Abstract
Advancement in metabolic engineering of microorganisms has enabled bio-based production of a range of chemicals, and such engineered microorganism can be used for sustainable production leading to reduced carbon dioxide emission there. One area that has attained much interest is microbial hydrocarbon biosynthesis, and in particular, alkanes and alkenes are important high-value chemicals as they can be utilized for a broad range of industrial purposes as well as ‘drop-in’ biofuels. Some microorganisms have the ability to biosynthesize alkanes and alkenes naturally, but their production level is extremely low. Therefore, there have been various attempts to recruit other microbial cell factories for production of alkanes and alkenes by applying metabolic engineering strategies. Here we review different pathways and involved enzymes for alkane and alkene production and discuss bottlenecks and possible solutions to accomplish industrial level production of these chemicals by microbial fermentation.
Keywords: Metabolic engineering; Alkanes/alkenes; Fatty acid biosynthesis; Cell factories; TRY (titer, rate, and yield)
Introduction
Environmental concerns and depletion of fossil fuels have raised interest in producing biofuels and bio-based chemicals with an environmental footprint that is lower than current production from fossil fuels [29, 48, 65]. Until the early 2000s, metabolic engineering was applied primarily to improve titer and productivity of industrial fermentation processes [45]. To date, integration of synthetic biology and systems biology into the field of metabolic engineering have resulted in remarkable advancement of the field [16, 20]. With these advanced technologies it has become possible to produce by engineered cell factories more diverse compounds, which can be used as pharmaceuticals, chemical building blocks, and fuels [47]. In addition, we have acquired better understanding of complex cellular networks from the massive biological data sets provided by systems biology approaches and these findings have brought new tools and techniques that can be used to engineer cell factories.
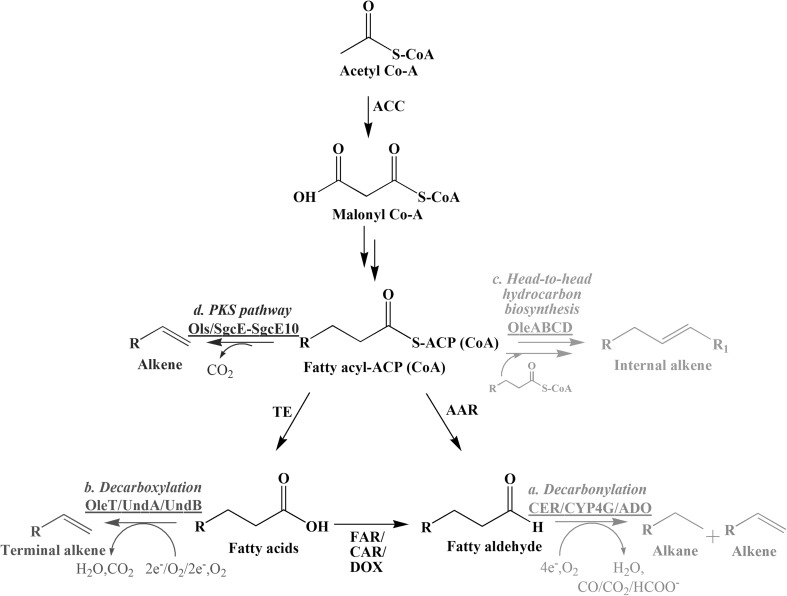
Alkanes and alkenes are a very important class of hydrocarbons, used as liquid transportation fuels and as plastics. However, to obtain alkanes and alkenes with the right properties, cracking of crude oils are necessary. The complexity of this process can cause technical difficulties in obtaining specific molecules as well as it can increase processing costs [33]. Many organisms synthesize alkanes and alkenes naturally for protection against environmental threats [27, 30, 54], but the production level and structures of the compounds are not ideal for direct utilization as drop-in fuels. Several pathways and enzymes involved in alkane and alkene biosynthesis from natural producers have been discovered (Fig. 1) and introduction of the biosynthetic pathways into heterologous microbial hosts has allowed for production of various structures of alkanes and alkenes (Table 1). Many efforts have been made to produce alkanes and alkenes in engineered microbial strains, and recent advancements shows promise for possible future industrial production [47]. However, the titer, rate, and yield (TRY) of alkanes and alkenes production by heterologous hosts is still too low to meet industrial requirements and there are still several challenges that need to be overcome before these molecules can be produced by microbial fermentation.
Fig. 1.
Alkane/alkene biosynthetic pathways and enzymes, which were utilized in previous reports. a Conversion of fatty aldehydes to alkanes/alkenes by AD enzymes, CER (plant), CYP4G (insects), and ADO (cyanobacteria), b terminal alkene production by decarboxylation enzymes, OleT, UndA, and UndB, c internal alkene biosynthesis by head-to-head hydrocarbon biosynthetic enzyme, OleABCD, d alkene production by PKS pathway enzymes, Ols and SgcE-SgcE10
Table 1.
Examples of alkane/alkene production by metabolically engineered microorganisms
| Enzyme | Strain | Products | Titer | References |
|---|---|---|---|---|
| AAR-AD | E. coli | Alkanes (C15, C17) | 7.7 mg/L | [18] |
| E. coli | Alkanes (C15, C17), Alkene (C17) | 255.6 mg/L | [57] | |
| E. coli | Alkanes (C15, C17) | 300 mg/L | [54] | |
| E. coli | Alkanes (C9, C12, C13, C14), Alkene (C13) | 580.8 mg/L | [15] | |
| S. cerevisiae | Alkanes (C13, C15, C17) | 22 μg/g of DW | [8] | |
| S. cerevisiae | Alkanes (C27–C31) | 86 μg/g of DW | [6] | |
| S. cerevisiae | Alkanes (C13, C15, C17), Alkenes (C15, C17) | 0.82 mg/L | [66] | |
| Synechocystis sp. PCC6803 | Alkanes (C15, C17), Alkene (C17) | 26 mg/L | [63] | |
| FAR-AD | E. coli | Alkanes (C13, iso-C13, C15, iso-C15, C16, C17), Alkenes (C13, C15, C16, C17) | 5 mg/L | [28] |
| CAR-AD | E. coli | Alkanes (C11, C13), Alkenes (C15, C17) | 2 mg/L | [2] |
| DOX-AD | S. cerevisiae | Alkanes (C14, C16) | 73. 5 μg/L | [24] |
| OleT | E. coli | Alkenes (C11, C13, C15, C15:2, C17:2) | 97.6 mg/L | [41] |
| S. cerevisiae | Alkenes (C11–C19) | 3.7 mg/L | [10] | |
| UndA | E. coli | Alkenes (C9–C13) | 6 mg/L | [53] |
| UndB | E. coli | Alkenes (C5–C17) | 55 mg/L | [52] |
| OleABCD | E. coli | Alkenes (27:3, 27:2, 29:2, 29:3) | 40 μg/L | [5] |
| Ols | Synechoccus sp. PCC7002 | Alkenes (C19, C19:2) | 4.2 mg/L/OD730 | [43] |
| S. globisporus | Alkene (15:7) | 129.3 mg/L | [14] | |
| E. coli | Alkene (C15) | 140 mg/L | [40] |
Here, we review the literature on alkane/alkene biosynthesis, and the associated fatty acid biosynthesis pathway and further discuss how the TRY challenge can be overcome to obtain efficient cell factories.
Pathways and enzymes of alkane/alkene biosynthesis
Decarbonylation of fatty aldehydes
Most pathways leading to alkanes/alkenes are via fatty aldehydes [36]. Aldehyde decarbonylases (ADs) have been discovered from various organisms including plants, insects, and cyanobacteria [6, 49] and they can convert fatty aldehydes to alkanes/alkenes with co-current production of carbon dioxide (CO2), carbon monoxide (CO), or formate depending on the organisms [42]. Some plants synthesize very-long-chain (VLC) alkanes to prevent water evaporation and protect themselves from environmental stresses [7]. The Arabidopsis thaliana aldehyde decarbonylase CER1 has been used to demonstrate alkane production in heterologous microbial systems [6, 15]. Insect cuticular layer is a mixture of alkanes/alkenes, and it acts as a barrier against environmental attacks as well as communication pheromones. Insect CYP4G1 is a P450 enzyme and co-expression of Drosophila CYP4G1 with cytochrome P450 reductase (CPR) enabled production of C23, C25 and C27 alkanes in S. cerevisiae [49]. Biosynthesis of alkanes has been identified in many cyanobacterial strains [54], but the reasons for alkane biosynthesis in cyanobacteria are unclear. ADs in cyanobacteria were initially reported to produce alkanes/alkenes and CO [54], but the actual co-product were later identified as formate by isotope tracer experiments. The cyanobacteria AD was therefore renamed as aldehyde deformylating oxygenase (ADO) [38].
There are two routes available for production of alkanes/alkenes via ADs, and these are from either fatty acyl-CoAs or from free fatty acids by the action of fatty acyl-acyl carrier protein (ACP) reductase (AAR) or fatty acid reductase (FAR), respectively (Fig. 1). Arabidopsis CER1 was revealed as an alkane biosynthetic enzyme through mutant and overexpression experiments in plants. However, alkanes were not detected when CER1 was solely expressed in yeast. Only through co-expression of Arabidopsis CER1–CER3 the production of VLC alkanes, chain-lengths C27–C31, was confirmed in yeast. In addition, alkane titer was increased and mainly resulted in production of nonacosane (C29) by 86 μg/mg of DW with co-expression of CYTB5s or/and LACS1, which acts as electron transfer components and long-chain acyl-CoA synthetase, respectively [6]. Even though CER1 produced VLC alkanes in yeast and plants, the enzyme was shown to enable production of C8–C14 alkanes in an E. coli strain that was engineered to produce short-chain fatty acids. The strain, that also encompassed several other engineering strategies, could produce short-chain alkanes up to 580.8 mg/L, and it was created by: (a) blocking the β-oxidation pathway by deleting the fadE gene which encodes acyl-CoA dehydrogenase, (b) increasing formation of short-chain FFAs by introducing modified thioesterase (TE), TesA with a L109P mutation and deleting the fadR gene, which is a transcriptional regulator, (c) expression of the fadD gene which encodes for fatty acyl-CoA synthetase for efficient conversion of FFAs to fatty acyl-CoAs, (d) expression of AAR from Clostridium acetobutylicum and CER1 from A. thaliana to produce short-chain alkanes from fatty acyl CoAs [15]. In an earlier study, cyanobacteria AAR activity was tested with two substrates, acyl-ACP and acyl-CoA in the presence of NADPH [54]. Both substrates were converted to fatty aldehydes by AAR, but acyl-ACP was the preferred substrate [54]. This enzyme has been frequently used for reconstruction of alkane/alkene biosynthetic pathways, and the expression of cyanobacteria AAR and ADO together in engineered strains results in production of predominantly C15 and C17 alkanes [8, 18, 54, 57, 63, 66]. However, the alkane titer in S. cerevisiae has not been reached the level of other hosts having co-expression of AAR-ADO. This was recently shown to be partly due to the presence of aldehyde dehydrogenase activity in yeast. Deletion of HFD1, encoding hexadecenal dehydrogenase (HFD), resulted in an increase in the alkane titer of S. cerevisiae by 22 μg/g of DW [8]. In addition, elimination of competing pathways (Δpox1 and Δadh5), increase of aldehyde supply (expression of carboxylic acid reductase (CAR) from Mycobacterium marinum), and enhancement of ADO expression (ADOs from Synechoccocus elongatus and Nostoc punctiforme) achieved an even highest alkane titer (0.82 mg/L) with an engineered S. cerevisiae [65], but it is still not comparable with the titers that can be obtained using E. coli (300 mg/L) [54].
Because fatty acids are abundant molecules in cells, they have been considered as desirable substrates for synthesis of alkanes/alkenes. Expression of the FAR complex from Photorhabdus luminescens [28], CAR from Mycobacterium marinum [2], and fatty acid α-dioxygenase (DOX) from Oryza sativa [24] enabled production of alkanes/alkenes when expressed together with AD. The FAR complex is encoded by reductase (luxC), synthetase (luxE), and transferase (luxD) gene operon, and it converts various chain-lengths of fatty acids into fatty aldehydes and expressed together with ADO from N. punctiforme it resulted in a more diverse range of alkane/alkene chain-lengths at a titer of 5 mg/L with E. coli compared with the use of the AAR-ADO pathway from N. punctiforme [28]. In addition, expression of FatB1, which is a C14 fatty acyl-ACP specific TE from Cinnamomum camphora, increased the C14 fatty acid and C13 alkane production in E. coli as well. The CAR enzyme has a broad substrate range (C4–C18), and addition of ATP and NADPH converts fatty acids to their corresponding fatty aldehydes, and co-expression of CAR and AD from Prochlorococcus marinus produce 2 mg/L of C11–C17 alkanes/alkenes in E. coli [2]. O. sativa DOX has the advantage as a fatty acid converting enzyme that it is using dioxygen as a co-factor, while AAR and CAR enzymes require NADPH and its substrate range is wider (C12–C18) than that of the cyanobacterial AAR (C16–C18).
Decarboxylation of fatty acids
Terminal alkenes, often referred to as olefins, are important compounds in the chemical industry as they are used for the production of detergents, lubricants, and polyethylene. Three different types of enzymes, OleTJE, UndA, and UndB are involved in direct enzymatic conversion of fatty acids to terminal alkenes, and heterologous expression of these enzymes enabled production of terminal alkenes in engineered microbial strains [51–53]. OleTJE is a cytochrome P450 enzyme belonging to the CYP152 family, and it was discovered from Jeotglicoccus sp. ATCC 8456. The CYP152 family OleTJE, forms alkenes by decarboxylation process rather than decarbonylation performed by the CYP4G family enzymes from insects. The CYP152 family members were reported to use only H2O2 as sole electron and oxygen donor, but recently H2O2-independent alkene biosynthesis was achieved using other biocatalytic co-factor systems: RhFRed, Fdr/Fdx, and CamAB [22, 41]. Recently the aldehyde decarboxylases, UndA and UndB, from Pseudomonas species were discovered, and both enzymes, when expressed in E. coli, resulted in production of terminal alkenes [52, 53]. UndA was found to be a non-heme iron oxidase, and the enzyme converted lauric acid (LA) to 1-undecene in the presence of Fe2+. In addition, it has a narrow substrate range, only medium-chain fatty acids (C10–C14) can be converted to ‘C-1’ corresponding terminal alkenes [53]. UndB was originally categorized as a fatty acid desaturase based on sequence homology, but found also to be an aldehyde decarboxylase. Compared with UndA, UndB shows a broad substrate range from C6 to C16, but it prefers C10–C14 fatty acids like UndA. The UndB homologue Pmen_4370 presented the highest conversion rate of undecanoic acid (C11) with E. coli strains co-expressing E. coli TE, UcFatB2 [52], but OleT provided the highest total titer of alkanes/alkenes in engineered E. coli strains (OleT: 97.6 mg/L, UndA: 6 mg/L UndB: 55 mg/L) [41, 52, 53].
Head-to-head hydrocarbon biosynthesis
Nearly half a century ago, two research groups observed long-chain alkene biosynthesis in Sarcina lutea, but they could not elucidate the biochemical and genetic data in detail [3, 4, 61]. In a recent study of long-chain alkene biosynthesis by Micrococcus luteus resulted in identification of the three genes (Mlut_13230–13250) encoding OleABCD involved in alkane biosynthesis. Mlut_13230 has a conserved active site residue which is found in fatty acid biosynthesis enzymes, and its function as a OleA homologue was revealed by its ability to convert acyl-CoA to unsaturated monoketones in vitro [5]. Mlut_13240 and 13250 have sequence similarities with oleD and fusion of oleB and oleC from oleABCD in Stenotrophomonas maltophilia and heterologous expression of oleABCD from M. lutea resulted in production of 40 μg/L of long-chain alkenes in E. coli. In Shewanella oneidensis Strain MR-1, one of the long-chain alkenes, 3,6,9,12,15,19,22,25,28-hentriacontanonaene was synthesized, and the compound is interestingly identified from many bacteria which were isolated from cold environments. The deletion of the oleABCD homologue in the MR-1 strain resulted loss of alkane production supporting that the enzyme complex is involved in alkane biosynthesis [60].
OleABCD enzymes were specified based on extensive sequence homology analysis as a combination of a superfamily of enzymes consisting of thiolase (OleA), α/β-hydrolase (OleB), AMP-dependent ligase/synthase (OleC), and short-chain dehydrogenase/reductase (OleD) [59]. The proposed pathway of OleABCD is initiated by OleA, which involves a non-decarboxylative Claisen condensation to generate a β-keto acid and then OleD produces β-hydroxy acid by NADPH-dependent reduction [26, 59]. After the process, OleC converts the β-hydroxy acid to an alkene by consuming ATP [25]. Even though the role of OleB is unclear so far, a fusion of oleB and oleC in some organisms, propose a linkage to the activity of OleC or it is presumed to perform scaffolding or regulatory function in the Ole complex [62].
Polyketide synthase (PKS) pathway
Polyketide and fatty acid biosynthetic pathways have very similar mechanisms, and both pathways are considered as promising for biofuel production. Generally, PKSs consist of 3-β-keto-acyl synthases (KS), acyl-transferases (AT), and acyl carrier proteins (ACP), β-keto reductases (KR), dehydratases (DH), enoyl reductases (ER), and finally a TE. The acyl substrates initiate the processes and malonyl-CoA extends the chain-length through elongation module, KS, AT, and ATP. After elongation the β-keto group is reduced to a β-hydroxyl group by KR, DH, and ER and then the TE domain catalyze decarboxylation and dehydration to release the alkene product. The Synechococcus sp. PCC 7002 synthesize C19 alkenes (1,14-nonadecadiene and 1-nonadecene) with terminal double bond, and to identify the involved enzyme, the sequence of Curacin A PKS was used to look for homologous potentially involved in alkane biosynthesis. CurM is the last module of Curacin A PKS from marine cyanobacteria Lyngbya majuscule, which forms the terminal double bond. Based on the sequence alignment result, one enzyme with 45 % amino acid homology to CurM was identified and it was named olefin synthase (Ols). In addition, Ols has several conserved domains of PKSs, loading domain (LD), ACP1, KS, AT, KR, ACP2, sulfotransferase (ST), and TE. To verify the involvement of Ols in long-chain alkene biosynthesis, an ols deletion mutant strain and an ols gene overexpression strain were constructed, and it was found that Ols deletion abolished alkene biosynthesis and strong expression of Ols increased alkene production by 4.2 mg/L/OD730 [43]. In another study, co-expression of enediyne PKS, SgcE and TE, SgcE10 from Streptomyces resulted in production of enediyne antibiotic C-1027 (37.5 mg/L) and 1,3,5,7,9,11,13-pentadecaheptaene (PDH, 129.3 mg/L) in engineered Streptomyces globisporus [14]. In addition, a SgcE-SgcE10 construct was introduced into E. coli, and it resulted in production of PDH. Through a chemical hydrogenation process, the cell culture extract product, PDH could be converted to pentadecane at a titer of 140 mg/L [40].
Increasing TRY
Since the first report of alkane production in E. coli [54], there have been increased interests for engineering microorganisms to develop efficient cell factories that can be used for for alkane/alkene production in industry. Despite extensive knowledge on the metabolism of E. coli and S. cerevisiae, the two preferred cell factory platforms, the productivity of alkanes/alkenes by these organisms is much below what is required for industrial production. Costs of goods sold (COGS), around $0.6 per liter is considered to be the economically profitable value of microbial biofuel production. However, it is challenging to produce hydrocarbons at this value. For example, to produce pentadecane (C15) in S. cerevisiae, there is a requirement for NADPH and ATP that will reduce the yield on glucose. However, using detailed metabolic modeling the theoretical yield for production of different hydrocarbons in yeast, including alkanes/alkenes, and from this analysis it was found that even though the molar yield is lower than for ethanol the energy yield is only 5–10 % lower [9]. Furthermore, from a techno-economic analysis it was found that alkanes/alkenes, if 90 % of the maximum theoretical yield can be obtained, can be produced cost efficiently compared with ethanol [9]. A major factor for this is that costs of separation of hydrocarbons is likely to be lower than the relatively expensive distillation used for ethanol production.
However, it is clear that to meet the industrial feasibility, the microorganisms needs to be optimized in terms of TRY, and in the following we review the status on engineering cell factories for improving the TRY of alkanes/alkenes.
Improving enzyme activities
Many enzymes have been discovered and applied to reconstruct the alkane/alkene biosynthesis in microorganisms, but they are commonly not very efficient and it is, therefore, difficult to reach high productivities. Since the last enzymatic steps such as decarboxylation and decarbonylation there is a loss of carbon in this step, and this impacts the product yield. This carbon loss is particular costly for production of short-chain alkanes/alkenes, whereas for longer chain alkanes/alkenes it is acceptable [9]. A major problem for increasing rate is, however, the low enzyme activity which will require high-level expression of the enzyme, something that may cause a significant protein burden for the cell. For instance, cyanobacteria ADO is a very slow enzyme and it cannot convert aldehydes efficiently to alkanes/alkenes, which generally results in accumulation of fatty alcohols as by-products [2, 8]. Fatty alcohols can be formed from fatty aldehydes by the action of unspecific alcohol dehydrogenases, of which there are many in both E. coli and S. cerevisiae. Therefore, researchers are trying to find better enzyme candidates from various resources by applying data derived from bioinformatics studies and sequence alignment analysis. In a previous study of OleT, several homologous genes of oleT JE presented different productivity and distribution of chain-lengths, and a codon-optimized version of the best enzyme enabled increased production of alkenes [10]. Structure-based engineering of enzymes can also change the active site and hereby improve the enzyme activity. A modified ADO from Procholorococcus marinus with point mutation A134F had an altered substrate specificity with enhanced activity towards short-chain aldehydes and could hereby improve about twofold total titer of propane (0.46 mg/L) in E. coli [32].
Enzyme activities can also be optimized by changing the environmental condition. This was demonstrated by analysis of engineered E. coli strain carrying ADs from Arabidopsis and cyanobacteria. These enzymes have different optimal temperatures and CER1 expression was found to be strongest at 30°C and could at this temperature result in an alkane/alkene titer of 580.8 mg/L [15], whereas cyanobacterial ADO expression lead to the highest titer (26 mg/L) and cell mass (OD600 = 19.0) when the temperature was adjusted to 24°C compared with three other temperatures evaluated (18°C, 5.3 mg/L, OD600 = 8.5; 24°C, 5.0 mg/L, OD600 = 9.3; and 37°C, 4.2 mg/L, OD600 = 7.3) [57]. In the case of OleT co-expressed with CamAB and formate dehydrogenase (FDH), the optimal temperature for enzyme activity was different depending on the FA chain-length (highest activity at 4°C for C4–C9 and C18, and room temperature for C10–C16) [22]. As titer is an accumulated metric it is closely correlated with cell growth and the alkane/alkene titer was therefore found to increase with the use of rich media for engineered strains of E. coli expressing Ado and S. cerevisiae expressing oleT [10, 57].
Improving precursor and co-factor supply
To increase the TRY of alkanes/alkenes, engineering of the fatty acid biosynthetic pathway is desirable as well as proper supply of acetyl-CoA, malonyl-CoA, and fatty acyl-CoA is important. Acetyl-CoA is a crucial molecule in microbial metabolism and it serves as one of the key precursors for fatty acid biosynthesis [35]. Because eukaryotic and prokaryotic cells have different acetyl-CoA metabolism, the strategies for precursor supply level increase depends on the cell factory platform used. In S. cerevisiae the acetyl-CoA metabolism is present in four different compartments (nucleus, mitochondria, cytosol and peroxisomes) and this makes it difficult to engineer acetyl-CoA metabolism in this cell factory. One strategy to enhance acetyl-CoA supply in the cytosol of S. cerevisiae was engineering of the pyruvate dehydrogenase (PDH) bypass in yeast, and this was achieved by expression of PDH from Enterococcus faecalis [34]. PDH is a very large multi-functional enzyme and it requires activation by lipoylation and as lipoic acid is synthesized in the mitochondria it was necessary to add lipoic acid to the medium [34, 46]. E. faecalis PDH has no mitochondrial targeting sequences unlike all eukaryotic PDHs, and it is relatively insensitive to high NADH but inhibited by high NADH/NAD+ ratios. Therefore, this PDH complex encoded by pdhA, pdhB, aceF, and lpd was able to function in the cytosol of S. cerevisiae and co-expression of E. faecalis lipolyation genes (lplA and lplA2) with the E. faecalis PDH enabled replacement of the native cytosol acetyl-CoA supply by ATP-independent mechanism [34]. In another strategy the ethanol degradation pathway was employed by overexpression of endogenous alcohol dehydrogenase (ADH2), acetaldehyde dehydrogenase (ALD6), and a mutant acetyl-CoA synthase (ACS with L641P) from Salmonella enterica [21]. Expression of these enzymes together with a wax ester synthase increased fatty acid ethyl ester (FAEE) production in S. cerevisiae [21]. Furthermore, this strategy also improved production of the plant sesquiterpen α-santalene in S. cerevisiae [13]. Compared to yeast, acetyl-CoA supply was designed differently in E. coli and cyanobacteria. In E. coli deletion of glucose fermentation pathway genes (ΔadhE, ΔackA-pta, ΔldhA, and ΔfrdC) redirected more carbon flux through acetaldehyde biosynthesis and overexpression of S. enterica EutE, a putative ALD/acetyl-CoA reductase, resulted in improvement of acetaldehyde production [68]. Improvement of alkane/alkene production was found in a Synechocystis mutant strain, which carried a deletion of the lactate biosynthesis gene, 2-hydroxyacid dehydrogenase (DDH), by overexpressing the AAR-ADO construct in the DDH site [63].
Malonyl-CoA is a key substrate for fatty acid biosynthesis and to malony-CoA production in is often increased by overexpression of acetyl-CoA carboxylase (ACC). It was reported that overexpression of endogenous ACC1 alone increased the production of 3-hydroxypropionic acid, which is derived from malonyl-CoA, by 65 % in S. cerevisiae [12]. Wax ester production were also increased by 30 % when ACC1 was overexpressed together with a wax ester synthase [55]. However, overexpression of ACC1 does not always increase formation of products derived from the fatty acid biosynthetic pathways [10, 17]. To increase the fatty acyl-CoA pool, disruption of the β-oxidation pathway has been implemented in S. cerevisiae and through the deletion of ∆faa1 and ∆faa4, the alkene titer was sevenfold increased [10]. In addition, deletion of additional genes involved in β-oxidation, ∆faa2, ∆pxa1, and ∆pox1 or ∆faa1, ∆faa2, ∆faa4, ∆fat1, ∆pxa1, and Δpox1 resulted in significant improvement of fatty acid production [37]. These strategies could reduce or completely eliminate competing pathways and hereby ensure more efficient production of fatty acids, and in particular ensure that there is no wasteful consumption of NADH and ATP, resulting in improved yield.
Together with precursor supply, proper co-factor supply can increase the productivity of alkanes/alkenes. Co-factor engineering has been used to ensure efficient OleT activity. OleT catalyze the decarboxylation of fatty acids to produce alkenes utilizing H2O2 as the sole electron and oxygen donor, and recently alternative systems was applied to replace the use of H2O2 [10, 22, 41]. In the presence of NADPH and oxygen, OleT used as redox partner the P450 reductase domain RhFRED from Rhodococcus sp. or the separate flavodoxin/flavodoxin reductase from E coli to produce alkenes [41]. Hem3, which is involved in heme biosynthesis was able to replace the use of H2O2 in alkene biosynthesis in S. cerevisiae, and it resulted in 8.7-fold increase of alkene titer by 427.7 μg/L [10]. Moreover, The CamAB components (CamA: NADH-putidaredoxin reductase and CamB: putidaredoxin) were able to convert fatty acids to alkenes when combined with a NAD(P)H regeneration system such as glucose dehydrogenase, phodphite dehydrogenase or FDH [22]. The ADs are required to utilize reducing components to increase alkane production and in yeast, VLC alkanes were improved when the CER1/CER3 were co-expressed with Cytochrome b5 (Cytb5) [6]. Similarly, co-expression of the insect CYP4G1 together with Drosophila CPR increased the production of alkanes in S. cerevisiae [49]. The important role of an efficient reducing system has been further supported by findings that expression of cyanobacteria ADO without a reducing system resulted in no alkane production and overexpression of reducing components from E. coli produced 2.7 μg/g of DW heptadecane in S. cerevisiae [8].
Toxicity
Toxicity is a crucial issue to be considered when high productivity of chemical production is required in microorganisms. Alkanes/alkenes are toxic molecules in microorganisms, and change cell membrane integrity and function which cause inhibition of growth or even cell death [56]. To overcome chemical toxicity in microbial hosts, various strategies have been implemented. For example, overexpression of heat shock proteins or small regulatory RNAs, two-phase culture system, engineering of regulatory gene or enzyme, and these strategies all enhanced the solvent tolerance [31]. However, each strategy only partly solves the problem, and more basic research on the toxicity of alkanes is required. One approach is to combine toxicology and genomics, by some referred to as toxicogenomics, to map cellular responses to toxicants [1]. Recently, based on mechanistic understanding of cellular functions, transporters have emerged as targets for engineering in order to alleviate chemical toxicity [23, 44]. To investigate alkane/alkene related transporters in S. cerevisiae, the cells were exposed to C9–C12 alkanes, and several induced plasma membrane efflux pumps were identified from transcriptome data [39]. Some of the efflux pump candidates were verified, and overexpression of Snq2p and Pdr5p improved alkane tolerance by reducing intracellular decane and undecane concentrations [39]. In another study, ABC transporters in Yarrowia lipolytica were studied to identify efficient efflux pumps. Different chain-lengths (C8–C12) alkanes were supplied for susceptibility assay, and expression of ABC2 and ABC3 transporters increased the tolerance towards C10 and C11 alkanes [11]. In particular, ABC2 was revealed as the best efflux pump among four ABC transporters evaluated, and the reason for improved alkane tolerance was explained by a decrease in the intracellular alkane level. However, the efflux pumps did not improve C8 and C9 alkane tolerance and in S. cerevisiae overexpression of the Snq2p and Pdr5p efflux pumps only increased tolerance towards C10 and C11 alkanes but not towards C9 alkane [39]. This shows that efflux pumps are quite specific and it is, therefore, suggested to broaden the tolerance ranges by performing several strategies such as directed evolution and enzyme engineering by structure-based study [11, 39].
Future outlook and conclusion
Alkanes/alkenes are industrially relevant chemicals, which can be used as starting materials in the chemical industry and as a liquid transportation fuel. Even though many organisms synthesize alkanes/alkenes naturally, industrial demands of these chemicals are currently only supplied by fossil fuels. Applications of the alkane/alkene biosynthesis in microorganisms can enable a future sustainable and green production of these important chemicals. Fatty acid biosynthesis provides several paths to re-construct alkane/alkene biosynthesis in heterologous microbial hosts. Efforts in microbial engineering for alkane/alkene production has shown that it is possible to engineer microorganisms for production of a diverse range of alkanes/alkenes. However, the TRY level of the compounds is far from the industrial requirements and further development is needed to obtain efficient cell factories.
For this intracellular biosensors may be valuable as they may allow for high-throughput screening and strain selection. Some of bacteria degrade alkanes/alkenes, and this has been exploited to create alkane biosensors. Alkane biosensors consist of three parts: alkane responsive transcriptional regulator, a promoter that is activated by regulator, and a reporter protein (e.g. GFP). Previous biosensors had problems for use in heterologous hosts because of limited detection range or proper function in a heterologous host [58, 67]. A new biosensor constructed as a chimera alkane response element (cARE), however, shows great promise [64, 65]. This was re-assembled using earlier biosensors, and enabled in situ detection of both mid- and long-chain alkanes by the fluorescence activated cell sorting (FACS) technology in E. coli [49, 65]. The use of a biosensor like this will enable faster screening of improved strains.
Even though it may be possible to produce alkanes/alkenes from sugar in a process similar to first generation ethanol production, our techno-economic analysis showed that using biomass as feedstock would be allow for further reduction of the production costs [9], and more importantly it will significantly reduce greenhouse gas emission, and even more than what is obtained with secondary bioethanol production. Therefore, it should be considered to reduce pre-treatment cost for lignocellulosic biomass and also engineer microorganisms for utilization of various carbon sources.
In conclusion, various natural alkane/alkene pathways have provided new enzymes that can be used for reconstructing alkane/alkene biosynthetic pathways in heterologous microbial hosts. Different combinations of these enzymes has allowed for increased production as well as enabled diversity of compound structures. Table 1 summarizes the titer of alkanes/alkenes in engineered strains, and it is observed that the AAR-AD pathway showed the highest titer among the reported biosynthetic pathways up to date. The enzymes showed different substrate specificities and activities. For example, UndB has broader substrate range than UndA [52], and also CAR from P. marinus synthesized several aldehydes while cyanobacteria AAR provide only two products [2, 54]. Besides, UndB displayed better conversion rate of fatty acids for alkene production compared with OleT and UndA [52]. It is, therefore, expected that discovery of new enzymes for alkane biosynthesis may allow for further improvement of alkane/alkene production. Furthermore, biosensors will probably promote the speed of enzyme screening and selection process, and biosensors will also enable faster evaluation of different strategies to improve the TRY of cell factories producing alkanes/alkenes and hereby reduce strain development time. In previous studies, engineered biosensors further enabled to control gene expression avoiding toxic intermediates accumulation in the cell [19], or it was possible to optimize individual reaction in biosynthetic pathways through real-time observation [50]. Together with such strategies, alkane/alkene biosensors will be possible breakthroughs to find the bottleneck steps faster, and also establish efficient cell factories by resolving current challenges in strain engineering.
Acknowledgments
We are happy and honored to contribute a paper in an issue of Journal of Industrial Microbiology and Biotechnology dedicated to Prof. Arnold Demain. His many fantastic contributions to industrial microbiology and biotechnology have served as a tremendous inspiration for our work. JN further thanks Arny for many excellent scientific discussions during my time at MIT in 1995–1996, but also at many occasions following that visit. We acknowledge funding from the Knut and Alice Wallenberg Foundation, the Novo Nordisk Foundation and Total New Energies.
References
- 1.Aardema MJ, MacGregor JT. Toxicology and genetic toxicology in the new era of “Toxicogenomics”: impact of “-omics” technologies. Mutat Res. 2002;499:13–25. doi: 10.1016/S0027-5107(01)00292-5. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar MK, Turner NJ, Jones PR. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc Natl Acad Sci. 2013;110:87–92. doi: 10.1073/pnas.1216516110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albro PW, Dittmer JC. Biochemistry of long-chain, nonisoprenoid hydrocarbons. I. Characterization of the hydrocarbons of Sarcina lutea and the isolation of possible intermediates of biosynthesis. Biochem. 1969;8:394–405. doi: 10.1021/bi00829a055. [DOI] [PubMed] [Google Scholar]
- 4.Albro PW, Huston CK. Lipids of Sarcina lutea II: hydrocarbon content of the lipid extracts. J Bacteriol. 1964;88:981–986. doi: 10.21236/ad0437968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beller HR, Goh E-B, Keasling JD. Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl Environ Microbiol. 2010;76:1212–1223. doi: 10.1128/AEM.02312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure J-D, Haslam RP, Napier JA, Lessire R, Joubès J. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell. 2012;24:3106–3118. doi: 10.1105/tpc.112.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourdenx B, Bernard A, Domergue F, Pascal S, Léger A, Roby D, Pervent M, Vile D, Haslam RP, Napier JA. Overexpression of ArabidopsisECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 2011;156:29–45. doi: 10.1104/pp.111.172320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buijs NA, Zhou YJ, Siewers V, Nielsen J. Long-chain alkane production by the yeast Saccharomyces cerevisiae. Biotechnol Bioeng. 2015;112:1275–1279. doi: 10.1002/bit.25522. [DOI] [PubMed] [Google Scholar]
- 9.Caspeta L, Nielsen J. Economic and environmental impacts of microbial biodiesel. Nat Biotech. 2013;31:789–793. doi: 10.1038/nbt.2683. [DOI] [PubMed] [Google Scholar]
- 10.Chen B, Lee D-Y, Chang MW. Combinatorial metabolic engineering of Saccharomyces cerevisiae for terminal alkene production. Metab Eng. 2015;31:53–61. doi: 10.1016/j.ymben.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Chen B, Ling H, Chang MW. Transporter engineering for improved tolerance against alkane biofuels in Saccharomyces cerevisiae. Biotechnol Biofuels. 2013;6:1–10. doi: 10.1186/1754-6834-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Bao J, Kim I-K, Siewers V, Nielsen J. Coupled incremental precursor and co-factor supply improves 3-hydroxypropionic acid production in Saccharomyces cerevisiae. Metab Eng. 2014;22:104–109. doi: 10.1016/j.ymben.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Daviet L, Schalk M, Siewers V, Nielsen J. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab Eng. 2013;15:48–54. doi: 10.1016/j.ymben.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Yin M, Horsman GP, Shen B. Improvement of the enediyne antitumor antibiotic C-1027 production by manipulating its biosynthetic pathway regulation in Streptomyces globisporus. J Nat Prod. 2011;74:420–424. doi: 10.1021/np100825y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi YJ, Lee SY. Microbial production of short-chain alkanes. Nature. 2013;502:571–574. doi: 10.1038/nature12536. [DOI] [PubMed] [Google Scholar]
- 16.Church GM, Elowitz MB, Smolke CD, Voigt CA, Weiss R. Realizing the potential of synthetic biology. Nat Rev Mol Cell Biol. 2014;15:289–294. doi: 10.1038/nrm3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Courchesne NMD, Parisien A, Wang B, Lan CQ. Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J Biotechnol. 2009;141:31–41. doi: 10.1016/j.jbiotec.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Coursolle D, Lian J, Shanklin J, Zhao H. Production of long chain alcohols and alkanes upon coexpression of an acyl-ACP reductase and aldehyde-deformylating oxygenase with a bacterial type-I fatty acid synthase in E. coli. Mol BioSyst. 2015;11:2464–2472. doi: 10.1039/C5MB00268K. [DOI] [PubMed] [Google Scholar]
- 19.Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, Keasling JD. Engineering dynamic pathway regulation using stress-response promoters. Nat Biotechnol. 2013;31:1039–1046. doi: 10.1038/nbt.2689. [DOI] [PubMed] [Google Scholar]
- 20.Dai Z, Nielsen J. Advancing metabolic engineering through systems biology of industrial microorganisms. Curr Opin Biotechnol. 2015;36:8–15. doi: 10.1016/j.copbio.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 21.De Jong BW, Shi S, Siewers V, Nielsen J. Improved production of fatty acid ethyl esters in Saccharomyces cerevisiae through up-regulation of the ethanol degradation pathway and expression of the heterologous phosphoketolase pathway. Microb Cell Fact. 2014;13:1–10. doi: 10.1186/s12934-014-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennig A, Kuhn M, Tassoti S, Thiessenhusen A, Gilch S, Bülter T, Haas T, Hall M, Faber K. Oxidative decarboxylation of short-chain fatty acids to 1-alkenes. Angew Chem Int Ed Engl. 2015;54:8819–8822. doi: 10.1002/anie.201502925. [DOI] [PubMed] [Google Scholar]
- 23.Dunlop MJ. Engineering microbes for tolerance to next-generation biofuels. Biotechnol Biofuels. 2011;4:1–9. doi: 10.1186/1754-6834-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foo JL, Susanto AV, Keasling JD, Leong SSJ, Chang MW. Whole-cell biocatalytic and de novo production of alkanes from free fatty acids in Saccharomyces cerevisiae. Biotechnol Bioeng. 2016 doi: 10.1002/bit.25920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frias JA, Goblirsch BR, Wackett LP, Wilmot CM. Cloning, purification, crystallization and preliminary X-ray diffraction of the OleC protein from Stenotrophomonas maltophilia involved in head-to-head hydrocarbon biosynthesis. Acta Crystallogr F. 2010;66:1108–1110. doi: 10.1107/S1744309110031751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frias JA, Richman JE, Erickson JS, Wackett LP. Purification and characterization of OleA from Xanthomonas campestris and demonstration of a non-decarboxylative claisen condensation reaction. J Biol Chem. 2011;286:10930–10938. doi: 10.1074/jbc.M110.216127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard RW, Blomquist GJ. Ecological, behavioral, and biochemical aspects of insect hydrocarbons. Annu Rev Entomol. 2005;50:371–393. doi: 10.1146/annurev.ento.50.071803.130359. [DOI] [PubMed] [Google Scholar]
- 28.Howard TP, Middelhaufe S, Moore K, Edner C, Kolak DM, Taylor GN, Parker DA, Lee R, Smirnoff N, Aves SJ, Love J. Synthesis of customized petroleum-replica fuel molecules by targeted modification of free fatty acid pools in Escherichia coli. Proc Natl Acad Sci. 2013;110:7636–7641. doi: 10.1073/pnas.1215966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang Y-S, Park JM, Choi S, Choi YJ, Cho JH, Lee SY. Engineering of microorganisms for the production of biofuels and perspectives based on systems metabolic engineering approaches. Biotechnol Adv. 2012;30:989–1000. doi: 10.1016/j.biotechadv.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 30.Jetter R, Kunst L. Plant surface lipid biosynthetic pathways and their utility for metabolic engineering of waxes and hydrocarbon biofuels. Plant J. 2008;54:670–683. doi: 10.1111/j.1365-313X.2008.03467.x. [DOI] [PubMed] [Google Scholar]
- 31.Jin H, Chen L, Wang J, Zhang W. Engineering biofuel tolerance in non-native producing microorganisms. Biotechnol Adv. 2014;32:541–548. doi: 10.1016/j.biotechadv.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Khara B, Menon N, Levy C, Mansell D, Das D, Marsh ENG, Leys D, Scrutton NS. Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase. ChemBioChem. 2013;14:1204–1208. doi: 10.1002/cbic.201300307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kissin YV. Chemical mechanisms of catalytic cracking over solid acidic catalysts: alkanes and alkenes. Catal Rev. 2001;43:85–146. doi: 10.1081/CR-100104387. [DOI] [Google Scholar]
- 34.Kozak BU, van Rossum HM, Luttik MAH, Akeroyd M, Benjamin KR, Wu L, de Vries S, Daran J-M, Pronk JT, van Maris AJA. Engineering acetyl coenzyme A supply: functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae. MBio. 2014 doi: 10.1128/mBio.01696-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krivoruchko A, Zhang Y, Siewers V, Chen Y, Nielsen J. Microbial acetyl-CoA metabolism and metabolic engineering. Metab Eng. 2015;28:28–42. doi: 10.1016/j.ymben.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Kunjapur AM, Prather KLJ. Microbial engineering for aldehyde synthesis. Appl Environ Microbiol. 2015;81:1892–1901. doi: 10.1128/AEM.03319-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leber C, Polson B, Fernandez-Moya R, Da Silva NA. Overproduction and secretion of free fatty acids through disrupted neutral lipid recycle in Saccharomyces cerevisiae. Metab Eng. 2015;28:54–62. doi: 10.1016/j.ymben.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Li N, W-c Chang, Warui DM, Booker SJ, Krebs C, Bollinger JM. Evidence for only oxygenative cleavage of aldehydes to alk(a/e)nes and formate by cyanobacterial aldehyde decarbonylases. Biochem. 2012;51:7908–7916. doi: 10.1021/bi300912n. [DOI] [PubMed] [Google Scholar]
- 39.Ling H, Chen B, Kang A, Lee J-M, Chang MW. Transcriptome response to alkane biofuels in Saccharomyces cerevisiae: identification of efflux pumps involved in alkane tolerance. Biotechnol Biofuels. 2013;6:1–10. doi: 10.1186/1754-6834-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Q, Wu K, Cheng Y, Lu L, Xiao E, Zhang Y, Deng Z, Liu T. Engineering an iterative polyketide pathway in Escherichia coli results in single-form alkene and alkane overproduction. Metab Eng. 2015;28:82–90. doi: 10.1016/j.ymben.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Wang C, Yan J, Zhang W, Guan W, Lu X, Li S. Hydrogen peroxide-independent production of α-alkenes by OleTJE P450 fatty acid decarboxylase. Biotechnol Biofuels. 2014;7:1–12. doi: 10.1186/1754-6834-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marsh ENG, Waugh MW. Aldehyde decarbonylases: enigmatic enzymes of hydrocarbon biosynthesis. ACS Catal. 2013;3:2515–2521. doi: 10.1021/cs400637t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendez-Perez D, Begemann MB, Pfleger BF. Modular synthase-encoding gene involved in α-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Appl Environ Microbiol. 2011;77:4264–4267. doi: 10.1128/AEM.00467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mukhopadhyay A. Tolerance engineering in bacteria for the production of advanced biofuels and chemicals. Trends Microbiol. 2015;23:498–508. doi: 10.1016/j.tim.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Nielsen J. Metabolic engineering. Appl Microbiol Biotechnol. 2001;55:263–283. doi: 10.1007/s002530000511. [DOI] [PubMed] [Google Scholar]
- 46.Nielsen J. Synthetic biology for engineering acetyl coenzyme A metabolism in yeast. MBio. 2014;5:e02153–e02114. doi: 10.1128/mBio.02153-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen J, Keasling JD. Engineering cellular metabolism. Cell. 2016;164:1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Peralta-Yahya PP, Zhang F, Del Cardayre SB, Keasling JD. Microbial engineering for the production of advanced biofuels. Nature. 2012;488:320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 49.Qiu Y, Tittiger C, Wicker-Thomas C, Le Goff G, Young S, Wajnberg E, Fricaux T, Taquet N, Blomquist GJ, Feyereisen R. An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc Natl Acad Sci. 2012;109:14858–14863. doi: 10.1073/pnas.1208650109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogers JK, Church GM. Genetically encoded sensors enable real-time observation of metabolite production. Proc Natl Acad Sci. 2016;113:2388–2393. doi: 10.1073/pnas.1600375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rude MA, Baron TS, Brubaker S, Alibhai M, Del Cardayre SB, Schirmer A. Terminal olefin (1-alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl Environ Microbiol. 2011;77:1718–1727. doi: 10.1128/AEM.02580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rui Z, Harris NC, Zhu X, Huang W, Zhang W. Discovery of a family of desaturase-like enzymes for 1-alkene biosynthesis. ACS Catal. 2015;5:7091–7094. doi: 10.1021/acscatal.5b01842. [DOI] [Google Scholar]
- 53.Rui Z, Li X, Zhu X, Liu J, Domigan B, Barr I, Cate JHD, Zhang W. Microbial biosynthesis of medium-chain 1-alkenes by a nonheme iron oxidase. Proc Natl Acad Sci. 2014;111:18237–18242. doi: 10.1073/pnas.1419701112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. Microbial biosynthesis of alkanes. Science. 2010 doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 55.Shi S, Valle-Rodríguez JO, Khoomrung S, Siewers V, Nielsen J. Functional expression and characterization of five wax ester synthases in Saccharomyces cerevisiae and their utility for biodiesel production. Biotechnol Biofuels. 2012;5:1–10. doi: 10.1186/PREACCEPT-1932279820621895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sikkema J, De Bont J, Poolman B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song X, Yu H, Zhu K. Improving alkane synthesis in Escherichia coli via metabolic engineering. Appl Microbiol Biotechnol. 2016;100:757–767. doi: 10.1007/s00253-015-7026-y. [DOI] [PubMed] [Google Scholar]
- 58.Sticher P, Jaspers MC, Stemmler K, Harms H, Zehnder AJ, van der Meer JR. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl Environ Microbiol. 1997;63:4053–4060. doi: 10.1128/aem.63.10.4053-4060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sukovich DJ, Seffernick JL, Richman JE, Gralnick JA, Wackett LP. Widespread head-to-head hydrocarbon biosynthesis in bacteria and role of OleA. Appl Environ Microbiol. 2010;76:3850–3862. doi: 10.1128/AEM.00436-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sukovich DJ, Seffernick JL, Richman JE, Hunt KA, Gralnick JA, Wackett LP. Structure, function, and insights into the biosynthesis of a head-to-head hydrocarbon in Shewanella oneidensis strain MR-1. Appl Environ Microbiol. 2010;76:3842–3849. doi: 10.1128/AEM.00433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tornabene TG, Gelpi E, Oró J. Identification of fatty acids and aliphatic hydrocarbons in Sarcina lutea by gas chromatography and combined gas chromatography-mass spectrometry. J Bacteriol. 1967;94:333–343. doi: 10.1128/jb.94.2.333-343.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wackett LP, Wilmot CM (2015) Chapter 2-hydrocarbon biosynthesis in microorganisms In: Direct microbial conversion of biomass to advanced biofuels. Elsevier, Amsterdam, pp 13–31. doi:10.1016/B978-0-444-59592-8.00002-6
- 63.Wang W, Liu X, Lu X. Engineering cyanobacteria to improve photosynthetic production of alka(e)nes. Biotechnol Biofuels. 2013;6:1. doi: 10.1186/1754-6834-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu W, Zhang L, Yao L, Tan X, Liu X, Lu X. Genetically assembled fluorescent biosensor for in situ detection of bio-synthesized alkanes. Sci Rep. 2015;5:10907. doi: 10.1038/srep10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan Y, Liao JC. Engineering metabolic systems for production of advanced fuels. J Ind Microbiol Biotechnol. 2009;36:471–479. doi: 10.1007/s10295-009-0532-0. [DOI] [PubMed] [Google Scholar]
- 66.Zhou Yongjin J, Buijs Nicolaas A, Zhu Zhiwei, Qin Jiufu, Siewers Verena, Nielsen Jens. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun. 2016 doi: 10.1038/ncomms11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang D, He Y, Wang Y, Wang H, Wu L, Aries E, Huang WE. Whole-cell bacterial bioreporter for actively searching and sensing of alkanes and oil spills. Microb Biotechnol. 2012;5:87–97. doi: 10.1111/j.1751-7915.2011.00301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu H, Gonzalez R, Bobik TA. Coproduction of acetaldehyde and hydrogen during glucose fermentation by Escherichia coli. Appl Environ Microbiol. 2011;77:6441–6450. doi: 10.1128/AEM.05358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]