Abstract
Stable isotope tracers have been invaluable assets in physiological research for over 80 years. The application of substrate‐specific stable isotope tracers has permitted exquisite insight into amino acid, fatty‐acid and carbohydrate metabolic regulation (i.e. incorporation, flux, and oxidation, in a tissue‐specific and whole‐body fashion) in health, disease and response to acute and chronic exercise. Yet, despite many breakthroughs, there are limitations to ‘substrate‐specific’ stable isotope tracers, which limit physiological insight, e.g. the need for intravenous infusions and restriction to short‐term studies (hours) in controlled laboratory settings. In recent years significant interest has developed in alternative stable isotope tracer techniques that overcome these limitations, in particular deuterium oxide (D2O or heavy water). The unique properties of this tracer mean that through oral administration, the turnover and flux through a number of different substrates (muscle proteins, lipids, glucose, DNA (satellite cells)) can be monitored simultaneously and flexibly (hours/weeks/months) without the need for restrictive experimental control. This makes it uniquely suited for the study of ‘real world’ human exercise physiology (amongst many other applications). Moreover, using D2O permits evaluation of turnover of plasma and muscle proteins (e.g. dynamic proteomics) in addition to metabolomics (e.g. fluxomics) to seek molecular underpinnings, e.g. of exercise adaptation. Here, we provide insight into the role of stable isotope tracers, from substrate‐specific to novel D2O approaches, in facilitating our understanding of metabolism. Further novel potential applications of stable isotope tracers are also discussed in the context of integration with the snowballing field of ‘omic’ technologies.
Keywords: exercise, metabolism, muscle, stable isotope
Abbreviations
- AA
amino acid
- D2O
deuterium oxide
- EAA
essential amino acid
- FSR
fractional synthetic rate
- i.v.
intravenous
- LC‐MS/MS
liquid chromatography tandem mass spectrometry
- 3‐MH
3‐methylhistidine
- MPB
muscle protein breakdown
- MPS
muscle protein synthesis
- MRI
magnetic resonance imaging
- Ra
rate of appearance
- Rd
rate of disappearance
- RET
resistance exercise training
What are stable isotope tracers and how are they applied in studies?
An isotope is a species of an element that differs in mass through the addition of one or more neutrons within the atomic nucleus; while naturally occurring they are far less abundant than their more common lighter isotopes. This difference in mass permits these isotopes to be detected, generally using the technique of mass spectrometry – a mass spectrometer separates atoms or molecules based on their mass and/or charge (after undergoing some form of ionization) as they pass through an electrical and/or magnetic field under vacuum; they are traditionally combined with chromatographic (gas chromatography or liquid chromatography) separation techniques, to permit the separation of complex mixtures of organic compounds prior to detection in the mass spectrometer. There are two forms of isotopic tracers: radio‐isotope tracers (e.g. 14C and 3H (tritium)) are radioactive and therefore potentially harmful; their use is now largely restricted to animal rather than human experiments. Stable isotope tracers (e.g. 18O, 15N, 13C and 2H) in contrast, are non‐radioactive, making them ideal for use in human research. Crucially, the chemistry of these elements and the compounds into which they are incorporated is essentially the same as that of the endogenous ones. As such, they can be used to ‘trace’ metabolic flux in pathways of interest and the fate of the tracer can be monitored (via its mass difference), providing vital information regarding rates and extent of its metabolism.
Stable isotope tracers are typically substrate specific. That is, stable isotopically labelled amino acids (AAs) provide information on amino acid and protein metabolism, labelled fatty acids will inform on fat metabolism, while glucose tracers will reflect carbohydrate metabolism and storage. Application of these tracers has been instrumental in yielding information on (1) the synthesis of polymers (i.e. proteins (Rennie et al. 1982; Halliday et al. 1988), triglycerides (Guo & Jensen, 1998) and glycogen (Romijn et al. 1993), (2) substrate uptake, release and intermediary metabolism across or within organs and tissues using arterial–venous balance techniques (Biolo et al. 1994), (3) substrate oxidation (Phillips et al. 1996) and energy expenditure (Goran et al. 1990), and (4) whole body and tissue/organ‐specific fuel metabolism (Rennie & Halliday, 1984). In terms of exercise physiology, these stable isotope tracer approaches have assisted in detailing the complex metabolic responses of tissues to exercise. Some of the most important discoveries include the delineation of protein synthetic and breakdown responses to exercise, i.e. when performed in the absence of nutrition, exercise is catabolic (Biolo et al. 1995; Phillips et al. 1999; Kumar et al. 2009), and that the essential amino acid (EAA) constituents of food are the key nutrients supporting exercise‐induced increases in muscle protein synthesis (MPS) (Pennings et al. 2011; Burd et al. 2011). Furthermore, the use of stable isotope tracers has also been key to determining shifts in substrate utilization and fuel use during exercise, e.g. highlighting reductions in lipid oxidation at increasing exercise intensities (Friedlander et al. 2007), and the important role of endurance training in increasing utilization of lipids to spare muscle glycogen (Phillips et al. 1996). To perform traditional stable isotope tracer experiments requires sterile infusates, intravenous (i.v.) cannulation(s) and multiple (usually muscle) tissue biopsies. Moreover, constant i.v. infusions require subjects be studied in a controlled clinical or laboratory environment – restricting measures to < 24 h and traditionally over 2–8 h – and requiring different stable isotope tracers for each substrate to be ‘measured or traced’ (e.g. [2H2]glucose for glucose metabolism, [U‐13C]palmitate for lipid metabolism and [1,2,13C2]leucine or [2H5]phenylalanine for protein metabolism) (Fig. 1). The time limited and invasive nature of these studies narrows their applications; moreover acute findings may not always translate into a chronic setting, e.g. acute postexercise MPS exhibited no correlation to ensuing skeletal muscle hypertrophy following resistance exercise training (RET) (Mitchell et al. 2014).
Figure 1. Application of substrate‐specific stable isotope tracers for measurement of metabolism.
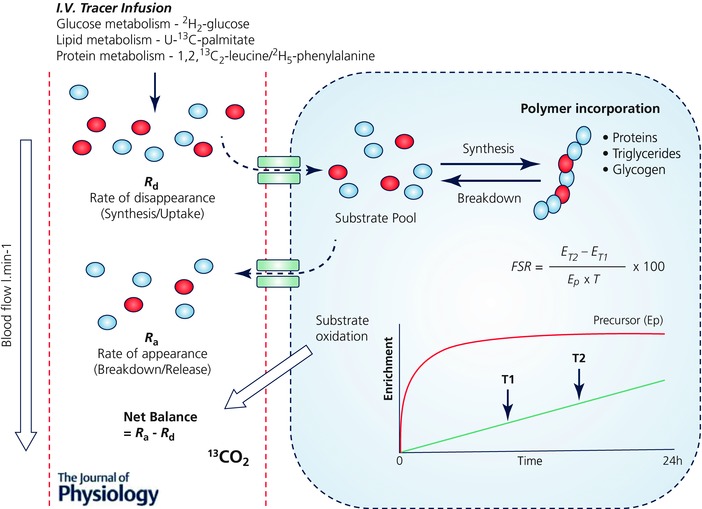
Schematic diagram summarizing the typical application of substrate‐specific tracers for measuring metabolism, utilizing both precursor–product and arterial–venous (A‐V) balance techniques. A single substrate‐specific tracer is required to be administered for the measurement of turnover of each substrate pool of interest (e.g. glucose metabolism using [2H2]glucose, lipid metabolism using [U‐13C]palmitate and protein/AA metabolism using [1,2,13C2]leucine). These substrate‐specific tracers are usually administered by i.v. infusion and as such are limited to measurement periods ≤ 24 h, but typically < 8 h. Measurement of the rate of disappearance (R d) of the stable isotope tracer from the arterial pool provides a proxy for synthesis, while the rate of appearance (R a) of the tracee in the venous pools (i.e. via dilution of the tracer) provides that of breakdown. Stable isotope tracers and A‐V balance calculations can therefore provide estimates of both synthesis and breakdown, as well as de novo synthesis of intermediary metabolites and substrate oxidation across tissues or limbs (when combined with blood flow and CO2 measurements). Furthermore, the rate of synthesis or incorporation into tissue pools (i.e. protein, glycogen, lipids) can be measured using the precursor–product approach. By sampling the tissue or pool at two time points in relation to the precursor enrichment (e.g. AA tracers incorporated into peptides via their corresponding amino acyl‐tRNA) provides a fractional synthesis rate (FSR). ET1, Enrichment in product at time T1; ET2, Enrichment in product at time T2; Ep, Enrichment in precursor pool.
However, despite these limitations, these substrate‐specific tracer techniques have been invaluable to our understanding of basic human metabolism and its responses to stress, nutrition, health and disease. The findings from these studies continue to provide us with a wealth of knowledge and mechanistic understanding of the intricacies of metabolism, and these approaches will continue to underpin advances in human metabolic research. It is important to note that with any in vivo technique, there will be inherent limitations to the methods and the following sections aim to highlight new tools that will compliment these traditional tracer techniques, thereby providing greater insight.
Emerging era of non‐substrate‐specific stable isotope tracers
Deuterium oxide (D2O, ‘heavy water’ or 2H2O) was one of the first stable isotope tracers to be used in metabolic research, by Schoenheimer (Schoenheimer & Rittenberg, 1936) and Ussing (1937, 1938, 1941). Nonetheless, the benefits of D2O as a stable isotope tracer only really began to be recognized in the late 20th century, with technical advances in analytical instrumentation and development of commercially available hybrid MS (i.e. gas chromatography–pyrolysis–isotope ratio mass spectrometry and liquid chromatography–tandem mass spectrometry (LC‐MS/MS); Kuksis & Myher, 1995; Begley & Scrimgeour, 1996). It was soon realized that D2O could provide an alternative approach and overcome some of the issues related to the use of traditional substrate‐specific stable isotope tracers (Dufner & Previs, 2003). Administered orally (Previs et al. 2004; Gasier et al. 2010, 2012; Robinson et al. 2011; MacDonald et al. 2013; Wilkinson et al. 2014, 2015; Decaris et al. 2015) as a single bolus (MacDonald et al. 2013; Wilkinson et al. 2014, 2015) or in the form of regular doses (Robinson et al. 2011; Brook et al. 2015; Decaris et al. 2015), D2O rapidly equilibrates with body water (∼20 min in rats (Dufner et al. 2005); 1–2 h in humans (IAEA, 2011)). This creates a homogeneous labelled precursor pool, which slowly turns over (half‐life ∼9–11 days; MacDonald et al. 2013; Wilkinson et al. 2014), thereby overcoming the need for i.v. stable isotope tracer administration (Dufner & Previs, 2003). Deuterium from the body water is exchanged through biological reductions during de novo (re)synthesis onto a myriad metabolic substrates at stable C‐H positions, allowing rates of flux and turnover (synthesis and breakdown) within these substrate pools to be calculated (Hellerstein, 2004). Thus, unlike substrate‐specific tracer methods, D2O provides an opportunity for measuring the turnover of multiple pools simultaneously (Fig. 2), overcoming the need for an individual stable isotope tracer for each different substrate pool. For example, deuterium is incorporated into AAs (traditionally alanine, glutamate or glutamine is monitored as they can be labelled at multiple carbon positions; Busch et al. 2006; Herath et al. 2011), allowing MPS rates to be probed from the incorporation of labelled AAs into tissue protein (Gasier et al. 2010). In addition through labelling of individual fatty acids, as well as glycerol via glucose, lipid turnover can be quantified (Diraison et al. 1997; Turner et al. 2003; Strawford et al. 2004), whilst labelling by D2O at different steps in de novo nucleotide synthesis can provide rates of DNA (and RNA) turnover (Busch et al. 2007; Voogt et al. 2007). Crucially, the relatively slow turnover of the body water pool permits measurement over longer periods, i.e. days, weeks or months (Robinson et al. 2011; Gasier et al. 2012; Wilkinson et al. 2014, 2015; Brook et al. 2015). This permits experiments to be extended beyond standard limits for traditional stable isotope tracer studies of ∼24 h, thereby providing a vital mechanism for monitoring chronic, cumulative metabolic rates of greater translational relevance.
Figure 2. Application of the non‐substrate‐specific stable isotope tracer D2O for the measurement of metabolic turnover.
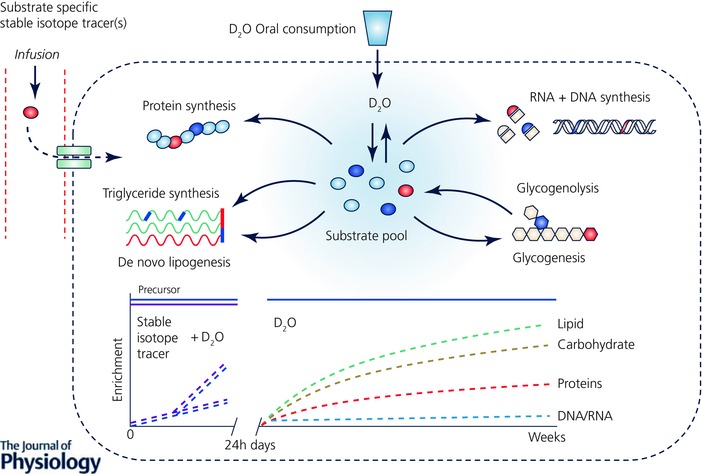
Schematic diagram summarizing how D2O can be used as a stable isotope tracer for measuring metabolism of multiple substrates in vivo. Upon oral administration (avoiding the need for invasive i.v. administration) D2O rapidly equilibrates with the body water pool and distributes equally throughout all tissues and organs. Deuterium from the body water is subsequently incorporated into multiple biological substrates such as glucose, AAs and fatty acids, which, unlike substrate‐specific stable isotope tracers, allows for the measurement of turnover of multiple substrate pools simultaneously. Due to the slow turnover of the body water pool (half‐life ∼9–11 days), D2O can provide measurement over periods as short as a few hours in duration (a time period typical for the use of substrate‐specific stable isotope tracers) up to days, weeks or even months depending on the substrate of interest (e.g. muscle protein turns over relatively slowly ∼1–2% day–1, whereas intramuscular triglycerides turnover at ∼1–2% h–1) and through maintenance of the precursor pool (D2O in the body water pool) with regular oral dosing of D2O, highlighting the benefit and additional value that D2O can provide complimentary to traditional substrate‐specific stable isotope tracers.
Since stable isotope tracers are intended to produce quantitative metabolic flux data, the validity of D2O was recently compared against substrate‐specific stable isotope tracers and was validated over hours, days and weeks. In a preliminary study we quantified MPS using a single oral D2O bolus alongside the ‘gold‐standard’ primed, continuous i.v. infusion of [13C6]phenylalanine within the same person in both fasted and fed (20 g EAA) states over 6 h (Wilkinson et al. 2015). D2O provided comparable mean fractional synthetic rate (FSR) values to those obtained using the traditional phenylalanine tracer, in both fasted and fed states, with a comparable magnitude and direction of change in FSR following feeding (Wilkinson et al. 2015). Whilst this shows validity of the technique in a short‐term laboratory setting, a major benefit of D2O is its potential over longer periods. To address this, we studied individuals over an 8 day period incorporating four bouts of unilateral resistance exercise; in doing so we found that MPS measured with D2O in the non‐exercised leg was equivalent to that normally reported with AA stable isotope tracer (1.2–1.5% day–1 or 0.05–0.06% h–1). Moreover, the exercised leg exhibited significantly greater rates of MPS in multiple muscle protein fractions over the 8 days consistent with early albeit ‘undetectable’ (i.e. by imaging techniques) muscle hypertrophy (Wilkinson et al. 2014). This further highlights the quantitative validity of the approach while exposing the capacity to gain insight into muscle protein fraction (e.g. contractile proteins/mitochondria) regulation on a long‐term basis. Following on from this we conducted a 6‐week RET study and showed that increased MPS over the first 3 weeks of RET matched early, plateauing muscle hypertrophy (Brook et al. 2015). Importantly, in contrast to findings with acute MPS (Mitchell et al. 2014), chronic MPS derived using D2O correlated to long term muscle hypertrophy due to RET (Brook et al. 2015), suggesting D2O can provide a predictive real life representation of exercise adaptation – a premise that was recently confirmed (Damas et al. 2016).
As with all tracer techniques, major questions are, what is the true precursor pool? How do we measure it? And can we define an appropriate surrogate precursor? In terms of protein synthesis, the true precursor pool is the amino acyl‐tRNA; however, this pool is extremely small and labile and difficult to analyse, and therefore a surrogate that best approximates the true labelling is often used, for example, venous plasma α‐ketoisocaproate for leucyl‐tRNA (Watt et al. 1991) or intracellular phenylalanine for phenylalanyl‐tRNA (Baumann et al. 1994). With respect to D2O, the rapid equilibrium throughout the body water pool allows for efficient exchange onto AA intracellularly, and as long as the rate of exchange onto the AA is faster than the rate of turnover of the product (e.g. alanine; Yang et al. 1984), the assumption is made that the labelling of the surrogate precursor, intracellular or plasma alanine, reflects that of the D2O body water labelling (corrected for the number of potential hydrogens labelled on the AA; Dufner & Previs, 2003; Gasier et al. 2010). Therefore we can use body water labelling to determine the true precursor for the alanyl‐tRNA. This assumption seems to hold for the case of plasma alanine (Dufner et al. 2005; Belloto et al. 2007), and intracellular alanine (Belloto et al. 2007). Significant dilutions to the alanine pool (i.e. through feeding) have been shown to equilibrate rapidly (Dufner et al. 2005; Belloto et al. 2007). As with many similar tracer techniques, the assumption is made that tracer recycling is minimal, particularly where the turnover of the pool is slow, such as in skeletal muscle. In addition with the rapid exchange of deuterium onto AA, any dilution through recycling may be overcome by this rapid exchange. This, however, is something that needs experimental confirmation, particularly with studies performed over longer periods, and groups are beginning to investigate these modelling assumptions further (Previs et al. 2015; Zhou et al. 2015).
Beyond protein metabolism, others have been able to use D2O to track synthesis rates of muscle DNA during prolonged exercise (Robinson et al. 2011), a technique with clear importance due to the controversial role of satellite cells in exercise adaptation (Gundersen, 2016). The same is theoretically true for RNA, meaning one can track the role of ribosomal biogenesis in human exercise adaptation. In addition, D2O has been used to trace cholesterol (and other sterols), plasma lipid metabolism (Castro‐Perez et al. 2011), and tissue triglyceride and glucose turnover (Strawford et al. 2004), e.g. in relation to energy substrate metabolism. Thus, D2O offers a full complement of tools for providing detailed translational insight into the metabolic basis of the physiological adaptation to exercise in both acute and chronic settings.
Stable isotope tracer and D2O ‘omics’: towards a more mechanistic era
Tissues consist of tens of thousands of proteins and many more metabolites that perform and regulate different functions at varying rates. Therefore measures of crudely fractionated proteins, e.g. sarcoplasmic/myofibrillar or ‘static’ metabolite abundances, provide limited information on individual proteins and metabolic fluxes. To gain insight into this, the recent explosion of systems biology has led to the combination of stable isotope tracers/D2O with ‘omics’, particularly metabolomics and proteomics. For instance, using D2O ingestion in humans, Hellerstein et al. devised a method to isolate peptides using nanoLC‐MS/MS and to quantify enrichment of deuterium within these peptides to calculate rates of turnover for individual proteins (Price et al. 2012). Unsurprisingly, despite their infancy, these methods are rapidly being utilized to provide unique information regarding tissue proteome dynamics in response to exercise. For example, Shankaran and colleagues were able to quantify FSRs for 273 individual muscle proteins in response to 3 weeks of sprint interval training, and highlight the distinct inter‐individual differences in FSR responses for each individual's proteome, suggestive that exercise responses are highly specific to each individual (Shankaran et al. 2016). Moreover, using these techniques the notion of a ‘virtual muscle biopsy’ has been described. Using D2O‐led dynamic proteomics, plasma proteins, such as the muscle‐specific creatine kinase and carbonic anhydrase 3 accurately represented rates of turnover of the same proteins (or crude fractions) sampled from muscle (Shankaran et al. 2016). This could prove useful in situations where muscle biopsies may be contraindicated, such as in young children or frail elderly, or where they are not easily obtainable such as during exercise.
Alongside novel proteomics, the emerging application of stable isotope tracers in metabolomics now enables ‘fluxomics’ (Fig. 3), whereby flux through metabolic pathways can be dynamically monitored (Niedenführ et al. 2014). A recent example of this was provided in an outbred rat model selected for high or low intrinsic or inborn aerobic capacity (Overmyer et al. 2015). Using a [U‐13C,15N]valine tracer, and monitoring the enrichment of 13C labelling in downstream metabolites of valine such as C3/C4 carnitines and β‐hydroxyisobutyrate, and the 15N labelling of transamination metabolites such as glutamate, it was shown that high aerobic capacity was associated with increased flux through branched chain amino acid catabolic pathways, which combined with more efficient fatty acid utilization spared muscle glycogen (Overmyer et al. 2015). These fluxomics techniques could become even more powerful when combined with novel tracers such as D2O, which has the potential to provide flux data from a multitude of different pathways simultaneously (through mechanisms described above (Fig. 3). Enabling energy substrate flux to be dynamically probed in an unbiased manner will be key to exercise physiology research as it can provide dynamic metabolic data in response to interventions such as exercise and diet, e.g. to quantify the mechanisms of energy substrate contributions to exercise. Whilst these techniques are still in their infancy, the application of dynamic proteomics and metabolomics alongside traditional ‘bulk’ metabolic tracer studies will provide tools to develop a more integrated and holistic understanding of physiological and health adaptations to exercise. It is noteworthy that all are indeed eminently doable in humans.
Figure 3. D2O and stable isotope tracer ‘omics’.
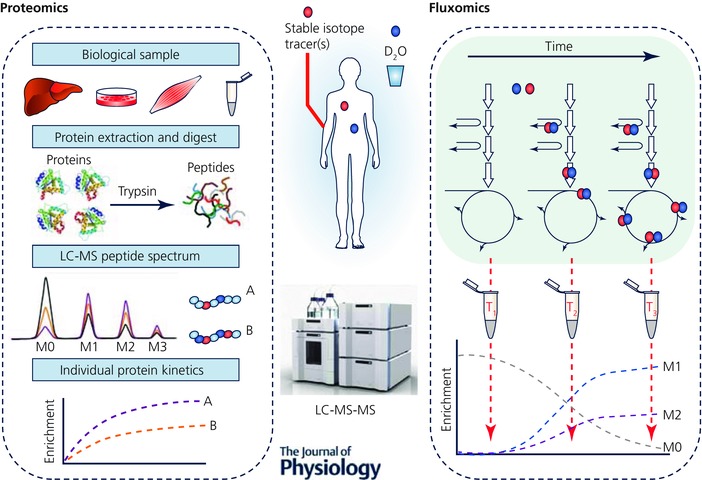
Summary of how D2O and substrate‐specific stable isotope tracers can be used in conjunction with omics techniques (proteomics and metabolomics) for measuring the turnover of individual proteins and the rates of flux through individual metabolic pathways (fluxomics) in vivo. Proteomics: bulk tissue protein is made up of thousands of individual proteins that have varying functions and turnover rates; using AA stable isotope tracers and D2O alongside novel nanoLC‐MS/MS techniques, the rates of turnover of individual proteins can be determined by looking for incorporation of isotopic label into isolated peptides. This will provide a more holistic insight into the regulation of tissue protein to interventions such as exercise. Fluxomics: complimentary to this, using metabolomics in conjunction with stable isotope tracers or D2O the flux of metabolites through different metabolic pathways can be measured, by monitoring the level of isotopic labelling in downstream metabolites. This will provide information on rates of flux through these metabolic pathways, an application particularly suited to exercise physiology where substrate fluxes will dynamically alter in response to different intensities, modes and durations of exercise. M0‐M2 represents the mass isotopomer distribution.
Emerging non‐invasive stable isotope tracer techniques: muscle protein breakdown and mass
Quantification of 3‐methylhistidine (3‐MH) in urine (a post‐translationally modified histidine residue that cannot be reincorporated into new peptides following breakdown) has been used as a proxy for muscle protein breakdown (MPB) for > 40 years (Bilmazes et al. 1978). Nonetheless, its abundance is very low, routinely being measured through 24 h urine collections, and it can be affected by dietary (meat) intake thereby influencing its accuracy (Marliss et al. 1979). However, via oral provision of 2H3‐labelled 3‐MH and monitoring its dilution by endogenously released 3‐MH in temporal blood or urine samples, a rate of whole body myofibrillar protein breakdown can be estimated in an approach that is not significantly influenced by diet (Sheffield‐Moore et al. 2014). Therefore, this provides a future opportunity to investigate the ‘missing part of the balance equation’ (most people tend to look only at MPS due to technical challenges of measuring MPB) and how MPB is modulated by exercise or nutritional interventions.
The quantification of whole body skeletal muscle mass, for example, typically requires expensive imaging technology (magnetic resonance imaging (MRI), computer tomography, dual‐energy X‐ray absorptiometry). However, it was recently shown that whole body muscle mass could be determined by measuring the enrichment of [2H3]creatinine in the urine following provision of a small oral bolus of [2H3]creatine; oral [2H3]creatine equilibrates within the muscle creatine pool, is converted to creatinine at a constant rate of ∼2% day–1 and excreted in urine, where the enrichment of [2H3]creatinine in the urine is representative of [2H3]creatine enrichment in the muscle. From this, creatine pool size and skeletal muscle mass can be estimated (Stimpson et al. 2012, 2013; Clark et al. 2014). While not a new technique (Kreisberg et al. 1970; Picou et al. 1976), this has since been validated providing accurate measures of whole body skeletal muscle which correlate with MRI measures in humans (Clark et al. 2014) and changes in muscle mass in growing animals (Stimpson et al. 2013).
Conclusions: the future
Substrate‐specific stable isotope tracers have provided great insight into protein, lipid and carbohydrate metabolism responses to acute/chronic exercise. Recent developments of novel stable isotope tracer techniques mean it is now possible to use a single non‐substrate‐specific tracer, D2O, to measure the turnover of multiple substrate pools simultaneously in ‘free living’ humans over hours to months, overcoming limitations associated with substrate‐specific tracers. When applied in conjunction with proteomics and metabolomics, D2O has the power to probe metabolism down to the level of the turnover of single proteins or flux through metabolic pathways. The combination of traditional substrate‐specific stable isotope tracers for measurement of acute tissue‐specific and whole body metabolism and D2O for chronic ‘free living’ measurements of multiple substrate turnover, single protein turnover and flux, alongside other minimally invasive stable isotope tracer techniques (e.g. MPB and muscle mass), means that in the future we can provide in humans a dynamic holistic picture of exercise metabolism in the minutest detail.
Additional information
Competing interests
The authors declare no competing interests.
Author contributions
All authors contributed to conception, writing and final editing of the manuscript. All authors have approved the final version of the manuscript and agree to be accountable for all aspects of the work. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
D.J.W. is a Post‐doctoral Research Fellow funded through the MRC‐ARUK Centre for Musculoskeletal Ageing Research. The MRC‐ARUK Centre for Musculoskeletal Ageing Research was funded by grants from the Medical Research Council (grant number MR/K00414X/1) and Arthritis Research UK (grant number 19891) awarded to the Universities of Nottingham and Birmingham. The authors’ work discussed in the review was funded through the Dunhill Medical Trust (grant number R264/1112), The Physiological Society and the Medical Research Council Confidence in Concept programme (grant number CIC12019).
Acknowledgements
The authors would like to acknowledge Dr Bethan Phillips for her insightful feedback following drafting of the manuscript.
Biographies
Daniel Wilkinson (Assistant Professor),

Matthew Brook (Post‐Doctoral Research Associate),
Ken Smith (Principal Research Fellow) and
Philip Atherton (Professor) work within the MRC‐ARUK Centre for Musculoskeletal Ageing Research at the University of Nottingham. Their research is involved in identification of the central mechanisms regulating metabolism in human musculoskeletal tissues, particularly those that govern the regulation of protein metabolism in response to ageing and disease. The group has extensive experience of using and developing stable isotope tracer techniques and applying them to human metabolic research. Recent developments within the group have focused on the application of novel D2O‐based stable isotope tracer techniques and how these can be integrated alongside ‘omic’ techniques, such as proteomics and metabolomics, to provide a better holistic understanding of the regulation of metabolism.
This review was presented at the symposium “New technologies providing insight to human physiological adaptation”, which took place at the meeting of The Biomedical Basis of Elite Performance in Nottingham, UK, 6–8 March 2016.
References
- Baumann PQ, Stirewalt WS, O'Rourke BD, Howard D & Nair KS (1994). Precursor pools of protein synthesis: a stable isotope study in a swine model. Am J Physiol Endocrinol Metab 267, E203–E209. [DOI] [PubMed] [Google Scholar]
- Begley I & Scrimgeour C (1996). On‐line reduction of H2O for δ2H and δ18O measurement by continuous‐flow isotope ratio mass spectrometry. Rapid Commun Mass Spectrom 10, 969–973. [Google Scholar]
- Belloto E, Diraison F, Basset A, Allain G, Abdallah P & Beylot M (2007). Determination of protein replacement rates by deuterated water: validation of underlying assumptions. Am J Physiol Endocrinol Metab 292, E1340–E1347. [DOI] [PubMed] [Google Scholar]
- Bilmazes C, Uauy R, Haverberg LN, Munro HN & Young VR (1978). Musle protein breakdown rates in humans based on Nτ‐methylhistidine (3‐methylhistidine) content of mixed proteins in skeletal muscle and urinary output of Nτ‐methylhistidine. Metabolism 27, 525–530. [DOI] [PubMed] [Google Scholar]
- Biolo G, Gastaldelli A, Zhang XJ & Wolfe RR (1994). Protein synthesis and breakdown in skin and muscle: a leg model of amino acid kinetics. Am J Physiol Endocrinol Metab 267, E467–E474. [DOI] [PubMed] [Google Scholar]
- Biolo G, Maggi SP, Williams BD, Tipton KD & Wolfe RR (1995). Increased rates of muscle protein turnover and amino acid transport after resistance exercise in humans. Am J Physiol Endocrinol Metab 268, E514–E520. [DOI] [PubMed] [Google Scholar]
- Brook M, Wilkinson D, Mitchell W, Lund J, Szewczyk NJ, Greenhaff P, Smith K & Atherton P (2015). Skeletal muscle hypertrophy is most active during early resistance exercise training responses, matching long term deuterium oxide (D2O)‐derived measures of muscle protein synthesis and mTORc1‐signaling. FASEB J 29, 4485–4496. [DOI] [PubMed] [Google Scholar]
- Burd NA, West DWD, Moore DR, Atherton PJ, Staples AW, Prior T, Tang JE, Rennie MJ, Baker SK & Phillips SM (2011). Enhanced amino acid sensitivity of myofibrillar protein synthesis persists for up to 24 h after resistance exercise in young men. J Nutr 141, 568–573. [DOI] [PubMed] [Google Scholar]
- Busch R, Kim Y‐K, Neese RA, Schade‐Serin V, Collins M, Awada M, Gardner JL, Beysen C, Marino ME, Misell LM & Hellerstein MK (2006). Measurement of protein turnover rates by heavy water labeling of nonessential amino acids. Biochim Biophys Acta 1760, 730–744. [DOI] [PubMed] [Google Scholar]
- Busch R, Neese RA, Awada M, Hayes GM & Hellerstein MK (2007). Measurement of cell proliferation by heavy water labeling. Nat Protoc 2, 3045–3057. [DOI] [PubMed] [Google Scholar]
- Castro‐Perez J, Previs SF, McLaren DG, Shah V, Herath K, Bhat G, Johns DG, Wang S‐P, Mitnaul L, Jensen K, Vreeken R, Hankemeier T, Roddy TP & Hubbard BK (2011). In vivo D2O labeling to quantify static and dynamic changes in cholesterol and cholesterol esters by high resolution LC/MS. J Lipid Res 52, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RV, Walker AC, O'Connor‐Semmes RL, Leonard MS, Miller RR, Stimpson SA, Turner SM, Ravussin E, Cefalu WT, Hellerstein MK & Evans WJ (2014). Total body skeletal muscle mass: estimation by creatine (methyl‐d3) dilution in humans. J Appl Physiol 116, 1605–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas F, Phillips SM, Libardi CA, Vechin FC, Lixandrão ME, Jannig PR, Costa LAR, Bacurau AV, Snijders T, Parise G, Tricoli V, Roschel H & Ugrinowitsch C (2016). Resistance training‐induced changes in integrated myofibrillar protein synthesis are related to hypertrophy only after attenuation of muscle damage. J Physiol 594, 5209–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaris ML, Emson CL, Li K, Gatmaitan M, Luo F, Cattin J, Nakamura C, Holmes WE, Angel TE, Peters MG, Turner SM & Hellerstein MK (2015). Turnover rates of hepatic collagen and circulating collagen‐associated proteins in humans with chronic liver disease. PLoS One 10, e0123311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diraison F, Pachiaudi C & Beylot M (1997). Measuring lipogenesis and cholesterol synthesis in humans with deuterated water: use of simple gas chromatographic/mass spectrometric techniques. J Mass Spectrom 32, 81–86. [DOI] [PubMed] [Google Scholar]
- Dufner DA, Bederman IR, Brunengraber DZ, Rachdaoui N, Ismail‐Beigi F, Siegfried BA, Kimball SR & Previs SF (2005). Using 2H2O to study the influence of feeding on protein synthesis: effect of isotope equilibration in vivo vs. in cell culture. Am J Physiol Endocrinol Metab 288, E1277–E1283. [DOI] [PubMed] [Google Scholar]
- Dufner D & Previs SF (2003). Measuring in vivo metabolism using heavy water. Curr Opin Clin Nutr Metab Care 6, 511–517. [DOI] [PubMed] [Google Scholar]
- Friedlander AL, Jacobs KA, Fattor JA, Horning MA, Hagobian TA, Bauer TA, Wolfel EE & Brooks GA (2007). Contributions of working muscle to whole body lipid metabolism are altered by exercise intensity and training. Am J Physiol Endocrinol Metab 292, E107–E116. [DOI] [PubMed] [Google Scholar]
- Gasier HG, Fluckey JD & Previs SF (2010). The application of 2H2O to measure skeletal muscle protein synthesis. Nutr Metab (Lond) 7, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasier HG, Fluckey JD, Previs SF, Wiggs MP & Riechman SE (2012). Acute resistance exercise augments integrative myofibrillar protein synthesis. Metabolism 61, 153–156. [DOI] [PubMed] [Google Scholar]
- Goran MI, Peters EJ, Herndon DN & Wolfe RR (1990). Total energy expenditure in burned children using the doubly labeled water technique. Am J Physiol Endocrinol Metab 259, E576–E585. [DOI] [PubMed] [Google Scholar]
- Gundersen K (2016). Muscle memory and a new cellular model for muscle atrophy and hypertrophy. J Exp Biol 219, 235–242. [DOI] [PubMed] [Google Scholar]
- Guo Z & Jensen MD (1998). Intramuscular fatty acid metabolism evaluated with stable isotopic tracers. J Appl Physiol 84, 1674–1679. [DOI] [PubMed] [Google Scholar]
- Halliday D, Pacy PJ, Cheng KN, Dworzak F, Gibson JN & Rennie MJ (1988). Rate of protein synthesis in skeletal muscle of normal man and patients with muscular dystrophy: a reassessment. Clin Sci (Lond) 74, 237–240. [DOI] [PubMed] [Google Scholar]
- Hellerstein MK (2004). New stable isotope‐mass spectrometric techniques for measuring fluxes through intact metabolic pathways in mammalian systems: introduction of moving pictures into functional genomics and biochemical phenotyping. Metab Eng 6, 85–100. [DOI] [PubMed] [Google Scholar]
- Herath K, Bhat G, Miller PL, Wang S‐P, Kulick A, Andrews‐Kelly G, Johnson C, Rohm RJ, Lassman ME, Previs SF, Johns DG, Hubbard BK & Roddy TP (2011). Equilibration of 2H labeling between body water and free amino acids: enabling studies of proteome synthesis. Anal Biochem 415, 197–199. [DOI] [PubMed] [Google Scholar]
- International Atomic Energy Agency (IAEA) (2011). Introduction to Body Composition Assessment Using the Deuterium Dilution Technique with Analysis of Saliva Samples by Fourier Transform Infrared Spectrometry . IAEA, Vienna.
- Kreisberg RA, Bowdoin B & Meador CK (1970). Measurement of muscle mass in humans by isotopic dilution of creatine‐14C. J Appl Physiol 28, 264–267. [DOI] [PubMed] [Google Scholar]
- Kuksis A & Myher JJ (1995). Application of tandem mass spectrometry for the analysis of long‐chain carboxylic acids. J Chromatogr B Biomed Sci Appl 671, 35–70. [DOI] [PubMed] [Google Scholar]
- Kumar V, Atherton P, Smith K & Rennie MJ (2009). Human muscle protein synthesis and breakdown during and after exercise. J Appl Physiol 106, 2026–2039. [DOI] [PubMed] [Google Scholar]
- MacDonald AJ, Small AC, Greig CA, Husi H, Ross JA, Stephens NA, Fearon KCH & Preston T (2013). A novel oral tracer procedure for measurement of habitual myofibrillar protein synthesis. Rapid Commun Mass Spectrom 27, 1769–1777. [DOI] [PubMed] [Google Scholar]
- Marliss EB, Wei CN & Dietrich LL (1979). The short‐term effects of protein intake on 3‐methylhistidine excretion. Am J Clin Nutr 32, 1617–1621. [DOI] [PubMed] [Google Scholar]
- Mitchell CJ, Churchward‐Venne TA, Parise G, Bellamy L, Baker SK, Smith K, Atherton PJ & Phillips SM (2014). Acute post‐exercise myofibrillar protein synthesis is not correlated with resistance training‐induced muscle hypertrophy in young men. PLoS One 9, e89431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenführ S, Wiechert W & Nöh K (2014). How to measure metabolic fluxes: a taxonomic guide for 13C fluxomics. Curr Opin Biotechnol 34C, 82–90. [DOI] [PubMed] [Google Scholar]
- Overmyer KA, Evans CR, Qi NR, Minogue CE, Carson JJ, Chermside‐Scabbo CJ, Koch LG, Britton SL, Pagliarini DJ, Coon JJ & Burant CF (2015). Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell Metab 21, 468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings B, Koopman R, Beelen M, Senden JMG, Saris WHM & van Loon LJC (2011). Exercising before protein intake allows for greater use of dietary protein‐derived amino acids for de novo muscle protein synthesis in both young and elderly men. Am J Clin Nutr 93, 322–331. [DOI] [PubMed] [Google Scholar]
- Phillips SM, Green HJ, Tarnopolsky MA, Heigenhauser GF, Hill RE & Grant SM (1996). Effects of training duration on substrate turnover and oxidation during exercise. J Appl Physiol 81, 2182–2191. [DOI] [PubMed] [Google Scholar]
- Phillips SM, Tipton KD, Ferrando AA & Wolfe RR (1999). Resistance training reduces the acute exercise‐induced increase in muscle protein turnover. Am J Physiol Endocrinol Metab. 276, E118–E124. [DOI] [PubMed] [Google Scholar]
- Picou D, Reeds PJ, Jackson A & Poulter N (1976). The measurement of muscle mass in children using [15N]creatine. Pediatr Res 10, 184–188. [DOI] [PubMed] [Google Scholar]
- Previs SF, Fatica R, Chandramouli V, Alexander JC, Brunengraber H & Landau BR (2004). Quantifying rates of protein synthesis in humans by use of 2H2O: application to patients with end‐stage renal disease. Am J Physiol Endocrinol Metab 286, E665–E672. [DOI] [PubMed] [Google Scholar]
- Previs SF, Herath K, Castro‐Perez J, Mahsut A, Zhou H, McLaren DG, Shah V, Rohm RJ, Stout SJ, Zhong W, Wang S‐P, Johns DG, Hubbard BK, Cleary MA & Roddy TP (2015). Effect of error propagation in stable isotope tracer studies: an approach for estimating impact on apparent biochemical flux. Methods Enzymol 561, 331–358. [DOI] [PubMed] [Google Scholar]
- Price JC, Holmes WE, Li KW, Floreani NA, Neese RA, Turner SM & Hellerstein MK (2012). Measurement of human plasma proteome dynamics with 2H2O and liquid chromatography tandem mass spectrometry. Anal Biochem 420, 73–83. [DOI] [PubMed] [Google Scholar]
- Rennie MJ, Edwards RH, Halliday D, Matthews DE, Wolman SL & Millward DJ (1982). Muscle protein synthesis measured by stable isotope techniques in man: the effects of feeding and fasting. Clin Sci (Lond) 63, 519–523. [DOI] [PubMed] [Google Scholar]
- Rennie MJ & Halliday D (1984). The use of stable isotope tracers as metabolic probes of whole‐body and limb metabolism. Proc Nutr Soc 43, 189–196. [DOI] [PubMed] [Google Scholar]
- Robinson MM, Turner SM, Hellerstein MK, Hamilton KL & Miller BF (2011). Long‐term synthesis rates of skeletal muscle DNA and protein are higher during aerobic training in older humans than in sedentary young subjects but are not altered by protein supplementation. FASEB J 25, 3240–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romijn JA, Coyle EF, Sidossis LS, Gastaldelli A, Horowitz JF, Endert E & Wolfe RR (1993). Regulation of endogenous fat and carbohydrate metabolism in relation to exercise intensity and duration. Am J Physiol Endocrinol Metab 265, E380–E391. [DOI] [PubMed] [Google Scholar]
- Schoenheimer R & Rittenberg D (1936). Deuterium as an indicator in the study of intermediary metabolism – VI. Synthesis and destruction of fatty acids in the organism. J Biol Chem 114, 381–396. [Google Scholar]
- Shankaran M, King CL, Angel TE, Holmes WE, Li KW, Colangelo M, Price JC, Turner SM, Bell C, Hamilton KL, Miller BF & Hellerstein MK (2016). Circulating protein synthesis rates reveal skeletal muscle proteome dynamics. J Clin Invest 126, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield‐Moore M, Dillon EL, Randolph KM, Casperson SL, White GR, Jennings K, Rathmacher J, Schuette S, Janghorbani M, Urban RJ, Hoang V, Willis M & Durham WJ (2014). Isotopic decay of urinary or plasma 3‐methylhistidine as a potential biomarker of pathologic skeletal muscle loss. J Cachexia Sarcopenia Muscle 5, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson SA, Leonard MS, Clifton LG, Poole JC, Turner SM, Shearer TW, Remlinger KS, Clark R V, Hellerstein MK & Evans WJ (2013). Longitudinal changes in total body creatine pool size and skeletal muscle mass using the D3‐creatine dilution method. J Cachexia Sarcopenia Muscle 4, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson SA, Turner SM, Clifton LG, Poole JC, Mohammed HA, Shearer TW, Waitt GM, Hagerty LL, Remlinger KS, Hellerstein MK & Evans WJ (2012). Total‐body creatine pool size and skeletal muscle mass determination by creatine‐(methyl‐d3) dilution in rats. J Appl Physiol 112, 1940–1948. [DOI] [PubMed] [Google Scholar]
- Strawford A, Antelo F, Christiansen M & Hellerstein MK (2004). Adipose tissue triglyceride turnover, de novo lipogenesis, and cell proliferation in humans measured with 2H2O. Am J Physiol Endocrinol Metab 286, E577–E588. [DOI] [PubMed] [Google Scholar]
- Turner SM, Murphy EJ, Neese RA, Antelo F, Thomas T, Agarwal A, Go C & Hellerstein MK (2003). Measurement of TG synthesis and turnover in vivo by 2H2O incorporation into the glycerol moiety and application of MIDA. Am J Physiol Endocrinol Metab 285, E790–E803. [DOI] [PubMed] [Google Scholar]
- Ussing H (1937). The exchange of H and D atoms between water and protein in vivo and in vitro. Skand Arch Physiol 78, 225–241. [Google Scholar]
- Ussing H (1938). The exchange of H and D atoms between water and protein in vivo and in vitro. Skand Arch Physiol 77, 107–122. [Google Scholar]
- Ussing H (1941). The rate of protein renewal in mice and rats studied by means of heavy hydrogen. Acta Physiol Scand 2, 209–221. [Google Scholar]
- Voogt JN, Awada M, Murphy EJ, Hayes GM, Busch R & Hellerstein MK (2007). Measurement of very low rates of cell proliferation by heavy water labeling of DNA and gas chromatography/pyrolysis/isotope ratio‐mass spectrometric analysis. Nat Protoc 2, 3058–3062. [DOI] [PubMed] [Google Scholar]
- Watt PW, Lindsay Y, Scrimgeour CM, Chien PA, Gibson JN, Taylor DJ & Rennie MJ (1991). Isolation of aminoacyl‐tRNA and its labeling with stable‐isotope tracers: Use in studies of human tissue protein synthesis. Proc Natl Acad Sci USA 88, 5892–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DJ, Cegielski J, Phillips BE, Boereboom C, Lund JN, Atherton PJ & Smith K (2015). Internal comparison between deuterium oxide (D2O) and L‐[ring‐13C6]phenylalanine for acute measurement of muscle protein synthesis in humans. Physiol Rep 3, e12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DJ, Franchi MV, Brook MS, Narici MV, Williams JP, Mitchell WK, Szewczyk NJ, Greenhaff PL, Atherton PJ & Smith K (2014). A validation of the application of D2O stable isotope tracer techniques for monitoring day‐to‐day changes in muscle protein subfraction synthesis in humans. Am J Physiol Endocrinol Metab 306, E571–E579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RD, Matthews DE, Bier DM, Lo C & Young VR (1984). Alanine kinetics in humans: influence of different isotopic tracers. Am J Physiol Endocrinol Metab 247, E634–E638. [DOI] [PubMed] [Google Scholar]
- Zhou H, Wang S‐P, Herath K, Kasumov T, Sadygov RG, Previs SF & Kelley DE (2015). Tracer‐based estimates of protein flux in cases of incomplete product renewal: evidence and implications of heterogeneity in collagen turnover. Am J Physiol Endocrinol Metab 309, E115–E121. [DOI] [PMC free article] [PubMed] [Google Scholar]


