjunction with the traditional targeted dereplication strategy is that it gives additional insight into the remaining unknown compounds present in the extract by simplifying the data. Once a potential structurally related compound based on mass defect is identified, its structure must be confirmed. However, with the absence of a standard, this might be a challenging task. Thus, to facilitate characterization within the context of an extract, the characteristic collision-induced dissociation (CID) fragmentation can be analyzed to tentatively assign a structure. In the literature, several studies have been reported that use a CID-MS/MS-based workflow for structural elucidation of natural products.35, 36, 37 Implementation of this strategy requires knowledge of the fragmentation patterns of compounds in the same structural class.
Compounds having the same core substructure generally exhibit similar mass fragmentation patterns, and thus can be identified based on their diagnostic fragment ions.26 For the case of ETP alkaloids, the recurrent fragmentation pattern was characterized based on the MS/MS CID fragmentation taken in positive mode (Supplementary Figure S5). In general, desulfurization of the precursor ion peak ([M-2S+H]+, arising from the polysulfide bridge in each of the dioxopiperazine rings, were observed in all CID-MS/MS analyses. Nearly all compounds exhibited dehydration of the [M-2S+H]+ ion, whereas the compound 11′-deoxyverticillin A (14), which possessed one hydroxy group, did not. This may indicate that the removal of a H2O was dependent on the number of hydroxy moieties in the compounds. After H2O loss, a fragment peak, indicating loss of one of the dioxopiperazine moieties was observed. For instance, in verticillin A, the fragment peak at m/z 615.18 was followed by a peak at m/z 465.08, corresponding to a difference of 150 Da and accounting for the loss of a dioxopiperazine moiety (C7H6N2O2). After this observed dissociation, dehydration and desulfurization occurred, most likely in the other dioxopiperazine moiety of the core.
With the examination of the CID diagnostic fragment ions of the ETP standards (Supplementary Figure S5), we sought to apply this to the rapid characterization of analogs of the ETPs in other complex mixtures, where laboratory reference standards were not available.
Analysis of verticillin production in G35
A culture of Colletotrichum fioriniae (G35) was identified previously by dereplication to produce 11 and 15.38 However, refermentation of the culture using the standard rice culture conditions failed to produce any structurally related analogs. In order to revive the ETP biosynthetic pathway, the OSMAC (one strain-many compounds) approach was applied.39 Using this strategy, 12 separate growth conditions providing different nutrients were tested to identify the most favorable condition that would lead to the revival of the biosynthetic pathway. Still, no analogs were dereplicated in any of the extracts.
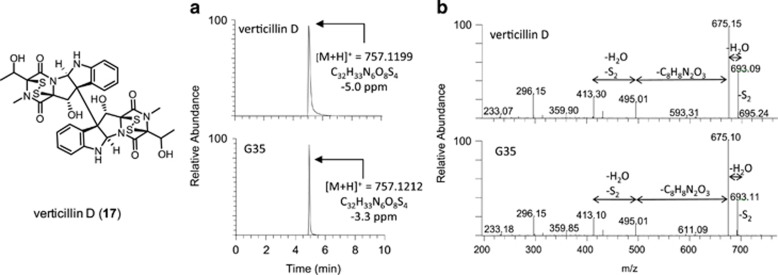
Upon analysis of the HRMS data with MDF, a culture of G35 grown in oatmeal agar (Difco, Sparks, MD, USA) showed a peak that had a corresponding value at m/z 757.1212 that matched the monoisotopic mass of protonated verticillin D (17) with +3.3 ppm mass accuracy (Figure 4). The peak with m/z 757.1212 was identified as an ETP alkaloid based on comparison of the characteristic losses observed in its MS/MS spectrum (Figure 4), exhibiting a distinctive fragmentation pattern similar to that of verticillin A (Supplementary Figure S5). In particular, the fragment ions at m/z 693.11 and m/z 675.10 indicated desulfurization and dehydration, respectively.
Figure 4.
Detection of verticillin D (17) in G35 grown in oatmeal agar after mass defect filtering. (a) Overlay of (+)-ESI-TIC of 17. (b) CID MS/MS (30 eV) spectra of 17. CID, collision-induced dissociation; ESI, electrospray ionization; TIC, total ion chromatogram.
Coincidentally, 17 was isolated from a fungal culture of a Clonostachys sp. (G600), and this presented an opportunity to verify the identity of the peak observed in G35. The HRMS and MS/MS data for 17 were analyzed and matched that of the compound with m/z 757.1212 from G35 (Figure 4). This result verified that 17 was indeed being produced in low quantities by G35 in oatmeal agar, an observation that would not be possible without MDF.
This result demonstrated the advantages of incorporating MDF as a processing technique in the dereplication platform. Verticillin D (17) was not part of the database when the samples were dereplicated and processed originally. As MDF is a post acquisition data mining technique that analyzes solely the HRMS data, samples can be reevaluated as more data become available.
Co-culture analysis using dereplication with MDF
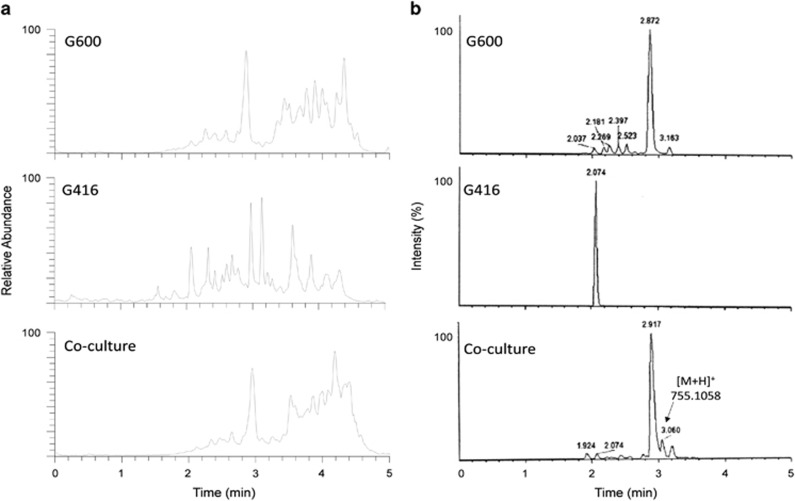
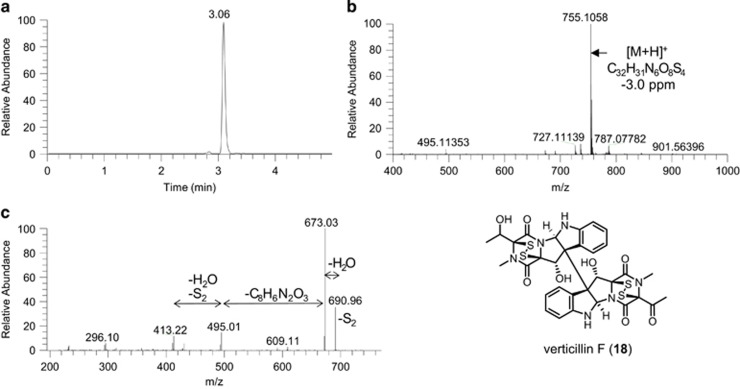
Analysis of the extract of G600 and a strain of Lindgomyces madisonensis (G416) grown in co-culture illustrates the use of dereplication with MDF to determine the production of induced secondary metabolites. In this case, the goal was to determine the presence of structurally related ETPs in the extract. As mentioned previously, G600 was determined to biosynthesize 17; G416, on the other hand, was known to produce mainly polyketides.40 The co-culture was analyzed, and the resulting mass defect profile was compared with separate cultures of G600 and G416 (Figure 5). As expected, 17 was identified and appeared to be the most abundant in both G600 and the co-culture (Supplementary Figure S7). Further inspection of the mass defect profile of the cultures indicated that the co-culture had a peak at m/z 755.1062 that was not detected in a monoculture of G600. The accurate mass for the detected molecular ion peak matched that of verticillin F (18), a dimeric ETP alkaloid, at −3.0 ppm mass accuracy (Figure 6). The MS/MS fragmentation for the peak indicated desulfurization of the precursor ion peak (m/z 690.16), followed by dehydration (m/z 673.03), possible loss of one of the dioxopiperazine moieties (m/z 495.01) and then desulfurization and dehydration (m/z 413.22), all consistent with the distinctive dissociation of ETPs (Figure 6). Based on this observation, the molecular ion peak identified could be tentatively assigned as verticillin F (18), even in the absence of a standard for confirmation.
Figure 5.
Analysis of the extract of G600 and G416 grown alone and in co-culture using dereplication with mass defect filtering to determine the production of induced secondary metabolites. (a) Overlay of (+)-ESI-TIC of the samples. (b) Overlay of (+)-ESI-TIC of the samples after mass defect filtering. ESI, electrospray ionization; TIC, total ion chromatogram.
Figure 6.
(a) (+)-ESI SIC of m/z 755.1058 within a ±5 ppm accuracy. (b) (+)-HRMS of peak at 3.06 min with predicted elemental composition matching the protonated mass of verticillin F with −3.0 ppm accuracy. (c) CID MS/MS (30 eV) spectrum of m/z 755 showing the distinct fragmentation pattern of an epipolythiodioxopiperazine (ETP) alkaloid and consistent with the structure of 18. CID, collision-induced dissociation; ESI, electrospray ionization; SIC, selected ion chromatogram.
Interestingly, the mass defect profile for a monoculture of G416 exhibited a peak at m/z 683.1545. However, the MS/MS profile for that peak did not display the characteristic pattern indicative of an ETP alkaloid (Supplementary Figure S8). This suggested that the detected peak was a false positive and illustrates how MS/MS data can be used to either verify or refute potential hits.
Conclusion
A method to dereplicate fungal extracts was further optimized. This methodology improves a previous dereplication strategy by combining two schemes: (1) targeted screening of compounds known to an in-house built database using a 5 min UPLC-PDA-HRMS-MS/MS where HESI was utilized and ACD/IntelliXtract software to facilitate the dereplication; (2) MDF of resulting accurate mass full-scan raw data for a targeted identification of selected known natural products. In particular, MDF is a data-mining technique that can be applied after acquisition, permitting the reevaluation of data when future compounds of interest are identified.
Experimental procedure
Fungal samples
All fungal samples used in this study were from either the Mycosynthetix (MSX) culture collection (Hillsborough, NC, USA) or the University of North Carolina at Greensboro, Department of Chemistry and Biochemistry, Fungal Culture Collection (Greensboro, NC, USA).
Isolation and identification of fungal strains
Dr Barry Katz isolated Mycosynthetix fungal strain MSX59553 in January 1992 from leaf litter,34 whereas strain MSX40080 was isolated from oak leaf litter in September 1988. Strain G600 was isolated from submerged wood from fresh water in Knight-Brown Nature Preserve in North Carolina, USA (N 36 18.861′, W 80 0.423′). Strain G35 was isolated and identified from surface sterilized seeds of Hydrastis canadensis (goldenseal) as Colletotrichum fioriniae (KX110401).38 In addition to examining the micromorphological characteristics, for molecular identification of new strains dereplicated in the present study (MSX59553 and G600), the internal transcribed spacer region of the nuclear RNA operon was sequenced and analyzed via BLAST search (National Center for Biotechnology Information, Bethesda, MD, USA). In addition, we constructed a Maximum Likelihood phylogenetic tree by incorporating internal transcribed spacer sequences from the newly sequenced strains along with internal transcribed spacer sequences used in a recent study on molecular identification of Clonostachys spp41 based on the outcome of results from BLAST search. Methods for sequencing of the strains, BLAST search and Maximum Likelihood phylogenetic analysis have been summarized previously.42, 43, 44, 45 Based on morphological and molecular data, MSX59553 was identified as Clonostachys rogersoniana,46 as it occurred in a strongly supported clade (99% PHYML bootstrap support) with other sequences of C. rogersoniana, including a reference strain (CBS 582.89). Culture G600 was identified as Clonostachys sp. The Maximum Likelihood tree along with micromorphological characters of the newly sequenced strains are presented in Supplementary Figures S9 and S10. The newly obtained sequences from this study have been deposited in GenBank (accession no. KX845687 and KX845688, respectively).
Fermentation and extraction of fungal cultures
Fungal cultures were grown as described previously.34, 47, 48 Once the screener cultures were generated, 60 ml of 1:1 CH3OH-CHCl3 was added into each flask, followed by shaking for ∼16 h at room temperature on an orbital shaker set at ∼100 r.p.m. The resulting mixtures were filtered under vacuum. To the filtrate, 90 ml of CHCl3 and 150 ml of H2O were added, and the mixture was stirred for 30 min and then transferred into a separatory funnel. The bottom layers were drawn off and dried in vacuo. The dried organic extracts were defatted by reconstituting in a mixture of 100 ml of 1:1 CH3OH-CH3CN and 100 ml of hexane, and then partitioned. The CH3OH-CH3CN layers were collected and concentrated in vacuo.
For G35, from the seed culture, an inoculum was transferred onto five 100 mm × 15 mm Petri dishes with solid oatmeal agar (Difco) mix. The cultures were then allowed to grow for 28 days at room temperature before they were extracted. Extraction of these samples were performed in the same way as the screener cultures.
UPLC-PDA-HRMS-MS/MS
Fungal extracts were dissolved with CH3OH-dioxane (1:1, v/v) to obtain a final concentration of 2 mg ml−1. UPLC-HESI-HRMS-MS/MS was performed on an Acquity UPLC system (Waters, Milford, MA, USA) equipped with a cooled autosampler kept at 10 °C, photodiode array detector (PDA), column manager and binary solvent manager, all coupled to an LTQ-Orbitrap XL mass spectrometry system (Thermo Finnigan, San Jose, CA, USA) with HESI.
UPLC separations were performed at 40 °C on a 50 mm × 2.1 mm i.d., 1.7 μm, Acquity BEH C18 column (Waters) equipped with a security guard precolumn. A linear CH3CN-H2O gradient of 15% CH3CN-H2O to 100% CH3CN in 4 min was applied at a constant flow rate of 0.6 ml min−1; then 100% CH3CN was maintained for 0.75 min before returning to the starting conditions in 0.25 min. All solvents used were acidified with 0.1% formic acid. PDA data were collected from 191 to 499 nm with resolution of 3.6 nm at a 5 point per s sampling rate.
The HESI source was operated in the positive ionization mode with the following conditions: vaporizer temperature set at 300 °C; capillary temperature set at 275 °C; 4.0 kV for the source voltage; 9.0 V for the capillary voltage; and 100.0 V for the tube lens; Nitrogen sheath gas flow rate set at 35.00 arb. Full-scan HRMS data were acquired from m/z 100 to 2000. A CID energy of 30 eV in the ion trap was used to obtain MS/MS fragmentation data. The system was controlled with Thermo Xcalibur software version 2.2 (ThermoFisher Scientific, San Jose, CA, USA).
The LC-MS raw data were processed by the add-in IntelliXtract feature of the ACD MS Manager (Advanced Chemistry Developments, Toronto, ON, Canada). MDF of the HRMS data was performed with Thermo Scientific Compound Discoverer software version 1.0 (Thermo Scientific, San Jose, CA, USA).
LOD and LOQ data analysis
Calibration curves were obtained by preparing twofold serial dilutions of the standards that were analyzed in triplicate via 3 μl injections. All LC-HRMS data were processed using Thermo Xcalibur to automatically integrate and calculate the MS signal peak area of each analyte over noise. Linearity of the calibration curves was assessed by least-squares analysis. The concentration of the analyte that gives the LOD (CLOD) and the concentration at LOQ (CLOQ) were calculated as 3 times the s.e. of the regression line (sy/x) divided by the slope (b) and 10 times sy/x divided by b, respectively. The standards, alamethicin F50 (1), trichokonin VI (2), equisetin (3), acremonidin A (4) and acuminatum B (5), were dissolved separately in a 1:1 CH3CN-dioxane mixture to produce a 15 726 μM stock solution of each. An aliquot from each standard solution was taken to prepare a 2621 μM multi-standard stock solution. A twofold serial dilution of the multistandard stock solution was prepared to make 20 standard solutions at concentrations ranging from 0.0025 to 1310 μM.
Scheme 1.
Proposed workflow for dereplication of fungal extracts including a 5 min ultraperformance liquid chromatography-photodiode array-high resolution tandem mass spectrometry (UPLC-PDA-HRMS-MS/MS) methodology. HRMS and MS/MS (collision-induced dissociation (CID) 30) data were acquired by LTQ Orbitrap using data-dependent scan. A postacquisition data analysis using IntelliXtract, an add-in feature of ACD MS Manager, cross-references the molecular ion peaks and retention times to identify compounds known to the in-house database. For targeted analysis of a specific class of compounds, a user-defined mass defect filtering criterion was designed based on accurate masses and mass defects centered around a core structure. The HRMS data were filtered with Compound Discoverer software to identify components related to the known compounds. The identity of the known compounds were verified based on HRMS, MS/MS fragmentation pattern, retention time and UV absorption maxima. The tentative identity of the discovered unknown compounds were assigned based on the interpretation of their CID-MS/MS (30 eV) fragmentation patterns.
Acknowledgments
This research was supported by Grant P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA.
Footnotes
Supplementary Information accompanies the paper on The Journal of Antibiotics website (http://www.nature.com/ja)
We dedicate this manuscript to the pioneering work of Professor Dr. Satoshi Ōmura. He has been an inspiration to all of us, both for his scientific skills and, perhaps more importantly, for his humanitarian endeavors.
The authors declare no conflict of interest.
Supplementary Material
References
- Burg, R. W. et al. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15, 361–367 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerton, J. R. et al. Avermectins, new family of potent anthelmintic agents: efficacy of the B1a component. Antimicrob. Agents Chemother. 15, 372–378 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, W. C., Burg, R. W., Fisher, M. H., Dybas, R. A. in Pesticide Synthesis Through Rational Approaches. The Discovery of Ivermectin and Other Avermectins (eds. Magee, P. S., Kohn, G. K. & Menn, J. J.5–20 American Chemical Society, (1984). [Google Scholar]
- Newman, D. J. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661 (2016). [DOI] [PubMed] [Google Scholar]
- Cragg, G. M. & Newman, D. J. Natural products: a continuuing source of novel drug leads. Biochim. Biophys. Acta 1830, 3670–3695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, A. L., Edrada-Ebel, R. & Quinn, R. J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 14, 111–129 (2015). [DOI] [PubMed] [Google Scholar]
- Hubert, J., Nuzillard, J.-M. & Renault, J.-H. Dereplication strategies in natural product research: how many tools and methodologies behind the same concept? Phytochem. Rev. 15, 1–41 (2015). [Google Scholar]
- Gaudencio, S. P. & Pereira, F. Dereplication: racing to speed up the natural products discovery process. Nat. Prod. Rep. 32, 779–810 (2015). [DOI] [PubMed] [Google Scholar]
- Ito, T. & Masubuchi, M. Dereplication of microbial extracts and related analytical technologies. J. Antibiot. 67, 353–360 (2014). [DOI] [PubMed] [Google Scholar]
- Kellogg, J. J. et al. Biochemometrics for natural products research: comparison of data analysis approaches and application to identification of bioactive compounds. J. Nat. Prod. 79, 376–386 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfender, J.-L., Marti, G., Thomas, A. & Bertrand, S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 1382, 136–164 (2015). [DOI] [PubMed] [Google Scholar]
- Ernst, M., Silva, D. B., Silva, R. R., Vencio, R. Z. N. & Lopes, N. P. Mass spectrometry in plant metabolomics strategies: from analytical platforms to data acquisition and processing. Nat. Prod. Rep. 31, 784–806 (2014). [DOI] [PubMed] [Google Scholar]
- Tchoumtchoua, J., Njamen, D., Mbanya, J. C., Skaltsounis, A. L. & Halabalaki, M. Structure-oriented UHPLC-LTQ Orbitrap-based approach as a dereplication strategy for the identification of isoflavonoids from Amphimas pterocarpoides crude extract. J. Mass Spectrom. 48, 561–575 (2013). [DOI] [PubMed] [Google Scholar]
- Kinghorn, A. D. Discovery of anticancer agents of diverse natural origin. Pure Appl. Chem. 81, 1051–1063 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Elimat, T. et al. Chemical diversity of metabolites from fungi, cyanobacteria, and plants relative to FDA-approved anticancer agents. ACS Med. Chem. Lett. 3, 645–649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy-Cordero, A. A. et al. Dereplication of macrocyclic trichothecenes from extracts of filamentous fungi through UV and NMR profiles. J. Antibiot. 63, 539–544 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica, V. P. et al. Dereplicating and spatial mapping of secondary metabolites from fungal cultures in situ. J. Nat. Prod. 78, 1926–1936 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Zhang, D., Ray, K. & Zhu, M. Mass defect filter technique and its applications to drug metabolite identification by high-resolution mass spectrometry. J. Mass Spectrom. 44, 999–1016 (2009). [DOI] [PubMed] [Google Scholar]
- Zhu, M. et al. Detection and characterization of metabolites in biological matrices using mass defect filtering of liquid chromatography/high resolution mass spectrometry data. Drug Metab. Dispos. 34, 1722–1733 (2006). [DOI] [PubMed] [Google Scholar]
- Xing, J., Zang, M., Zhang, H. & Zhu, M. The application of high-resolution mass spectrometry-based data-mining tools in tandem to metabolite profiling of a triple drug combination in humans. Anal. Chim. Acta 897, 34–44 (2015). [DOI] [PubMed] [Google Scholar]
- Pähler, A. & Brink, A. Software aided approaches to structure-based metabolite identification in drug discovery and development. Drug Discov. Today Technol. 10, e207–e217 (2013). [DOI] [PubMed] [Google Scholar]
- Sleno, L. The use of mass defect in modern mass spectrometry. J. Mass Spectrom. 47, 226–236 (2012). [DOI] [PubMed] [Google Scholar]
- Ekanayaka, E. A. P., Celiz, M. D. & Jones, A. D. Relative mass defect filtering of mass spectra: a path to discovery of plant specialized metabolites. Plant Physiol. 167, 1221–1232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. et al. Comprehensive qualitative ingredient profiling of Chinese herbal formula Wu-Zhu-Yu decoction via a mass defect and fragment filtering approach using high resolution mass spectrometry. Molecules 21, 664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, T. et al. Rapid identification of ophiopogonins and ophiopogonones in Ophiopogon japonicus extract with a practical technique of mass defect filtering based on high resolution mass spectrometry. J. Chromatogr. A 1227, 234–244 (2012). [DOI] [PubMed] [Google Scholar]
- El-Elimat, T. et al. High-Resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J. Nat. Prod. 76, 1709–1716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W., Bennett, B. D. & Rabinowitz, J. D. Analytical strategies for LC-MS-based targeted metabolomics. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 871, 236–242 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, J. S. Ionization and transmission efficiency in an electrospray ionization-mass spectrometry interface. J. Am. Soc. Mass Spectrom 18, 1582–1590 (2007). [DOI] [PubMed] [Google Scholar]
- Liu, F. et al. Verticillin A overcomes apoptosis resistance in human colon carcinoma through DNA methylation-dependent upregulation of BNIP3. Cancer Res. 71, 6807–6816 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschall, A. V. et al. H3K9 trimethylation silences Fas expression to confer colon carcinoma immune escape and 5-fluorouracil chemoresistance. J. Immunol. 195, 1868–1882 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Miao, Z.-H., Zhao, W.-M. & Ding, J. The p53 pathway is synergized by p38 MAPK signaling to mediate 11,11′-dideoxyverticillin-induced G2/M arrest. FEBS Lett. 579, 3683–3690 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang, Y. X. et al. 11,11′-Dideoxyverticillin: a natural compound possessing growth factor receptor tyrosine kinase-inhibitory effect with anti-tumor activity. Anticancer Drugs 16, 515–524 (2005). [DOI] [PubMed] [Google Scholar]
- Figueroa, M. et al. Cytotoxic epipolythiodioxopiperazine alkaloids from filamentous fungi of the Bionectriaceae. J. Antibiot. 65, 559–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. et al. A novel dereplication strategy for the identification of two new trace compounds in the extract of Gastrodia elata using UHPLC/Q-TOF-MS/MS. J. Chromatogr. B 988, 45–52 (2015). [DOI] [PubMed] [Google Scholar]
- Maslen, S. L., Goubet, F., Adam, A., Dupree, P. & Stephens, E. Structure elucidation of arabinoxylan isomers by normal phase HPLC–MALDI-TOF/TOF-MS/MS. Carbohydr. Res. 342, 724–735 (2007). [DOI] [PubMed] [Google Scholar]
- Schmidt, G. Viscosalines B 1,2 and E 1,2: challenging new 3-alkyl pyridinium alkaloids from the marine sponge Haliclona viscosa. Chem. Eur. J 18, 8180–8189 (2012). [DOI] [PubMed] [Google Scholar]
- Egan, J. M. et al. Antimicrobial fungal endophytes from the botanical medicine goldenseal (Hydrastis canadensis. Phytochem. Lett. 17, 219–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode, H. B., Bethe, B., Hofs, R. & Zeeck, A. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem. 3, 619–627 (2002). [DOI] [PubMed] [Google Scholar]
- Paguigan, N. D., Raja, H. A., Day, C. S. & Oberlies, N. H. Acetophenone derivatives from a freshwater fungal isolate of recently described Lindgomyces madisonensis (G416). Phytochemistry 126, 59–65 (2016). [DOI] [PubMed] [Google Scholar]
- Abreu, L. M., Moreira, G. M., Ferreira, D., Rodrigues-Filho, E. & Pfenning, L. H. Diversity of Clonostachys species assessed by molecular phylogenetics and MALDI-TOF mass spectrometry. Fungal Biol. 118, 1004–1012 (2014). [DOI] [PubMed] [Google Scholar]
- Raja, H. A. et al. Phylogenetic and chemical diversity of fungal endophytes isolated from Silybum marianum (L) Gaertn. (milk thistle). Mycology 6, 8–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja, H. A. et al. Minutisphaerales (Dothideomycetes, Ascomycota): a new order of freshwater ascomycetes including a new family, Minutisphaeraceae, and two new species from North Carolina, USA. Mycologia 107, 845–862 (2015). [DOI] [PubMed] [Google Scholar]
- Raja, H. A., Baker, T. R., Little, J. G. & Oberlies, N. H. DNA barcoding for identification of consumer-relevant mushrooms: a partial solution for product certification? Food Chem. 214, 383–392 (2017). [DOI] [PubMed] [Google Scholar]
- Raja, H., Schoch, C. L., Hustad, V., Shearer, C. & Miller, A. Testing the phylogenetic utility of MCM7 in the Ascomycota. MycoKeys 1, 63 (2011). [Google Scholar]
- Schroers, H. J. A monograph of Bionectria (Ascomycota, Hypocreales, Bionectriaceae) and its Clonostachys anamorphs. Stud. Mycol. 2001, 1–211 (2002). [Google Scholar]
- El-Elimat, T. et al. Greensporones: resorcylic acid lactones from an aquatic Halenospora sp. J. Nat. Prod. 77, 2088–2098 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, A. et al. New diketopiperazine dimer from a filamentous fungal isolate of Aspergillus sydowii. Magn. Reson. Chem. 53, 616–619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.