Abstract
The speed of sequencing of microbial genomes and metagenomes is providing an ever increasing resource for the identification of new robust biocatalysts with industrial applications for many different aspects of industrial biotechnology. Using ‘natures catalysts’ provides a sustainable approach to chemical synthesis of fine chemicals, general chemicals such as surfactants and new consumer-based materials such as biodegradable plastics. This provides a sustainable and ‘green chemistry’ route to chemical synthesis which generates no toxic waste and is environmentally friendly. In addition, enzymes can play important roles in other applications such as carbon dioxide capture, breakdown of food and other waste streams to provide a route to the concept of a ‘circular economy’ where nothing is wasted. The use of improved bioinformatic approaches and the development of new rapid enzyme activity screening methodology can provide an endless resource for new robust industrial biocatalysts.This mini-review will discuss several recent case studies where industrial enzymes of ‘high priority’ have been identified and characterised. It will highlight specific hydrolase enzymes and recent case studies which have been carried out within our group in Exeter.
Keywords: Thermophilic enzymes, Esterases, Lactonases, Epoxide hydrolases, Carbonic anhydrases
Introduction
Many hydrolase enzymes including proteases, lipases, esterases and cellulases are already used commercially in industrial biocatalysis as documented in recent review chapters [56, 63, 66]. However, there is still an increasing need for the new enzymes to compliment the existing ‘Tool Box’ and to make these commercially available. The enzyme biocatalysts required for industrial processes need to withstand conditions that they are not usually exposed to within their natural environments such as high temperatures, extremes of pH, high salt, high substrate concentrations and the presence of organic solvents.
Extremophiles are organisms that have evolved to survive under many of these conditions and their intracellular and exported proteins often possess the desired attributes required for industrial applications.
This mini-review will concentrate on specific novel enzymes from characterised thermophilic archaeal and bacterial species isolated from marine and terrestrial ‘hot’ environments. In addition, enzymes sourced from metagenomes containing DNA and RNA from un-culturable microorganisms and their viruses will be included. Thermophilic enzymes from the archaea often offer additional novelty when compared to those from the thermophilic bacteria. The archaeal enzymes have evolved under different evolutionary pressure and generally represent more primitive enzymes. Different archaeal species have novel metabolic pathways that are not found in other microorganisms. For example, some species have modified versions of the Embden Meyerhof and Entner Doudoroff pathway involving a large number of novel enzymes [64] and have unusual pentose degradation pathways. Several archaeal enzymes are promiscuous regarding their activity to related substrates when compared to the equivalent enzymes isolated from bacteria or eukaryotes [13, 41, 57, 71]. This property to act on different structurally related substrates may be advantageous for some industrial processes.
The stability of an enzyme is dependent on maintenance of a functional structure which relies on a number of molecular interactions [12]. This property is also the case with a thermostable protein where the free energy of stabilisation is slightly higher than that of its mesophilic counterpart [32]. The overall stability of all proteins is marginal and they are easily denatured by a variety of changes in temperature and pH. The active form of a protein is held together using a combination of non-covalent forces including hydrogen bonds, ion pairs, hydrophobic bonds and Van der Waals interactions. When these interactions are disrupted, for example by elevated temperatures, both mesophilic and thermophilic proteins unfold into inactive but kinetically stable structures. Once unfolded in this manner the protein is prone to aggregation and chemical modification. Chemical modifications of the protein can also occur under other conditions and include cysteine oxidation, deamination of asparagine and glutamine residues and peptide bond hydrolysis. The unfolding of the protein when exposed to elevated temperatures may be reversible for smaller proteins, but is usually irreversible with larger proteins.
Some extremophilic microorganisms are able to grow at extremes of pH and are called acidophiles and alkalophiles, respectively. It is only the proteins exported from the cell that have to be stable under the pH conditions of the extreme growth environment. The proteins inside the cell do not have to withstand these extreme conditions as the intracellular pH is maintained around pH 5.0–6.0.
The use of more thermostable proteins, apart from being more cost effective with regard to enzyme use, can offer the option to run the process at elevated temperatures where non-natural substrates are more soluble. The temperature for operation of the industrial process needs to be balanced against the overall economics of the biocatalytic conversion. An example of a process which is commercially operated at 50 °C is the production of L-amino acids and L-amino acid analogues using a thermophilic archaeal L-aminoacylase enzyme that has been cloned and over-expressed from the archaeon Thermococcus litoralis [72]. This enzyme is used for the multi-ton commercial production of L-amino acids by Chirotech/Dow Pharma. More recently, the process operated by Chirotech/Dr Reddy’s [4] uses the addition of a racemase enzyme to convert the isomer not used by the enzyme to the isomer which is used, enabling a more efficient process with 100% conversion of a racemic substrate to one desired product [28].
What makes a protein more stable to elevated temperatures has been identified by studying the biochemical and structural features of a range of purified thermophilic proteins [45]. The structural features to increase thermostability as used by ‘nature’ include an increase in ionic interactions within the monomeric protein structure and at subunit interfaces for multi-subunit proteins. These offer most stabilisation when they occur in clusters as seen in many thermophilic enzymes including a thermophilic alcohol dehydrogenase enzyme from the hyperthermophilic archaeon Aeropyrum pernix which can be used industrially for chiral alcohol production [24]. The α-helices in the protein can be ‘capped’ by introduction of an acidic amino acid to neutralise the charge at the amino end of the helix and a basic amino acid to neutralise the charge at the acidic end of the helix. Many thermophilic proteins are also stabilised by hydrophobic interactions within the protein interior and at the subunit interfaces. This is often a feature of enzymes from thermophilic organisms such as the omega transaminase from Sulfolobus solfataricus [58] and pyroglutamyl carboxyl peptidase from T. litoralis [67]. The thermophilic proteins often have increased internal packing such as additional secondary structures and C-terminal extensions which can pack into the protein to fill unnecessary voids as seen in the glyceraldehyde 3-phosphate dehydrogenase from the thermophilic archaeon S. solfataricus [31]. Most thermophilic proteins have shorter surface loops and often the internal loops can be stabilised by metal ions [45]. An increased content of proline residues is seen in some thermophilic bacteria such as Thermus species which have a high G-C content in their DNA. Generally there is a reduction in amino acids that are unstable at high temperatures such as asparagines and cysteines except where they play an important catalytic role. Some hyperthermophilic aerobic archaea such as the A. pernix species use the introduction of a covalent disulfide bond into the protein to offer the necessary stability at high temperatures [24, 46].
Esterase enzymes
The esterase enzymes are widely used industrial enzymes. They are able to carry out hydrolysis of an ester bond by a hydrolytic reaction using the classic ‘catalytic triad’ of three amino acids Ser, His, Glu or Asp [8]. They are relatively stable enzymes in organic solvents and have been widely used in both the hydrolytic and the reverse synthetic direction to carry out esterification and transesterification reactions. A general review on lipases was published by Bornscheuer [10]. Most lipolytic enzymes belong to the α/β hydrolase protein fold superfamily [53]. The catalytic serine residue in the α/β hydrolase fold esterases is usually located in a tight nucleophilic elbow with the consensus sequence Gly-X-Ser-X-Gly, although deviations from this consensus have been reported [9, 51].
However, esterase activity has also been reported for enzymes with a β lactamase fold [74] and an α/β/α hydrolase fold [73] or as a side activity for the zinc containing carbonic anhydrase enzymes [30]. The ESTHER database [42] divides the α/β hydrolase enzymes into over 140 families and superfamilies which are further assigned to groups C, H, L, and X.
Esterases are used for pharmaceutical intermediates due to the stereo-selectivity of their reactions, but also are finding many other uses as bulk enzymes in the healthcare, laundry and food industries and for degradation of domestic and agricultural biomass. There is always a demand for esterase enzymes with increased stability and selectivity for a variety of applications in industrial biotechnology. As part of a recent EU project ‘HOTZYME’ new esterase enzymes with different structural features and specificities have been identified and characterised. The first of these was found in the genome of a well characterised thermophilic archaeon, Archaeoglobus fulgidus [70]. The Arch. fulgidus is an anaerobic heterotrophic sulphate-reducing archaeon that grows at temperatures between 60 and 95 °C, with an optimal growth at 83 °C. It was isolated at Volcano Island in Italy 30 years ago. It was the first sulphur metabolising organism to have its genome sequence determined [40]. Some esterases and a lipase were earlier characterised from this organism including AFEST [47] which is a member of a hormone-sensitive lipase family and can be used for the biocatalytic conversion of poly δ-valerolactone being thermostable up to 90 °C. The first esterase Est-AF [16] and directed mutants of this enzyme have been designed for the industrial production of the (S)-enantiomer of ketoprofen that acts as an anti-inflammatory drug [37, 38].
The new carboxyl esterase, AF-Est2, belongs to the α/β hydrolase 6 family and the X group of the ESTHER classification. It has been cloned, over-expressed in Escherichia coli and biochemically and structurally characterised [61]. The AF-Est2 has very good solvent and pH stability and is very thermostable, showing no loss of activity after incubation for 30 min at 80 °C making it a good addition to the industrial esterase enzyme ‘tool box’. The structure of the esterase at high-resolution (PDB: 5FRD, Fig. 1a) revealed Coenzyme A (CoA) bound in the vicinity of the active site; however, this enzyme has no CoA thioesterase activity as described for related enzymes [5]. The pantetheine group of CoA partially obstructs the active site alcohol pocket of AF-Est2 suggesting that this ligand has a role in regulation of the enzyme activity. A comparison of the structure of AF-Est2 with the human carboxyl esterase 1 [5] which has CoA thioesterase activity, reveals that the cofactor is bound to different parts of the core domain in these two enzymes and approaches the active site from opposite directions.
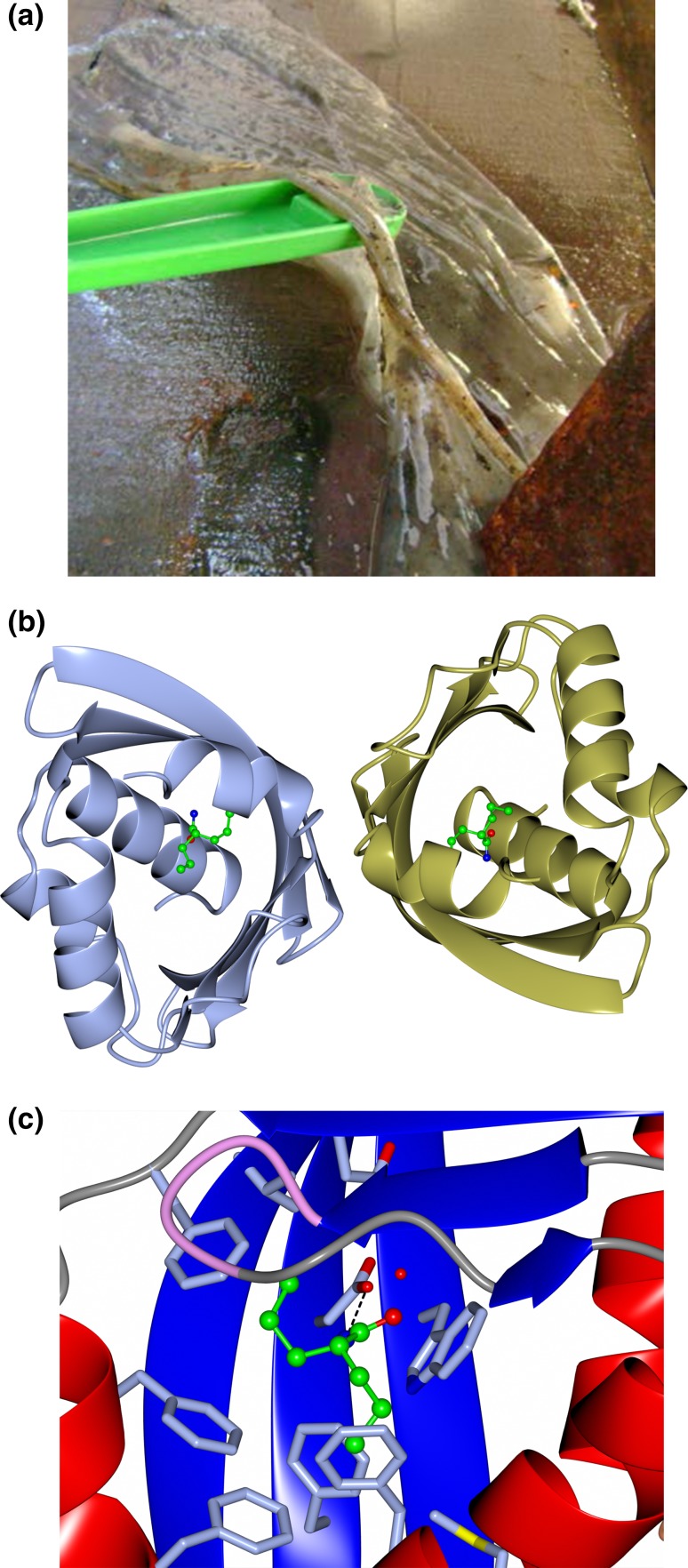
Fig. 1.
A cartoon representation of the crystal structures of the three thermophilic esterase enzymes described above. The helices are shown as red cylinders and the sheets as blue arrows. Other selected atoms are shown as ball and stick representation. a The Archaeoglobus archaeal esterase, AF-Est2, showing the larger core and smaller cap domains, with the bound CoA molecule and catalytic residues shown as a stick model PDB code 5FRD. b The T. terrifontis bacterial esterase, TtEst, showing D-malate which maps the alcohol-binding pocket of the active site PDB code 4UHE. c The second bacterial esterase from T. terrifontis, TtEst, showing the open active site and minimal cap domain. The substrate butyrate is bound shown in ball and stick representation which maps the carboxyl-binding pocket of the active site PDB code 5AOB. Images were generated using CCP4 MG [48]
Two other interesting thermophilic esterases have been characterised from a recently discovered thermophilic Planctomycetes species of bacterium named Thermogutta terrifontis [59, 60]. The T. terrifontis has been isolated from a terrestrial hot spring on Kunashir Island, Russia [68]. It is the first thermophilic member of the phylum Planctomycetes. This organism grows up to 70 °C, with an optimum growth temperature of 55–60 °C. Planctomycetes represents a deep distinct phylum in the bacterial domain and includes a large group of microorganisms with unique features not found in other bacteria such as reproduction by budding, presence of intra-cytoplasmic membranes which divide the cell into compartments, presence of a primitive nucleoid and the lack of peptidoglycan in the cell wall.
The first T. terrifontis esterase, TtEst, has been cloned and over-expressed in E. coli [59]. The enzyme is highly thermostable retaining 95% of its activity after incubation for 1 h at 80 °C. It has been biochemically characterised and shown to have activity towards small p-nitrophenyl (pNP) carboxylic esters with optimal activity for pNP-propionate. The structure of the enzyme has been determined without ligands bound in the active site (Fig. 1b) and in complex with a substrate analogue, D-malate and the product acetate (PDBs: 4UHC (native), 4UHD (acetate bound), 4UHE (malate bound), 4UHF (L37A mutant with butyrate bound)).
The bound ligands in the structure have allowed the identification of the carboxyl and alcohol-binding pockets in the enzyme active site. The results have also contributed to an understanding of substrate specificity and the subtle structural differences between esterase and lactonase enzymes. A comparison has been made of the alcohol-binding pocket in TtEst with the equivalent pocket in two other structurally related enzymes, a lactonase and a γ-lactamase. The 3-oxoadipate-enol lactonase from Burkholderia xenovorans (PcaD) (PDB code 2XUA) [3] has only 29% sequence identity to TtEst and the Aureobacterium species (−) γ-lactamase (Agl) (PDB code 1HKH) [44] with 30% sequence identity has shown that the catalytic triad residues and the position of the oxyanion hole are conserved between the enzymes. The PcaD and Agl show that the TtEst pocket that forms part of the active site has a much more polar and charged environment allowing the binding of organic acids such as D-malate where the distant carboxyl is coordinated by Arg139 and Tyr105. The PcaD and Agl enzymes have more hydrophobic substrate binding pockets, with the residues Arg139 and Tyr105 in TtEst replaced by Trp135 and Ile129 in PcaD and Trp204 and Leu125 in Agl. For PcaD and Agl, the active site is suited for the binding of structures such as lactone and γ-lactam rings. Similarly, the Pseudomonas fluorescens esterase with 30% sequence identity (PDB code 3HEA) [78] has an alcohol pocket which is lined with several hydrophobic phenylalanine side chains that should have affinity for the lactone ring. This would explain its lactonase activity towards caprolactone [11] in addition to its esterase activity. This enzyme and the Agl enzyme both have perhydrolase activity. They are able to produce hypochlorous and hyperbromous acid in a side reaction at acidic pH [6] and were originally named as non-cofactor chloroperoxidases [27].
Mutant enzymes have been constructed to extend the substrate range of the TtEst esterase to accept the larger butyrate and valerate pNP-esters. The crystal structure of the Leu37Ala mutant shows the butyrate product bound in the carboxyl pocket of the active site. The structure shows an expansion of the pocket that binds the substrate carboxyl group which allows the observed activity towards pNP-butyrate [59].
The second T. terrifontis esterase, TtEst2, shows substantial differences when compared with the TtEst1. A large difference is observed with the absence of the usual ‘cap’ domain found in most other esterase enzymes which results in an open substrate binding site that is solvent accessible (Fig. 1c) [60]. This TtEst2 enzyme belongs to the α/β-hydrolase family 3 in the Pfam classification [21]. It has been characterised biochemically and shown to have activity towards small p-nitrophenyl (pNP) carboxylic esters with optimal activity for pNP-acetate. The enzyme is not as thermostable as TtEst1, but still retains 75% of its activity after incubation for 30 min at 70 °C. The structure of this enzyme has been determined in the native form and in complex with the carboxylic acid products propionate, butyrate and valerate. The PDB codes 5AO9 (native), 5AOa (propionate bound), 5AOb (butyrate bound), 5AOc (valerate bound) have been deposited. The structures with bound ligands have allowed the identification of the carboxyl-binding pocket in the enzyme active site. Since this enzyme has a very open active site compared to other esterase enzymes it has the potential to be active towards larger substrates.
Lactonases
The specific cleavage of a lactone ring is an important activity of interest to many pharmaceutical companies. Lactones have been shown to be important communication molecules between microbial cells and the topic has been reviewed [75].
The lactonase enzymes identified to date fall into three structurally diverse groups: the enol lactonases, the gluconolactonases and the quorum-sensing lactonases. Phosphotriesterase-like lactonases were identified in the archaeal species S. solfataricus and S. acidocaldarius [1, 49, 50, 54]. These enzymes catalyse the hydrolytic cleavage of the intramolecular ester bond in lactones and acyl-homoserine lactones to give the corresponding hydroxyacylic acids. This class of lactonases also have a promiscuous phosphotriesterase activity towards organophosphate compounds and have the potential to be used for remediation of contaminated soils. However, the natural role for these lactonases is to break down lactones that are thought to play a role in quorum sensing between microorganisms and are involved in biofilm formation [14, 39] and also in the expression of virulence factors that are of interest in medicinal and biotechnological applications [25]. Therefore, the enzymatic degradation of lactones could be used to interrupt quorum-sensing signalling pathways and to control microbial communication so that they cannot form communities.
A lactonase of this class has been identified, cloned, over-expressed and characterised [34] from Vulcanisaeta moutnovskia a hyperthermoacidophilic crenarchaeon isolated from a solfataric field in Kamchatka (Russia) [23, 55]. This lactonase was studied with view to its substrate specificity for biocatalytic applications of interest to the pharmaceutical industries. The lactonase converted lactones and acyl-homoserine lactones with comparable activities. A promiscuous, lower activity was observed with organophosphates and minor activity was observed with carboxyl esters. The catalytic activity was strictly dependant on bivalent cations (Cd2+ > Ni2+ > Co2+ > Mn2+ > Zn2+) and was most active at pH 8.0, and at 80 °C. The enzyme has high activity towards linear γ-lactones with hydrophobic side chains of variable lengths from γ-butyrolactone (no side chain) and γ-valerolactone which has a methyl side chain and γ-dodecalactone which had a seven-carbon side chain. It was shown that the enzyme has activity to whiskey lactone and δ-dodecalactone. No measurable activity was seen for mevalonolactone or δ-decalactone. The lactonase enzyme had increased activity towards the D form of these substrates [34].
The recent structure of the V. moutnovskia lactonase has been carried out in complex with a long chain fatty acid which maps the position of the substrate binding pocket [26]. This enzyme belongs to the amidohydrolase enzyme superfamily which has a (β/α)8-barrel structural fold. The two bound cobalt ions in this enzyme that are essential for activity are located in at the C-terminus of the β barrel in the crystal structure. In the proposed catalytic cycle, the metal ions activate a bridging water molecule through proton abstraction. The resulting hydroxide ion then performs a nucleophilic attack on the C1 of the lactone ring resulting in hydrolysis [26].
The high thermal stability of this class of lactonase enzymes as well as their broad substrate specificity for different lactones makes them interesting new enzyme for the biocatalytic ‘tool box’.
The enol lactonases are members of the α/β hydrolase superfamily and are closely related to the esterases. The 3-oxoadipate-enol lactonase from B. xenovorans (PcaD) (PDB code 2XUA) [7] has been mentioned above. A thermophilic member of this family has been identified, cloned and studied from the archaeon Carboxydothermus hydrogenoformans [62].
Epoxide hydrolases from extremophilic metagenomes
An important enzyme activity of interest to the pharmaceutical industry is the ability to catalyse the hydrolysis of an epoxide ring by addition of a molecule of water to form a vicinal diol as a product [36, 76]. Enzymes found in nature that can carry out this reaction play an important role in the detoxification of reactive xenobiotics or endogenous metabolites and in the formation of biologically active mediators. The so-called ‘epoxide hydrolases’ are already used for the production of optically pure epoxides and diols which are important synthons for the preparation of fine chemicals and drugs, for example the chiral precursors of β-blockers [35, 52]. These enzymes are found in two classes which have different structure and mechanisms. The most studied class has the α/β hydrolase fold which is characteristic of the esterase enzymes described above. The other less studied class is called the limonene epoxide hydrolases (LEHs) after the substrate, limonene, that they were shown to hydrolyse. The LEH enzyme active site contains three residues (Asp, Arg, and Asp) that have been proposed to act in a concerted fashion to activate a water molecule which is able to open the epoxide ring without the formation of a covalently bound alkyl-enzyme intermediate [2, 29]. A recent review has described the importance and industrial interest of these hydrolase enzymes [77].
Recently, as part of a thermophilic metagenomic project two new thermostable epoxide hydrolases of the limonene class have been discovered. The metagenomes were isolated in Russia and China from hot terrestrial environments growing at 46 and 55 °C, respectively, and at neutral pH. The microbial mat which was sampled at the Russian Tomsk site is shown in Fig. 2a. A bioinformatic approach was used to identify the genes coding for these industrially important enzymes using the mesophilic Rhodococcus LEH as a search. The two new thermophilic LEHs have been cloned and over-expressed in E. coli and the resultant proteins characterised both biochemically and structurally [19]. The atomic coordinates and structure factors of the crystal structures obtained have been deposited in the PDB: 5AIF (Tomsk-LEH native structure), 5AIG (Tomsk-LEH valpromide complex), 5AIH (CH55-LEH native structure) and 5AII (CH55-LEH PEG complex). The overall structure of the Tomsk-LEH from the Russian-sourced metagenome is shown in Fig. 2b and the active site showing the catalytic water is shown in Fig. 2c. The new LEH enzymes have attracted industrial interest since they are more thermostable than the available LEH and have different stereo-preference for the different isomers of limonene-1,2-epoxide. They have already been used in pilot-scale biotransformations for efficient epoxide hydrolase-catalysed resolutions of (+)- and (−)-cis/trans-limonene oxides which have important industrial applications [20].
Fig. 2.
a The microbiological mat at the Tomsk sampling site, Parabel, Tomsk Region, West Siberia, Russia, which provided the metagenome DNA sample where the LEH was identified. Picture kindly provided by Prof. Elizaveta Bonch-Osmolovskaya. b A cartoon representation of the dimeric Tomsk-LEH structure in complex with an inhibitor valpromide shown in ball and stick representation which is bound at the active site. PDB code 5IG. c A close-up representation of the active site of Tomsk-LEH with the active site residues and inhibitor highlighted. The red sphere represents the active site water molecule. Images were generated using CCP4 MG [48]
Carbonic anhydrases
Carbonic anhydrase enzymes (CAs) catalyse the reversible hydration of carbon dioxide to bicarbonate. The bovine and human enzymes of the α-carbonic anhydrase class have been widely studied and are known to be very efficient catalysts of CO2 hydration. Most of these enzymes also have a side activity as an esterase although the mechanism involves a zinc ion and not the catalytic triad found in most esterases. The carbonic anhydrase reaction mechanism involves an attack of a zinc-bound hydroxide ion onto a CO2 molecule which is bound in a hydrophobic pocket of the enzyme. The resulting zinc-coordinated bicarbonate ion is removed from the metal ion by water. There is then a rate-limiting step where an intramolecular proton is transferred from the zinc-bound water molecule to a histidine amino which serves as a proton shuttle between the metal centre and buffer molecules in the reaction medium [43].
Carbonic anhydrase has industrial applications as a catalyst for CO2 capture from the environment and from industrial waste streams. However, the biocatalyst used has to be stable to the temperature and other harsh conditions that it is exposed to during the process.
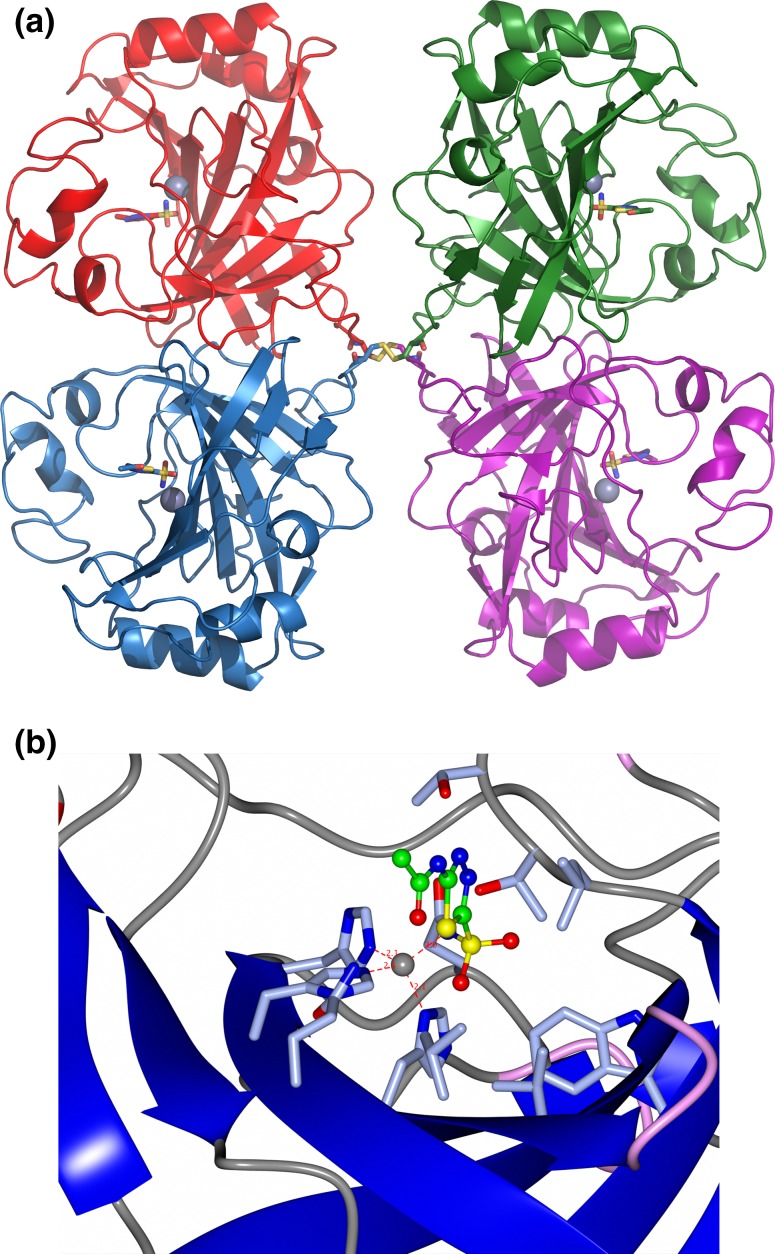
The thermophilic bacteria appear to contain high activity α-carbonic anhydrase enzymes which are related to the bovine and human carbonic anhydrase enzymes, whereas the archaea have carbonic anhydrases that have different structures and mechanisms. There are six distinct families of CAs (α, β, γ, δ, ζ, and η) [22, 69]. The amino acid sequences are conserved between each family; however, there is no sequence or structural similarity between the different families. The CA activity always requires the presence of a catalytic zinc ion which is coordinated to either histidine or cysteine amino acids depending on the class of the enzyme [65]. A stable robust α-carbonic anhydrase has been identified in the thermophilic bacterium Thermovibrio ammonificans. The enzyme has been cloned and over-expressed in E. coli. This protein has been characterised both biochemically and structurally [33]. The crystal structure of this enzyme has been determined in its native form and in two complexes with bound inhibitors. The overall structure of the enzyme is unique since it forms a tetrameric structure rather than the dimer reported for related enzymes. The Thermovibrio carbonic anhydrase is stabilised by a unique core in the centre of the tetramer which is formed by two intersubunit disulfide bonds and a single lysine residue from each monomer (Fig. 3a). The structure of this central core region protects the intersubunit disulfide bonds from reduction. The catalytic zinc ion coordinated to histidine residues that is observed in the active site of the Thermovibrio enzyme is shown in Fig. 3c.
Fig. 3.
a A cartoon representation of the tetramer structure of the α-carbonic anhydrase from T. ammonificans showing each subunit in a different colour. The active site zinc is shown as a sphere in each subunit together with an inhibitor in ball and stick representation bound at each active site. The disulfide bonds at the centre of the tetramer which add stability to the enzyme are shown in yellow. b A close-up representation of the active site of the enzyme showing the catalytic zinc molecule and the inhibitor acetazolamide bound to the active site. Side chain residues co-ordinating the zinc ion and residues making up the active site are highlighted. PDB code 4COQ. Images were generated using CCP4 MG [48] and PyMOL [15]
Another thermophilic bacterial α-carbonic anhydrase has been described from Sulfurihydrogenibium yellowstonense. This carbonic anhydrase is also thermostable and is a dimer stabilised by ionic networks [17]. A review of the different types of thermophilic carbonic anhydrase enzymes has recently been published [18].
Summary
This mini-review has provided a selection of examples demonstrating the use of ‘Natures Catalysts’ to provide a ‘tool box’ of biocatalysts for sustainable applications in industrial biotechnology. This has been highlighted using different thermophilic hydrolase enzymes identified in thermophilic genomes and metagenomes from both bacterial and archaeal sources. These enzymes are robust to the conditions required for their different industrial applications. It is expected that the number of these enzymes used commercially will increase due to their inherent stability and novel specificities. The enzymes can be cloned and over-expressed in easily growing hosts such as E. coli which can provide sufficient quantities of the purified enzymes for different biocatalytic applications. For larger scale industrial applications, where the enzyme is required in kg quantities, an alternative fungal host system that exports the enzyme into the growth media could be developed.
The stability of the biocatalyst is an important issue since to be economically viable the enzyme needs to be reused for several biocatalytic cycles. Immobilisation of the enzyme can often increase its stability and will also allow it to be easily recovered for reuse. The cost of the enzyme biocatalyst is usually the most expensive component of the industrial biotransformation and this must be matched to the value of the end product or process.
The use of enzymes is expected to grow in the next year with bio-based materials, chemicals and sustainable processes predicted to rise globally to over 7.4 million metric tons in 2018 (Lux Research analysts). The time required to source the best enzyme catalyst and its optimisation for the desired process, is still a limiting factor for a cost-effective biocatalytic route. There is, however, a large natural resource available to search for novel enzymes to add to the currently available ‘tool box’.
The adoption of new commercial biocatalytic processes will be important to achieve the goal of a sustainable ‘circular economy’ and to address important global challenges.
Acknowledgements
The author would like to thank the University of Exeter, the Wellcome Trust, the BBSRC, EPSRC and Technology Strategy Board, UK, for sponsoring research in the JAL group at the Exeter Biocatalysis Centre. The BBSRC funded ERA-IB grant BB/L002035/1 and the University of Exeter are thanked for support. Thanks are also given to the EU Framework 7 grant ‘HOTZYME’ entitled Systematic Screening of Organisms from Hot Environments, Grant Number 265933.
References
- 1.Afriat L, Roodveldt C, Manco G, Tawfik DS. The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry. 2006;45:13677–13686. doi: 10.1021/bi061268r. [DOI] [PubMed] [Google Scholar]
- 2.Arand M, Hallberg BM, Zou J, Bergfors T, Oesch F, van der Werf MJ, deBont JAM, Jones TA, Mowbray SL. Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site. EMBO J. 2003;22:2583–2592. doi: 10.1093/emboj/cdg275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bains J, Kaufman L, Farnell B, Boulanger MJ. A product analog bound form of 3-oxoadipate-enol-lactonase (PcaD) reveals a multifunctional role for the divergent cap domain. J Mol Biol. 2011;406:649–658. doi: 10.1016/j.jmb.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Baxter S, Royer S, Grogan G, Brown F, Holt-Tiffin K, Taylor I, Fotheringham I, Campopiano D. An improved racemase/acylase biotransformation for the amino acids. J Am Chem Soc. 2012;134:19310–19313. doi: 10.1021/ja305438y. [DOI] [PubMed] [Google Scholar]
- 5.Bencharit S, Edwards CC, Morton CL, Howard-Williams EL, Kuhn P, Potter PM, Redinbo MR. Multisite promiscuity in the processing of endogenous substrates by human carboxylesterase 1. J Mol Biol. 2006;363:201–214. doi: 10.1016/j.jmb.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhardt P, Hult K, Kazlauskas RJ. Molecular basis of perhydrolase activity in serine hydrolases. Angew Chem Int Ed. 2005;44:2742–2746. doi: 10.1002/anie.200463006. [DOI] [PubMed] [Google Scholar]
- 7.Bigley AN, Raushel FM. Catalytic mechanisms for phosphotriesterases. Biochim Biophys Acta. 2013;1834:443–453. doi: 10.1016/j.bbapap.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blow DM, Birktoft JJ, Hartley BS. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature. 1969;221:337–340. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- 9.Bourne PC, Isupov MN, Littlechild JA. The atomic-resolution structure of a novel bacterial esterase. Structure. 2000;8:143–151. doi: 10.1016/S0969-2126(00)00090-3. [DOI] [PubMed] [Google Scholar]
- 10.Bornscheuer UT. Microbial carboxyl esterases: classification, properties and application in biocatalysis . FEMS Microbiol Rev. 2002;26:73–81. doi: 10.1111/j.1574-6976.2002.tb00599.x. [DOI] [PubMed] [Google Scholar]
- 11.Cheeseman JD, Tocilj A, Park S, Schrag JD, Kazlauskas RJ. Structure of an aryl esterase from Pseudomonas fluorescens. Acta Crystallogr D. 2004;60:1237–1243. doi: 10.1107/S0907444904010522. [DOI] [PubMed] [Google Scholar]
- 12.Daniel RM, Danson MJ, Hough DW, Lee CK, Peterson ME, Cowan DA. Enzyme stability and activity at high temperatures. In: Siddiqui KS, Thomas T, editors. Protein adaptation in extremophiles. New York: Nova science Publishers; 2008. pp. 1–34. [Google Scholar]
- 13.Danson MJ, Hough DW. Promiscuity in the Archaea. The enzymology of metabolic pathways. Biochemist. 2005;27:17–21. [Google Scholar]
- 14.De Kievit TR. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2009;11:279–288. doi: 10.1111/j.1462-2920.2008.01792.x. [DOI] [PubMed] [Google Scholar]
- 15.DeLano WL. The PyMOL molecular graphics system. San Carlos: DeLano Scientific; 2002. [Google Scholar]
- 16.De Simone G, Menchise V, Manco G, Mandrich L, Sorrentino N, Lang D, Rossi M, Pedone D. The crystal structure of a hyper-thermophilic carboxylesterase from the archaeon Archaeoglobus fulgidus. J Mol Biol. 2001;314:507–518. doi: 10.1006/jmbi.2001.5152. [DOI] [PubMed] [Google Scholar]
- 17.Di Fiore A, Capasso C, De Luca V, Monti SM, Carginale V, Supuran CT, Scozzafava A, Pedone C, Rossi M, De Simone G. X-ray structure of the first ‘extremo-α-carbonic anhydraseʼ, a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr D. 2013;69:1150–1159. doi: 10.1107/S0907444913007208. [DOI] [PubMed] [Google Scholar]
- 18.Di Fiore A, Alterio V, Monti SM, De Simone G, D’Ambrosio K. Thermostable carbonic anhydrases in biotechnological applications . Int J Mol Sci. 2015;16:15456–15480. doi: 10.3390/ijms160715456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrandi EE, Sayer C, Isupov MN, Annovazzi C, Marchesi C, Iacobone G, Peng X, Bonch-Osmolovskaya E, Wohlgemuth R, Littlechild JA, Monti D. Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomics libraries. FEBS J. 2015;282:2879–2894. doi: 10.1111/febs.13328. [DOI] [PubMed] [Google Scholar]
- 20.Ferrandi E, Marchesi C, Annovazzi C, Riva S, Monti D, Wohlgemuth R. Efficient epoxide hydrolase-catalyzed resolutions of (+)- and (−)-cis/trans-limonene oxides. ChemCatChem. 2015;7:3171–3178. doi: 10.1002/cctc.201500608. [DOI] [Google Scholar]
- 21.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guler OO, Capasso C, Supuran CT. A magnificent enzyme superfamily: carbonic anhydrases, their purification and characterization. J Enzyme Inhib Med Chem. 2015;31(5):669–689. doi: 10.3109/14756366.2015.1059333. [DOI] [PubMed] [Google Scholar]
- 23.Gumerov VM, Mardanov AV, Beletsky AV, Prokofeva MI, Bonch-Osmolovskaya EA, Ravin NV, Skryabin KG. Complete genome sequence of Vulcanisaeta moutnovskia strain 768-28, a novel member of the hyperthermophilic crenarchaeal genus Vulcanisaeta. J Bacteriol. 2011;193:2355–2356. doi: 10.1128/JB.00237-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guy JE, Isupov MN, Littlechild JA. The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix. J Mol Biol. 2003;331:1041–1051. doi: 10.1016/S0022-2836(03)00857-X. [DOI] [PubMed] [Google Scholar]
- 25.Hiblot J, Gotthard G, Chabriere E, Elias M. Structural and enzymatic characterization of the lactonase SisLac from Sulfolobus islandicus. PLoS One. 2012;7:e47028. doi: 10.1371/journal.pone.0047028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiblot J, Bzdrenga J, Champion C, Chabriere E, Elias M. Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia. Sci Rep. 2015;5:8372. doi: 10.1038/srep08372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann B, Tölzer S, Pelletier I, Altenbuchner J, van Pée KH, Hecht HJ. Structural investigation of the cofactor-free chloroperoxidases. J Mol Biol. 1998;279:889–900. doi: 10.1006/jmbi.1998.1802. [DOI] [PubMed] [Google Scholar]
- 28.Holt K (2005) Biocatalysis and chemocatalysis—a powerful combination for the preparation of enantiomerically pure α-amino acids. ChemInform 36(42). doi:10.1002/chin.200542274
- 29.Hopmann KH, Hallberg BM, Himo F. Catalytic mechanism of limonene epoxide hydrolase, a theoretical study. J Am Chem Soc. 2005;127:14339–14347. doi: 10.1021/ja050940p. [DOI] [PubMed] [Google Scholar]
- 30.Höst G, Mårtensson LG, Jonsson BH. Redesign of human carbonic anhydrase II for increased esterase activity and specificity towards esters with long acyl chains. Biochim Biophys Acta. 2006;1764:1601–1606. doi: 10.1016/j.bbapap.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Isupov MN, Fleming TM, Dalby AR, Crowhurst GS, Bourne PC, Littlechild JA. Crystal structure of the glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus. J Mol Biol. 1999;291:651–660. doi: 10.1006/jmbi.1999.3003. [DOI] [PubMed] [Google Scholar]
- 32.Jaenicke R, Bohm G. The stability of proteins in extreme environments. Curr Opin Struct Biol. 1998;8:738–748. doi: 10.1016/S0959-440X(98)80094-8. [DOI] [PubMed] [Google Scholar]
- 33.James P, Isupov MN, Sayer C, Saneei V, Berg S, Lioliou M, Kotlar HK, Littlechild JA. The structure of a tetrameric α-carbonic anhydrase from Thermovibrio ammonificans reveals a core formed around intermolecular disulfides that contribute to its thermostability. Acta Crystallogr. 2014;D70:2607–2618. doi: 10.1107/S1399004714016526. [DOI] [PubMed] [Google Scholar]
- 34.Kallnik V, Bunescu A, Sayer C, Bräsen C, Wohlgemuth R, Littlechild J, Siebers B. Characterization of a phosphotriesterase-like lactonase from the hyperthermoacidophilic crenarchaeon Vulcanisaeta moutnovskia. J Biotechnol. 2014;190:11–17. doi: 10.1016/j.jbiotec.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 35.Kong X-D, Ma Q, Zhou J, Zeng B-B, Xu J-H. A smart library of epoxide hydrolase variants and the top hits for synthesis of (S)-β-blocker precursors. Angew Chem Int Ed. 2014;53:6641–6644. doi: 10.1002/anie.201402653. [DOI] [PubMed] [Google Scholar]
- 36.Kotik M, Archelas A, Wohlgemuth R. Epoxide hydrolases and their application in organic synthesis. Curr Org Chem. 2012;16:451–482. doi: 10.2174/138527212799499840. [DOI] [Google Scholar]
- 37.Kim SB, Lee W, Ryu YW. Cloning and characterization of thermostable esterase from Archaeoglobus fulgidus. J Microbiol. 2008;46:100–107. doi: 10.1007/s12275-007-0185-5. [DOI] [PubMed] [Google Scholar]
- 38.Kim J, Kim S, Yoon S, Hong E, Ryu Y. Improved enantioselectivity of thermostable esterase from Archaeoglobus fulgidus toward (S)-ketoprofen ethyl ester by directed evolution and characterization of mutant esterases. Appl Microbiol Biotechnol. 2015;99:6293–6301. doi: 10.1007/s00253-015-6422-7. [DOI] [PubMed] [Google Scholar]
- 39.Kirisits MJ, Parsek M. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell Microbiol. 2006;8:1841–1849. doi: 10.1111/j.1462-5822.2006.00817.x. [DOI] [PubMed] [Google Scholar]
- 40.Hans-Peter Klenk, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 41.Lamble HJ, Heyer NI, Bull SD, Hough DW, Danson MJ. Metabolic pathway promiscuity in the archaeon Sulfolobus solfataricus revealed by studies on glucose dehydrogenase and 2-keto-3-deoxygluconate aldolase. J Biol Chem. 2003;278:34066–34072. doi: 10.1074/jbc.M305818200. [DOI] [PubMed] [Google Scholar]
- 42.Lenfant N, Hotelier T, Velluet E, Bourne Y, Marchot P, Chatonnet A. ESTHER, the database of the α/β hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 2013;41:D423–D429. doi: 10.1093/nar/gks1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindskog S. Structure and mechanism of carbonic anhydrase. Pharmacol Ther. 1997;74:1–20. doi: 10.1016/S0163-7258(96)00198-2. [DOI] [PubMed] [Google Scholar]
- 44.Line K, Isupov MN, Littlechild JA. The crystal structure of a (−) gamma-lactamase from an Aureobacterium species reveals a tetrahedral intermediate in the active site. J Mol Biol. 2004;338:519–532. doi: 10.1016/j.jmb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Littlechild J, James P, Novak H, Sayer C. Thermophilic microbes in environmental and industrial biotechnology, biotechnology of thermophiles. 2. New York: Springer Publishers; 2013. Mechanisms of thermal stability adopted by thermophilic proteins and their use in white biotechnology; pp. 481–507. [Google Scholar]
- 46.Mallick P, Boutz DR, Eisenberg D, Yeates TO. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds Proc. Natl Acad Sci USA. 2002;99:9679–9684. doi: 10.1073/pnas.142310499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manco G, Giosuè E, D’Auria S, Herman P, Carrea G, Rossi M. Cloning, overexpression, and properties of a new thermophilic and thermostable esterase with sequence similarity to hormone-sensitive lipase subfamily from the archaeon Archaeoglobus fulgidus. Arch Biochem Biophys. 2000;373:182–192. doi: 10.1006/abbi.1999.1497. [DOI] [PubMed] [Google Scholar]
- 48.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4 mg molecular-graphics software. Acta Cryst D. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merone L, Mandrich L, Rossi M, Manco G. A thermostable phosphotriesterase from the archaeon Sulfolobus solfataricus: cloning, overexpression and properties. Extremophiles. 2005;9:297–305. doi: 10.1007/s00792-005-0445-4. [DOI] [PubMed] [Google Scholar]
- 50.Merone L, Mandrich L, Rossi M, Manco G. Enzymes with phosphotriesterase and lactonase activities in Archaea. Curr Chem Biol. 2008;2:237–248. [Google Scholar]
- 51.Nardini M, Dijkstra BW. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/S0959-440X(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 52.Nestl BM, Hammer SC, Nebel BA, Hauer B. New generation of biocatalysts for organic synthesis. Angew Chem Int Ed. 2014;53:3070–3095. doi: 10.1002/anie.201302195. [DOI] [PubMed] [Google Scholar]
- 53.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 54.Porzio E, Merone L, Mandrich L, Rossi M, Manco G. A new phosphotriesterase from Sulfolobus acidocaldarius and its comparison with the homologue from Sulfolobus solfataricus. Biochimie. 2007;89:625–636. doi: 10.1016/j.biochi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 55.Prokofeva MI, Kublanov IV, Nercessian O, Tourova TP, Kolganova TV, Lebedinsky AV, Bonch-Osmolovskaya EA, Spring S, Jeanthon C. Cultivated anaerobic acidophilic/acidotolerant thermophiles from terrestrial and deep-sea hydrothermal habitats. Extremophiles. 2005;9:437–448. doi: 10.1007/s00792-005-0461-4. [DOI] [PubMed] [Google Scholar]
- 56.Rao LV, Chandel AK, Chandrasekhar G, Rodhe AV, Sridevi J. Environmental and industrial biotechnology, biotechnology of thermophiles. 2. New york: Springer Publishers; 2013. Cellulases of thermophilic microbes in thermophilic microbes; pp. 730–771. [Google Scholar]
- 57.Reher M, Fuhrer T, Bott M, Schönheit P. The nonphosphorylative Entner–Doudoroff pathway in the thermoacidophilic euryarchaeon Picrophilus torridus involves a novel 2-Keto-3-deoxygluconate–Specific Aldolase. J Bacter. 2010;192:964–974. doi: 10.1128/JB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sayer C, Bommer M, Ward JM, Isupov M, Littlechild JA. Crystal structure and substrate specificity of the thermophilic serine:pyruvate aminotransferase from Sulfolobus solfataricus. Acta Crystallogr D. 2012;68:763–772. doi: 10.1107/S0907444912011274. [DOI] [PubMed] [Google Scholar]
- 59.Sayer C, Isupov M, Bonch-Osmolovskaya E, Littlechild JA. Structural studies of a thermophilic esterase from a new planctomycetes species thermogutta terrifontis. FEBS J. 2015;282:2846–2857. doi: 10.1111/febs.13326. [DOI] [PubMed] [Google Scholar]
- 60.Sayer C, Szabo Z, Isupov MN, Ingham C, Littlechild JA. The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis reveals an open active site due to a minimal cap domain. Front Microbiol. 2015;11:1294. doi: 10.3389/fmicb.2015.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sayer C, Finnigan W, Isupov M, Levisson M, Kengen S, van der Oost J, Harmer N, Littlechild JA. Structural and biochemical characterisation of Archaeoglobus fulgidus esterase reveals a bound CoA molecule in the vicinity of the active site. Sci Rep. 2016;6:25542. doi: 10.1038/srep25542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sayer C, Isupov M, Littlechild J (2017) Structural studies of a thermophilic enol lactonase from the archaeon Carboxydothermus hydrogenoformans Extremophiles, submitted
- 63.Sharma R, Thakur V, Sharma M, Birkland Nils-Kare. Thermophilic Microbes in environmental and industrial biotechnology, biotechnology of thermophiles. 2. New York: Springer Publishers; 2013. Biocatalysis through thermostable lipases: adding flavour to chemistry; pp. 905–928. [Google Scholar]
- 64.Siebers B, Schönheit P. Unusual pathways and enzymes of central carbohydrate metabolism in Archaea . Curr Opin Microbiol. 2005;8:695–705. doi: 10.1016/j.mib.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 65.Silverman DN, Lindskog S. The catalytic mechanism of carbonic anhydrase: implications of a rate-limiting protolysis of water. Acc Chem Res. 1988;21:30–36. doi: 10.1021/ar00145a005. [DOI] [Google Scholar]
- 66.Sinha R, Khara SK. Thermophilic microbes in environmental and industrial biotechnology, biotechnology of thermophiles. 2. New York: Springer Publishers; 2013. Thermostable proteases; pp. 859–880. [Google Scholar]
- 67.Singleton M, Isupov M, Littlechild J. X-ray structure of pyrrolidone carboxyl peptidase from the hyperthermophilic archaeon Thermococcus litoralis. Structure. 1999;7:237–244. doi: 10.1016/S0969-2126(99)80034-3. [DOI] [PubMed] [Google Scholar]
- 68.Slobodkina GB, Kovaleva OL, Miroshnichenko ML, Slobodkin AI, Kolganova TV, Novikov AA, et al. Thermogutta terrifontis gen. nov., sp. nov. and Thermogutta hypogea sp. nov., novel thermophilic anaerobic representatives of the phylum Planctomycetes. Int J Syst Evol Microbiol. 2014;65:760–765. doi: 10.1099/ijs.0.000009. [DOI] [PubMed] [Google Scholar]
- 69.Smith KS, Jakubzick C, Whittam TS, Ferry JG. Carbonic anhydrase is an ancient enzyme widespread in prokaryotes . Proc Natl Acad Sci. USA. 1999;96:15184–15189. doi: 10.1073/pnas.96.26.15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stetter KO, Lauerer G, Thomm M, Neuner A. Isolation of extremely thermophilic sulfate reducers: evidence for a novel branch of archaebacteria. Science. 1987;236:822–824. doi: 10.1126/science.236.4803.822. [DOI] [PubMed] [Google Scholar]
- 71.Theodossis A, Walden H, Westwick EJ, Connaris H, Lamble HJ, Hough DW, Danson MJ, Taylor GL. The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner–Doudoroff pathway in Sulfolobus solfataricus. J Biol Chem. 2004;279:43886–43892. doi: 10.1074/jbc.M407702200. [DOI] [PubMed] [Google Scholar]
- 72.Toogood HS, Hollingsworth EJ, Brown RC, Taylor IN, Taylor SJ, McCague R, Littlechild JA. A thermostable L-aminoacylase from Thermococcus litoralis: cloning, overexpression, characterization, and applications in biotransformations. Extremophiles. 2002;6:111–122. doi: 10.1007/s007920100230. [DOI] [PubMed] [Google Scholar]
- 73.Upton C, Buckley JT. A new family of lipolytic enzymes? Trends Biochem Sci. 1995;20:178–179. doi: 10.1016/S0968-0004(00)89002-7. [DOI] [PubMed] [Google Scholar]
- 74.Wagner UG, Petersen EI, Schwab H, Kratky C. EstB from Burkholderia gladioli: a novel esterase with a β-lactamase fold reveals steric factors to discriminate between esterolytic and β-lactam cleaving activity. Protein Sci. 2002;11:467–478. doi: 10.1110/ps.33002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 76.Widersten M, Gurell A, Lindberg D. Structure-function relationships of epoxide hydrolases and their potential use in biocatalysis. Biochim Biophys Acta. 2010;1800:316–326. doi: 10.1016/j.bbagen.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 77.Wohlgemuth R. Epoxide hydrolysis. In: Faber K, Fessner W-D, Turner NJ, editors. Science of synthesis, biocatalysis in organic synthesis 2. Stuttgart: Georg Thieme Verlag KG; 2015. pp. 529–555. [Google Scholar]
- 78.Yin DL, Bernhardt P, Morley KL, Jiang Y, Cheeseman JD, Purpero V, Schrag JD, Kazlauskas RJ. Switching catalysis from hydrolysis to perhydrolysis in P. fluorescens esterase. Biochemistry. 2010;49:1931–1942. doi: 10.1021/bi9021268. [DOI] [PMC free article] [PubMed] [Google Scholar]