Abstract
GRAS genes encode plant-specific transcription factors that play important roles in plant growth and development. However, little is known about the GRAS gene family in apple. In this study, 127 GRAS genes were identified in the apple (Malus domestica Borkh.) genome and named MdGRAS1 to MdGRAS127 according to their chromosomal locations. The chemical characteristics, gene structures and evolutionary relationships of the MdGRAS genes were investigated. The 127 MdGRAS genes could be grouped into eight subfamilies based on their structural features and phylogenetic relationships. Further analysis of gene structures, segmental and tandem duplication, gene phylogeny and tissue-specific expression with ArrayExpress database indicated their diversification in quantity, structure and function. We further examined the expression pattern of MdGRAS genes during apple flower induction with transcriptome sequencing. Eight higher MdGRAS (MdGRAS6, 26, 28, 44, 53, 64, 107, and 122) genes were surfaced. Further quantitative reverse transcription PCR indicated that the candidate eight genes showed distinct expression patterns among different tissues (leaves, stems, flowers, buds, and fruits). The transcription levels of eight genes were also investigated with various flowering related treatments (GA3, 6-BA, and sucrose) and different flowering varieties (Yanfu No. 6 and Nagafu No. 2). They all were affected by flowering-related circumstance and showed different expression level. Changes in response to these hormone or sugar related treatments indicated their potential involvement during apple flower induction. Taken together, our results provide rich resources for studying GRAS genes and their potential clues in genetic improvement of apple flowering, which enriches biological theories of GRAS genes in apple and their involvement in flower induction of fruit trees.
Keywords: apple, GRAS, synteny, expression analysis, flower induction
Introduction
Transcription factors function as trans-acting factors, combining with specific cis- elements in eukaryotic gene promoter regions to regulate plant growth and development.
GRAS genes encode a family of plant-specific transcription factors. The GRAS name derives from the first letters of the first identified three members, including GAI (gibberellic acid insensitive), RGA (repressor of GA1-3 mutant), and SCR (scarecrow) (Bolle, 2004). In plants, proteins in the GRAS family consist of 400–700 amino acids with a conserved GRAS carboxyl terminus comprising several typical structures, including seven leucine-repeat domains (LHRI), a VHIID domain, seven leucine-repeat II domains (LHRII), and PFYRE and SAW motifs (Hirsch et al., 2009). Bacteria also harbor GRAS-like proteins, which may function as methylases (Zhang et al., 2012). In recent years, Many GRAS genes have also been identified in a variety of plant species, including Prunus mume, Populus, grape, tomato, Chinese cabbage, and castor bean (Tian et al., 2004; Song et al., 2014; Huang et al., 2015; Lu et al., 2015; Grimplet et al., 2016; Xu W. et al., 2016). The GRAS family, which has 33 members in Arabidopsis thaliana and 57 members in rice (Oryza sativa), can be divided into eight subfamilies: SCL3, SHR, PAT1, LISCL, DELLA, SCR, LS, and HAM (Hirsch et al., 2009).
Recent studies have revealed that the N-termini of GRAS proteins play important roles in their specific functions. The N-termini contain intrinsically disordered regions (IDRs); thus, GRAS proteins are also known as intrinsically disordered proteins (IDPs) (Sun et al., 2011). Several studies have focused on the structures and functions of the IDRs in the N-termini of GRAS proteins (Sun et al., 2010, 2011, 2012). Different subfamilies have different types and numbers of molecular recognition features in their N-termini (Sun et al., 2010, 2011, 2012). For instance, the DELLA subfamily has three molecular recognition features in its IDR at the N terminus: DELLA, VHYNP, and L(R/K) XI. Protein motifs at the C-terminus include the LHR I, HIID, LHRII, PFYRE, and SAW motifs (Pysh et al., 1999).
The functions of GRAS family genes have been studied in recent years; there is a large body of evidence that the GRAS genes family play important roles in various biological processes in many different plant species. They are involved in diverse processes related to root meristem development, axillary bud outgrowth, phytochrome signaling pathways, nodular signal transduction, and stress adaptation, especially in the gibberellins (GA)-signaling pathway (Ait-ali et al., 2003; Greb et al., 2003; Heckmann et al., 2006; Torres-Galea et al., 2006; Koizumi et al., 2012; Park et al., 2013). DELLA proteins are negative regulators of the GA signal, and have been shown to repress growth in many different plant species including Arabidopsis (AtGAI, AtRGA, AtRGL1, AtRGL2, and AtRGL3), O. sativa (OsSLR1), Zea mays (ZmD8), Malus domestica (MdRGL2a), and Vitis vinifera (VvGAI) (Foster et al., 2007; Vandenbussche et al., 2007; Aleman et al., 2008; Dai and Xue, 2010). Various studies have shown that the GA-GID1-DELLA complex and ubiquitin ligase E3 play important roles in GA signaling, and that SCR functions in the development of roots and above-ground organs in Arabidopsis (Zhang et al., 2011; Koizumi et al., 2012). PhHAM (Petunia hybrid), a petunia hairy meristem gene, was shown to be necessary to maintain the apical meristem in petunia, and the petunia recessive mutant phham (ham-B4281) produced fewer flowers than wild type (Stuurman et al., 2002). SCL13 was shown to positively regulate phyB, and the scl13 mutant showed impaired sensitivity in Arabidopsis (Torres-Galea et al., 2006). Several studies have shown that some GRAS genes are up-regulated under stress conditions in Arabidopsis, rice and tobacco (Day et al., 2003; Torres-Galea et al., 2006; Czikkel and Maxwell, 2007). Among all the biological processes, their flowering related characterizations and functions have been researched in some model plants. On the one hand, several GRAS genes are associated with the flowering phenotype. For example, in tomato, Ls (Later Suppressor), defined as another GRAS gene, its mutant ls showed lower numbers of inflorescence (Schumacher et al., 1999; Bolle, 2004). In Arabidopsis, the LS subfamily gene AtLAS and the SHR subfamily gene AtSHR were involved in shoot apical meristem. The Ls subfamily gene AtSCL26 and the HAM subfamily genes AtSCL6 and AtSCL27 were highly expressed in flowers (Bolle, 2004); the rga and gai mutant also showed earlier flowering in Arabidopsis (Galvao et al., 2012). On the other hand, GRAS genes can integrate or regulate flowering related genes such as SPL (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE), LFY (LEAFY), and AP1 (APETALA1) as well as FT (FLOWERING LOCUS T). They all acted positive roles and intergrated various signals in flower induction, of which FT and SPL affected flowering time; while LFY and AP1 belonged to flower meristem identify genes. Several GRAS genes can be recruited and integrated with SPL proteins to regulate LFY and AP1 expression in Arabidopsis (Yamaguchi et al., 2014). Additionally, the PAT and SCL subfamilies of GRAS genes were both reported involved in photosignal in many plants, it influenced the expression of phyA (phytochrome A), a phytochrome A signal transduction gene, which caused to the change of the circadian clock and expression of FT and other flowering genes (Bolle et al., 2000; Torres-Galea et al., 2006; Li et al., 2014). However, little is known about the GRAS family of transcription factors in woody fruit trees, such as its expression in response to flower induction in apple.
Apple is one of the most widely cultivated and economically important fruit trees in the temperate regions of the world. In apple production, flower induction and flower buds formation sustain two growing seasons. Flower induction is a key stage, and always limits fruit yield (Guitton et al., 2012). Apple (Malus domestica Borkh.) cv. “Fuji” is one of the most popular cultivars, and it accounts for 65% planting areas in China. “Fuji” has trouble in flowering induction. Thus, characteristic the molecular mechanism of apple flower induction is necessary. Flower induction was affected by environmental and internal factors (Guitton et al., 2012; Xing et al., 2014, 2015; Fan et al., 2016). Sugar and hormones were involved in flower induction (Guitton et al., 2012; Xing et al., 2014, 2015; Fan et al., 2016; Li et al., 2016; Zhang et al., 2016). Among all kinds of hormones, including auxin, cytokinins, abscisic acid, GA and other hormones. GA was defined the most important hormones in regulating flower induction and GA pathway was recognized as one of the flowering pathways (Guitton et al., 2012; Porri et al., 2012; Li et al., 2016; Zhang et al., 2016). 6-BA and sucrose, which can promote flower bud differentiation, were also important and widely used (Xing et al., 2015; Li et al., 2016). Several flowering-related genes families have been identified and characterized in the apple genome, including SPL, IDD, and MADX-box (Li et al., 2013; Kumar et al., 2016; Fan et al., 2017). GRAS genes were reported to partially or entirely involve in flower induction in various plants (Hynes et al., 2003; Foster et al., 2007; Vandenbussche et al., 2007; Galvao et al., 2012; Yamaguchi et al., 2014). Little is known about MdGRAS genes and their potential involvement in apple flower induction. The recently published genome sequence of apple (Velasco et al., 2010) has made it possible to characterize the structures, functions, evolution of apple GRAS genes and their response to flower induction. Moreover, it is also available to utilize candidate MdGRAS genes for further genetic improvement aiming at flowering induction.
In this study, we performed a genome-wide survey to identify GRAS genes in the apple genome. A systematic analysis including gene classification, gene characterization, gene structures and gene phylogenies were determined. We further investigated their expression levels of all the 127 putative MdGRAS genes among different tissues and varieties during apple growth and development with ArrayExpress database. Moreover, eight MdGRAS genes were further analyzed.
We investigated their expression levels of the candidate eight genes among different tissues (stems, leaves, flowers, fruits, and buds), different flowering-related treatments (GA3, 6-BA and sugar) and different flowering varieties (Yanfu No. 6 and Nagafu No. 2). Changes in response to these flowering-related treatments indicated their potential involvement in apple flower induction. Our results provided basic clues for further analyses of MdGRAS in apple growth and development.
Materials and methods
Identification of GRAS genes in apple
Hidden Markov Model (HMM) searches (Finn et al., 2011) were performed to identify GRAS genes in the apple genome. The HMM profiles (PF03514) were downloaded from the Pfam database (http://pfam.sanger.ac.uk). Additionally, AtGRAS protein sequences were used as queries for BLASTP search against the Apple Genome Database (https://www.rosaceae.org/) with an e-value < 1e-4 to identify further possible members of GRAS gene as previous investigation (Li et al., 2011; Jia et al., 2013; Cui et al., 2015; Xu J. N. et al., 2016). Arabidopsis GRAS genes were downloaded from TAIR (http://www.arabidopsis.org/). Finally, genes with the same accession number were removed. All the identified GRAS genes were further manually checked for the GRAS domain.
Gene structures and locations determination
Annotations for candidate GRAS genes were obtained from the Genome Database for Rosaceae (GDR) (http://www.rosaceae.org), and chromosome locations were retrieved from these annotations. MapDraw was used to show the accurate locations of the genes on each chromosome. The online software Gene Structure Display Server (GSDS: http://gsds.cbi.pku.edu.cn) was used to generate the exon-intron structures of all the GRAS genes.
Chemical characteristics, elements in the promoters and phylogenetic analysis
Prosite ExPaSy (http://web.expasy.org/protparam/) was used to predict the potential chemical characteristics of the MdGRAS genes. A phylogenetic tree was generated using MEGA 6.06 with the Maximum Likelihood (ML) method and 1000 bootstrap replications. The upstream of 1.5 Kb were used for cis-elements in the promoters of the candidate MdGRAS genes. PlantCARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used for searching regulatory elements.
Tandem duplication and synteny analysis
Tandem duplication and synteny analysis were conducted using Circos v. 0.63 (http://circos.ca/). Tandem duplication of the MdGRAS genes was identified according to their physical locations on individual chromosomes in the apple genome. Adjacent homologous GRAS genes on the same apple chromosome with no more than one intervening gene were considered tandem duplicates. The Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/) was used to identify syntenic blocks.
Plant material
In this study, 72 uniform 6-year-old “Fuji”/T337/Malus robusta Rehd. apple trees were randomly divided into three groups: 18 were treated with GA3, 18 were treated with sucrose, 18 were treated with 6-BA and 18 were sprayed with water as a control. These tress were grown at the experimental orchard of Northwest A& F University in Yangling (108°04′ E, 34°16′ N), China. And the orchard was well managed. Each group consisted of three blocks, with three replicates. The experiment was conducted from 30 to 70 days after full bloom (DAFB) in 2015. (1) GA3 treatment was performed as described by Zhang et al. (2016) with a slight modification: 700 mg L−1 GA3 (Sigma, Deisenhofen, Germany) was sprayed once on a clear morning at 30 DAFB (May 9). (2) 300 mg L−1 6-BA (Sigma, Deisenhofen, Germany) was also sprayed on a clear morning at 30 DAFB (May 9). (3)For sugar treatment, trees were sprayed twice on clear mornings at 30 and 37 DAFB (May 9 and May 16) with 15,000 mg L−1 and 20,000 mg L−1 sucrose. All treatments were performed on the whole plant and were applied with a low-pressure hand-wand sprayer. Terminal buds on current-year spurs (<5 cm), which were chosen according to our previous studies (Xing et al., 2014, 2015; Li et al., 2016; Zhang et al., 2016), were collected into liquid nitrogen at 30, 40, 50, 60, and 70 DAFB, and then stored at −80°C for use in gene expression analyses.
Additionally, buds from two apple varieties (“Yanfu No. 6” and “Nagafu No. 2”) with contrasting flowering rates were collected at 30, 40, 50, 60, and 70 DAFB from 18 uniform 6-year-old trees in 2015. “Yanfu No. 6” is a spontaneous mutant of “Nagafu No. 2.” It develops more flower buds than “Nagafu No. 2.” “Yanfu No. 6” always exhibits a higher proportion of spurs, shorter internodes, bigger buds and more flowers. Terminal buds on current-year spurs (<5 cm) were collected as described above.
Different organs were also collected for tissue-specific expression analysis. Flowers were collected at full blossom on April 9 in 2015. Stems were collected from new shoots with a diameter of 2–3 mm. Mature leaves were collected from the adjacent buds. Fruits were collected with a diameter of 2–3 cm. All tissues were immediately frozen in liquid nitrogen and stored at −80°C for gene expression analysis. Additionally, the online ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/, E-GEOD-42873) was also employed to investigate their expression patterns among different apple varieties and tissues.
Flowering rate analysis
Flowering rate was also investigated among different exogenous treatment and two different flowering varieties. One hundred twenty terminal buds on short shoots which were less than 5 cm and no fruits on them were tagged randomly at 15 DAFB. The number of flowers was counted of the tagged shorts as finally flowering rate. And it was investigated at full blossom on April 10 in 2016.
RNA extraction and cDNA synthesis
Total RNA was extracted from plant tissue samples using the cetyltrimethyl ammonium bromide (CTAB) method with slight modifications (Gambino et al., 2008). Briefly, 900 μL extraction buffer (2% CTAB, 2.5% PVP-40, 2 M NaCl, 100 mM Tris-HCl [pH 8.0], 25 mM EDTA [pH 8.0], and 2% β-mercaptoethanol) was preheated at 65°C and added to 2-mL microcentrifuge tubes just before use. Samples containing 200 mg of bud tissue stored at −80°C were ground to a powder, added to the tubes, and mixed with extraction buffer. After shaking and inverting each tube vigorously for 5 min and incubating at 65°C for 30 min, an equal volume of chloroform:isoamyl alcohol (24:1, v/v) was added. Each tube was shaken and inverted vigorously and then centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was collected into a new tube and re-extracted with an equal volume of chloroform:isoamyl alcohol (24:1, v/v). The resulting supernatant was then transferred into a new 2-mL tube and LiCl (3 M final concentration) was added. The mixture was incubated at −20°C for 4 h and the RNA was selectively pelleted by LiCl after centrifugation at 18,000 × g for 20 min at 4°C. The pellet was resuspended in 500 μL of SSTE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 1% SDS, and 1 M NaCl) preheated to 65°C and an equal volume of chloroform:isoamyl alcohol. The mixture was then centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was transferred to a new microcentrifuge tube, and the RNA was precipitated with 2.5 volumes of cold ethanol at −80°C for at least 30 min and centrifuged at 1,200 × g for 20 min at 4°C. Finally, the pellets were washed with 70% ethanol and resuspended in diethylpyrocarbonate-treated water.
Total RNA integrity was verified by running the samples on 2% agarose gels. First-strand cDNA was synthesized from 1 μg of total RNA using a PrimeScript RT Reagent kit with gDNA Eraser (Takara Bio, Shiga, Japan) following the manufacturer's instructions.
Expression analysis by qRT-PCR
The expression levels of all identified MdGRAS genes were analyzed by qRT-PCR using primer pairs designed with Primer 5.0 (Table S1). The real-time PCR assay mix (20 μL) consisted of 2-μL cDNA samples (diluted 1:8), 10 μL 2 × SYBR Premix ExTaq II (Takara Bio), 0.8 μL of each primer (10 μM) (Table 1), and 6.4 μL distilled deionized H2O. Each PCR assay was run on an iCycler iQ Real Time PCR Detection System (Bio-Rad) with an initial denaturation at 95°C for 3 min, followed by 40 cycles at 94°C for 15 s, 62°C for 20 s, and 72°C for 20 s. The resulting fragments were subjected to melting-curve analysis to verify the presence of gene-specific PCR products. The melting-curve analysis was performed directly after real-time PCR and included an initial step of 94°C for 15 s, followed by a constant increase from 60° to 95°C at a 2% ramp rate. The apple EF-1α gene (GenBank accession no. DQ341381) was used as an internal control to normalize all mRNA expression levels. The 2−ΔΔCt method was used to calculate the relative amount of template present in each PCR amplification mixture (Livak and Schmittgen, 2001).
Table 1.
List of MdGRAS genes and their chemical characteristics.
| Gene name | Gene locusa | Location | CDS (bp) | Peptide (aa) | Strand | MWb (kDa) | GRAVYc | PId | IIe | AIf |
|---|---|---|---|---|---|---|---|---|---|---|
| MdGRAS1 | MDP0000135256 | chr1:16537486.16539435 | 930 | 309 | − | 33.36 | −0.011 | 9.33 | 42.29 | 96.60 |
| MdGRAS2 | MDP0000252300 | chr1:16538919.16544966 | 2,550 | 849 | − | 93.62 | −0.547 | 8.10 | 52.08 | 75.54 |
| MdGRAS3 | MDP0000119183 | chr1:18011215.18015301 | 1,356 | 451 | − | 50.28 | −0.263 | 6.33 | 43.93 | 82.20 |
| MdGRAS4 | MDP0000825698 | chr2:3727360.3729003 | 1,644 | 547 | − | 61.13 | −0.316 | 10.01 | 59.37 | 81.72 |
| MdGRAS5 | MDP0000134341 | chr2:3744608.3746247 | 1,572 | 523 | − | 57.30 | −0.156 | 5.70 | 45.66 | 85.64 |
| MdGRAS6 | MDP0000256486 | chr2:12924073.12925887 | 1,815 | 604 | − | 66.48 | −0.378 | 5.13 | 52.06 | 78.86 |
| MdGRAS7 | MDP0000139971 | chr2:13285923.13292998 | 2,331 | 776 | − | 86.60 | −0.126 | 6.08 | 45.09 | 90.70 |
| MdGRAS8 | MDP0000247514 | chr2:17606743.17608173 | 1,431 | 476 | − | 53.23 | −0.069 | 5.67 | 43.06 | 90.99 |
| MdGRAS9 | MDP0000165587 | chr2:18027038.18028561 | 1,524 | 507 | − | 56.88 | −0.392 | 5.22 | 48.32 | 69.29 |
| MdGRAS10 | MDP0000859087 | chr2:25937193.25938899 | 1,707 | 568 | − | 63.23 | −0.446 | 5.02 | 44.23 | 74.00 |
| MdGRAS11 | MDP0000159341 | chr3:4321397.4322773 | 1,377 | 458 | + | 51.96 | −0.272 | 6.07 | 54.41 | 92.64 |
| MdGRAS12 | MDP0000194661 | chr3:7771083.7773134 | 2,052 | 683 | − | 77.70 | −0.645 | 5.52 | 53.63 | 74.64 |
| MdGRAS13 | MDP0000796348 | chr3:7777819.7780444 | 2,160 | 719 | − | 82.05 | −0.547 | 6.90 | 42.05 | 75.80 |
| MdGRAS14 | MDP0000194663 | chr3:7781197.7788706 | 5,787 | 1928 | − | 218.53 | −0.568 | 5.83 | 49.45 | 72.05 |
| MdGRAS15 | MDP0000247940 | chr3:7827530.7831637 | 2,715 | 904 | + | 101.59 | −0.540 | 5.38 | 50.48 | 71.38 |
| MdGRAS16 | MDP0000306154 | chr3:20678219.20679634 | 1,365 | 454 | − | 50.87 | −0.125 | 6.16 | 40.19 | 92.64 |
| MdGRAS17 | MDP0000793615 | chr3:25279993.25282044 | 2,052 | 683 | + | 75.01 | −0.418 | 5.64 | 52.31 | 77.42 |
| MdGRAS18 | MDP0000205622 | chr3:25473680.25476670 | 2,007 | 668 | − | 74.17 | −0.399 | 5.61 | 48.56 | 80.90 |
| MdGRAS19 | MDP0000835454 | chr3:32975889.32976838 | 876 | 291 | − | 32.90 | −0.713 | 5.40 | 41.66 | 68.38 |
| MdGRAS20 | MDP0000300311 | chr3:33027149.33029331 | 735 | 244 | − | 27.06 | 0.038 | 8.86 | 43.86 | 90.66 |
| MdGRAS21 | MDP0000259453 | chr3:33044014.33045333 | 1,188 | 395 | − | 43.95 | −0.304 | 5.87 | 40.29 | 84.43 |
| MdGRAS22 | MDP0000420150 | chr3:33053754.33054086 | 333 | 110 | + | 12.55 | 0.305 | 4.53 | 25.59 | 119.73 |
| MdGRAS23 | MDP0000141505 | chr3:33061403.33061960 | 558 | 185 | + | 21.06 | 0.094 | 6.82 | 48.488 | 92.70 |
| MdGRAS24 | MDP0000465513 | chr3:33061932.33063815 | 1,884 | 627 | − | 70.91 | −0.263 | 5.50 | 41.06 | 81.31 |
| MdGRAS25 | MDP0000232659 | chr3:33105946.33112977 | 3,024 | 1007 | − | 113.57 | −0.535 | 7.10 | 50.65 | 73.76 |
| MdGRAS26 | MDP0000147173 | chr4:915116.916492 | 1,377 | 458 | − | 51.30 | −0.285 | 5.12 | 45.39 | 83.01 |
| MdGRAS27 | MDP0000396037 | chr4:3510983.3512255 | 993 | 330 | + | 36.34 | −0.308 | 5.33 | 48.90 | 67.45 |
| MdGRAS28 | MDP0000945004 | chr4:4693449.4695500 | 2,052 | 683 | − | 74.98 | −0.407 | 5.69 | 52.03 | 77.85 |
| MdGRAS29 | MDP0000241048 | chr4:12671528.12673775 | 1,668 | 555 | − | 61.88 | −0.387 | 5.98 | 49.78 | 79.89 |
| MdGRAS30 | MDP0000228483 | chr4:16466303.16467886 | 1,584 | 527 | − | 59.59 | −0.320 | 4.82 | 50.99 | 76.94 |
| MdGRAS31 | MDP0000317825 | chr4:16469757.16471367 | 1,611 | 536 | − | 60.75 | −0.309 | 4.67 | 53.32 | 75.67 |
| MdGRAS32 | MDP0000419045 | chr4:18100227.18103427 | 2,490 | 829 | − | 88.67 | −0.308 | 5.82 | 59.22 | 73.84 |
| MdGRAS33 | MDP0000254936 | chr5:411761.422370 | 3,600 | 1199 | + | 133.82 | −0.316 | 6.64 | 39.77 | 79.76 |
| MdGRAS34 | MDP0000151144 | chr5:3168395.3171022 | 2,400 | 799 | − | 87.03 | −0.267 | 6.03 | 57.31 | 76.92 |
| MdGRAS35 | MDP0000242829 | chr5:10558101.10559735 | 1,635 | 544 | + | 60.88 | −0.244 | 4.83 | 46.59 | 83.75 |
| MdGRAS36 | MDP0000230994 | chr5:15148360.15151129 | 1,572 | 523 | − | 58.94 | −0.320 | 6.24 | 48.46 | 82.26 |
| MdGRAS37 | MDP0000566079 | chr5:22506172.22506504 | 330 | 110 | − | 12.50 | −0.232 | 6.04 | 44.89 | 80.64 |
| MdGRAS38 | MDP0000243351 | chr6:9882603.9884659 | 1,824 | 607 | + | 66.98 | −0.509 | 5.19 | 53.22 | 72.49 |
| MdGRAS39 | MDP0000915100 | chr6:9882928.9884982 | 2,055 | 694 | + | 75.47 | −0.403 | 5.56 | 51.47 | 78.03 |
| MdGRAS40 | MDP0000322003 | chr6:9923402.9927244 | 981 | 326 | + | 36.85 | −0.214 | 4.91 | 43.91 | 88.50 |
| MdGRAS41 | MDP0000124576 | chr6:16659904.16660845 | 567 | 188 | − | 21.11 | −0.162 | 6.36 | 33.54 | 82.39 |
| MdGRAS42 | MDP0000128964 | chr6:16680332.16681280 | 804 | 267 | + | 29.94 | −0.114 | 7.19 | 29.99 | 84.72 |
| MdGRAS43 | MDP0000206202 | chr6:23034606.23035559 | 954 | 317 | − | 35.85 | −0.707 | 5.68 | 39.04 | 67.41 |
| MdGRAS44 | MDP0000159531 | chr7:22551974.22553587 | 1,614 | 537 | − | 59.25 | −0.215 | 5.34 | 55.11 | 85.72 |
| MdGRAS45 | MDP0000315706 | chr7:24383528.24394942 | 2,817 | 938 | − | 103.28 | −0.432 | 6.41 | 47.36 | 75.69 |
| MdGRAS46 | MDP0000275704 | chr8:10371640.10374024 | 2,385 | 794 | − | 86.86 | −0.327 | 6.05 | 55.01 | 76.76 |
| MdGRAS47 | MDP0000287321 | chr8:18091272.18096730 | 1,827 | 608 | + | 67.24 | −0.270 | 9.15 | 46.58 | 87.42 |
| MdGRAS48 | MDP0000441376 | chr9:562753.565977 | 2,091 | 696 | − | 77.38 | −0.406 | 5.73 | 64.97 | 73.22 |
| MdGRAS49 | MDP0000863616 | chr9:6708558.6709889 | 1,332 | 443 | − | 49.06 | −0.131 | 5.19 | 49.04 | 93.05 |
| MdGRAS50 | MDP0000804652 | chr9:8891282.8892943 | 1,662 | 553 | − | 62.32 | −0.346 | 5.74 | 49.48 | 78.66 |
| MdGRAS51 | MDP0000921788 | chr9:9424971.9428298 | 2,073 | 690 | − | 77.57 | −0.312 | 8.85 | 47.18 | 80.71 |
| MdGRAS52 | MDP0000203826 | chr9:22347041.22348471 | 1,431 | 476 | + | 54.20 | −0.347 | 6.31 | 41.21 | 89.33 |
| MdGRAS53 | MDP0000181482 | chr9:29373931.29376390 | 1,938 | 645 | + | 70.41 | −0.258 | 4.76 | 39.14 | 81.19 |
| MdGRAS54 | MDP0000727836 | chr10:1113520.1115259 | 1,740 | 579 | − | 64.46 | −0.448 | 5.03 | 40.79 | 75.80 |
| MdGRAS55 | MDP0000169115 | chr10:5639948.5641363 | 1,416 | 471 | + | 53.93 | −0.406 | 6.31 | 44.39 | 89.87 |
| MdGRAS56 | MDP0000512470 | chr10:5640084.5641499 | 1,416 | 471 | + | 53.93 | −0.406 | 6.31 | 44.39 | 89.87 |
| MdGRAS57 | MDP0000307619 | chr10:18265808.18267481 | 1,464 | 487 | − | 54.88 | −0.243 | 6.07 | 46.99 | 86.12 |
| MdGRAS58 | MDP0000288971 | chr10:22109238.22112956 | 831 | 276 | − | 31.19 | −0.097 | 5.27 | 47.65 | 91.45 |
| MdGRAS59 | MDP0000162468 | chr10:26355418.26356833 | 1,416 | 471 | + | 53.93 | −0.406 | 6.31 | 44.39 | 89.87 |
| MdGRAS60 | MDP0000443053 | chr10:29148732.29150108 | 1,377 | 458 | − | 51.67 | −0.211 | 5.81 | 52.99 | 90.74 |
| MdGRAS61 | MDP0000607785 | chr10:32638991.32646762 | 1,821 | 606 | + | 67.87 | −0.369 | 6.58 | 43.64 | 76.73 |
| MdGRAS62 | MDP0000715138 | chr11:2058625.2059440 | 717 | 238 | − | 26.65 | −0.444 | 5.12 | 46.87 | 68.49 |
| MdGRAS63 | MDP0000389025 | chr11:4337648.4339036 | 1,389 | 462 | + | 52.60 | −0.243 | 6.03 | 58.05 | 94.35 |
| MdGRAS64 | MDP0000264347 | chr11:8307859.8309883 | 2,025 | 674 | + | 76.62 | −0.616 | 5.40 | 48.74 | 74.32 |
| MdGRAS65 | MDP0000287039 | chr11:8329524.8331601 | 1,233 | 410 | + | 47.21 | −0.377 | 7.20 | 38.27 | 78.71 |
| MdGRAS66 | MDP0000851230 | chr11:8330639.8332645 | 2,007 | 668 | − | 76.04 | −0.609 | 5.96 | 45.35 | 73.13 |
| MdGRAS67 | MDP0000289783 | chr11:8333605.8335590 | 1,986 | 661 | − | 75.26 | −0.570 | 6.47 | 54.65 | 70.67 |
| MdGRAS68 | MDP0000287040 | chr11:8334218.8339685 | 4,134 | 1377 | + | 155.67 | −0.517 | 6.02 | 52.42 | 72.45 |
| MdGRAS69 | MDP0000289784 | chr11:8336934.8339074 | 2,100 | 699 | − | 78.44 | −0.478 | 5.90 | 47.91 | 73.96 |
| MdGRAS70 | MDP0000287041 | chr11:8340645.8342651 | 2,007 | 668 | + | 76.04 | −0.609 | 5.96 | 45.35 | 73.13 |
| MdGRAS71 | MDP0000250943 | chr11:8344394.8346472 | 2,076 | 692 | − | 78.26 | −0.562 | 5.89 | 46.69 | 71.03 |
| MdGRAS72 | MDP0000827722 | chr11:8398699.8400897 | 2,199 | 732 | + | 82.40 | −0.519 | 5.80 | 44.56 | 72.45 |
| MdGRAS73 | MDP0000453000 | chr11:8405492.8408822 | 3,054 | 1017 | + | 114.99 | −0.545 | 5.86 | 46.75 | 72.50 |
| MdGRAS74 | MDP0000227056 | chr11:8417502.8420917 | 2,607 | 868 | + | 97.64 | −0.580 | 6.17 | 52.53 | 65.66 |
| MdGRAS75 | MDP0000236041 | chr11:8419367.8421957 | 2,313 | 770 | + | 88.11 | −0.491 | 8.69 | 45.15 | 77.70 |
| MdGRAS76 | MDP0000581417 | chr11:8434126.8436324 | 2,199 | 732 | − | 82.40 | −0.519 | 5.80 | 44.56 | 72.45 |
| MdGRAS77 | MDP0000822651 | chr11:10148161.10149699 | 1,539 | 512 | − | 57.41 | −0.381 | 5.33 | 47.06 | 68.22 |
| MdGRAS78 | MDP0000301368 | chr11:20713600.20715024 | 1,425 | 474 | + | 52.98 | −0.156 | 5.95 | 40.62 | 94.49 |
| MdGRAS79 | MDP0000809337 | chr11:25863922.25865673 | 1,752 | 583 | + | 64.77 | −0.378 | 5.94 | 51.14 | 84.32 |
| MdGRAS80 | MDP0000321018 | chr11:25863923.25875124 | 3,474 | 1157 | + | 130.01 | −0.289 | 6.43 | 45.30 | 87.14 |
| MdGRAS81 | MDP0000768445 | chr11:34920904.34922793 | 1,889 | 629 | + | 71.39 | −0.276 | 5.81 | 48.55 | 80.59 |
| MdGRAS82 | MDP0000119093 | chr12:19433725.19435980 | 2,256 | 751 | + | 84.11 | −0.499 | 7.09 | 48.76 | 74.51 |
| MdGRAS83 | MDP0000884524 | chr12:24351825.24353366 | 1,542 | 513 | + | 56.34 | −0.119 | 5.36 | 34.40 | 92.22 |
| MdGRAS84 | MDP0000148294 | chr12:24959928.24961310 | 1,383 | 460 | − | 52.22 | −0.303 | 4.90 | 58.00 | 83.91 |
| MdGRAS85 | MDP0000235134 | chr12:24972396.24973970 | 1,575 | 525 | − | 59.08 | −0.337 | 4.67 | 53.02 | 75.41 |
| MdGRAS86 | MDP0000926466 | chr12:24983309.24984916 | 1,608 | 535 | − | 60.29 | −0.344 | 4.72 | 52.16 | 75.08 |
| MdGRAS87 | MDP0000666991 | chr12:25017893.25019506 | 1,614 | 537 | + | 61.11 | −0.347 | 4.69 | 51.34 | 71.55 |
| MdGRAS88 | MDP0000397638 | chr12:26835765.26838962 | 2,475 | 824 | − | 88.38 | −0.296 | 6.01 | 55.30 | 76.52 |
| MdGRAS89 | MDP0000266986 | chr13:2853393.2854769 | 1,377 | 458 | + | 51.63 | −0.180 | 5.67 | 53.61 | 94.78 |
| MdGRAS90 | MDP0000464809 | chr13:7394453.7398805 | 2,856 | 951 | + | 105.52 | −0.238 | 6.01 | 49.95 | 83.71 |
| MdGRAS91 | MDP0000122441 | chr13:13603697.13605022 | 1,326 | 441 | − | 49.16 | −0.013 | 8.38 | 47.79 | 87.82 |
| MdGRAS92 | MDP0000245252 | chr13:13603924.13605249 | 1,326 | 441 | − | 49.11 | −0.013 | 8.25 | 46.59 | 86.94 |
| MdGRAS93 | MDP0000522931 | chr13:13606211.13607461 | 1,251 | 416 | − | 46.86 | −0.077 | 5.19 | 37.37 | 90.41 |
| MdGRAS94 | MDP0000245253 | chr13:13606548.13608350 | 1,803 | 600 | − | 67.20 | −0.225 | 4.97 | 35.14 | 80.88 |
| MdGRAS95 | MDP0000173434 | chr13:25599457.25600123 | 597 | 198 | + | 21.97 | 0.356 | 6.14 | 44.59 | 119.19 |
| MdGRAS96 | MDP0000138367 | chr13:25626475.25627141 | 495 | 164 | − | 18.47 | 0.405 | 5.84 | 33.17 | 120.67 |
| MdGRAS97 | MDP0000274120 | chr13:26280448.26284950 | 3,743 | 980 | + | 107.63 | −0.315 | 6.78 | 49.41 | 80.11 |
| MdGRAS98 | MDP0000308434 | chr13:30511943.30513601 | 1,659 | 552 | − | 61.15 | −0.647 | 6.07 | 40.73 | 66.61 |
| MdGRAS99 | MDP0000431628 | chr14:22609508.22611103 | 1,596 | 531 | − | 58.82 | −0.149 | 5.78 | 56.26 | 87.29 |
| MdGRAS100 | MDP0000258655 | chr14:22631424.22637535 | 777 | 258 | + | 29.07 | −0.279 | 8.86 | 50.64 | 85.81 |
| MdGRAS101 | MDP0000259403 | chr14:22631424.22637537 | 777 | 258 | + | 29.07 | −0.279 | 8.86 | 50.64 | 85.81 |
| MdGRAS102 | MDP0000319851 | chr14:28611108.28615187 | 2,319 | 772 | 87.26 | −0.146 | 8.63 | 43.98 | 95.08 | |
| MdGRAS103 | MDP0000171602 | chr14:28838647.28840187 | 1,506 | 501 | + | 56.15 | −0.388 | 5.37 | 46.69 | 66.99 |
| MdGRAS104 | MDP0000687763 | chr14:28859934.28861253 | 1,320 | 440 | − | 49.36 | −0.292 | 5.92 | 43.43 | 76.30 |
| MdGRAS105 | MDP0000840369 | chr14:28859987.28861525 | 1,539 | 512 | − | 57.41 | −0.381 | 5.33 | 47.06 | 68.22 |
| MdGRAS106 | MDP0000196258 | chr14:29246984.29249224 | 747 | 248 | + | 27.99 | −0.505 | 7.52 | 41.95 | 59.76 |
| MdGRAS107 | MDP0000662303 | chr15:11412854.11414497 | 1,644 | 547 | − | 59.78 | −0.097 | 5.53 | 42.31 | 92.07 |
| MdGRAS108 | MDP0000575908 | chr15:19584108.19585916 | 1,809 | 602 | + | 66.40 | −0.371 | 5.11 | 49.76 | 77.21 |
| MdGRAS109 | MDP0000315270 | chr15:23947574.23950023 | 2,181 | 726 | + | 79.31 | −0.283 | 5.26 | 48.40 | 85.76 |
| MdGRAS110 | MDP0000295058 | chr15:25739441.25740609 | 1,035 | 344 | + | 38.97 | −0.104 | 6.01 | 43.81 | 87.53 |
| MdGRAS111 | MDP0000187914 | chr15:25748188.25749720 | 1,533 | 510 | − | 57.15 | −0.072 | 6.04 | 45.81 | 89.67 |
| MdGRAS112 | MDP0000241137 | chr15:25750590.25752014 | 1,425 | 474 | + | 52.94 | −0.046 | 5.69 | 43.48 | 89.51 |
| MdGRAS113 | MDP0000263576 | chr17:336468.348106 | 4,695 | 1564 | − | 175.11 | −0.600 | 5.36 | 53.34 | 69.93 |
| MdGRAS114 | MDP0000441088 | chr17:844678.846561 | 1,884 | 627 | + | 70.97 | −0.272 | 5.51 | 39.89 | 79.60 |
| MdGRAS115 | MDP0000784909 | chr17:4316837.4319083 | 2,247 | 747 | − | 81.54 | −0.295 | 5.56 | 59.76 | 77.30 |
| MdGRAS116 | MDP0000827871 | chr17:7181440.7182768 | 1,329 | 442 | − | 49.03 | −0.127 | 5.30 | 46.49 | 91.49 |
| MdGRAS117 | MDP0000152670 | chr17:9016383.9018038 | 1,656 | 551 | − | 61.95 | −0.355 | 6.39 | 53.78 | 78.62 |
| MdGRAS118 | MDP0000284679 | chr17:9255749.9257927 | 1,917 | 638 | + | 70.63 | −0.136 | 5.32 | 52.66 | 85.00 |
| MdGRAS119 | MDP0000205236 | chr17:9258368.9260546 | 1,917 | 638 | + | 70.63 | −0.136 | 5.32 | 52.66 | 85.00 |
| MdGRAS120 | MDP0000289554 | chr17:10806972.10811830 | 1,926 | 641 | − | 70.67 | −0.198 | 9.28 | 48.25 | 90.41 |
| MdGRAS121 | MDP0000269044 | chr17:10818117.10822975 | 1,926 | 641 | − | 70.67 | −0.198 | 9.28 | 48.25 | 90.41 |
| MdGRAS122 | MDP0000669451 | chr17:22659094.22660848 | 1,755 | 584 | − | 63.71 | −0.281 | 4.91 | 45.60 | 79.73 |
| MdGRAS123 | MDP0000950387 | chr17:22676488.22677480 | 993 | 330 | − | 36.58 | −0.099 | 5.50 | 41.12 | 84.61 |
| MdGRAS124 | MDP0000319342 | unanchored:17969995.17971509 | 1,515 | 504 | − | 55.32 | −0.205 | 5.13 | 34.83 | 85.40 |
| MdGRAS125 | MDP0000237978 | unanchored:26304735.26306654 | 1,920 | 639 | − | 70.24 | −0.287 | 5.25 | 46.96 | 76.51 |
| MdGRAS126 | MDP0000640034 | unanchored:50237359.50239266 | 1,908 | 635 | + | 69.73 | −0.274 | 5.26 | 47.40 | 77.92 |
| MdGRAS127 | MDP0000291579 | unanchored:106037882.106039530 | 1,593 | 530 | + | 60.97 | −0.425 | 9.28 | 46.99 | 83.91 |
Gene ID in the apple (Malus × domestica) genome (Malus domestica Genome database v1.0);
MW: Molecular weight;
GRAVY: Grand average of hydropathicity;
PI: Isoelectric point;
II: Instability index;
AI: Aliphatic index.
Statistical analysis
Data were analyzed using variance (ANOVA) at the 5% level with the SPSS 11.5 software package (SPSS, Chicago, IL, USA). Figures were made using Origin 8.0 (Microcal Software Inc., Northampton, MA, USA).
Results
Identification and characterization analysis of MdGRAS genes
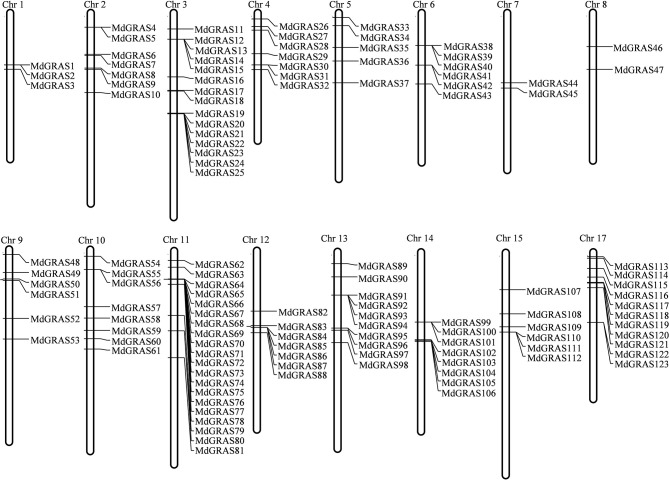
To identify members of the MdGRAS gene family, both HMM profiles and BLASTP were used with the default parameters. After manual checking, a total of 127 candidate MdGRAS genes were identified (Table 1). The genes were named according to their chromosomal locations. The 127 candidate MdGRAS genes were distributed on 16 chromosomes of the apple genome (Figure 1). Chromosome 11 contained the most MdGRAS genes (15.75%), followed by chromosome 3 (11.81%), while chromosome 16 contained no MdGRAS genes (0%) (Figure S1). Additionally, four genes (MDP0000950387, MDP0000319342, MDP0000640034, and MDP0000640034) could not be located on any of the chromosomes and were named MdGRAS124 to MdGRAS127. Only two genes were distributed on each of chromosomes 7 and 8. The open reading frame (ORF) lengths of the MdGRAS genes ranged from 333 to 5,787 bp, encoding peptides ranging from 110 to 1,928 amino acids. We used ExPaSy to predict the characterization of MdGRAS proteins. The predicted molecular weights of the GRAS proteins ranged from 12.50 to 218.53 kDa. The grand average of hydropathicity was less than 0 for all GRAS proteins. The instability index of most of the 127 MdGRAS proteins was greater than 40, except for MdGRAS22, MdGRAS33, MdGRAS41, MdGRAS42, MdGRAS43, MdGRAS53, and MdGRAS65, indicating that these proteins were unstable.
Figure 1.
Chromosome map of MdGRAS genes in apple.
Phylogenetic and gene structure of the GRAS gene family
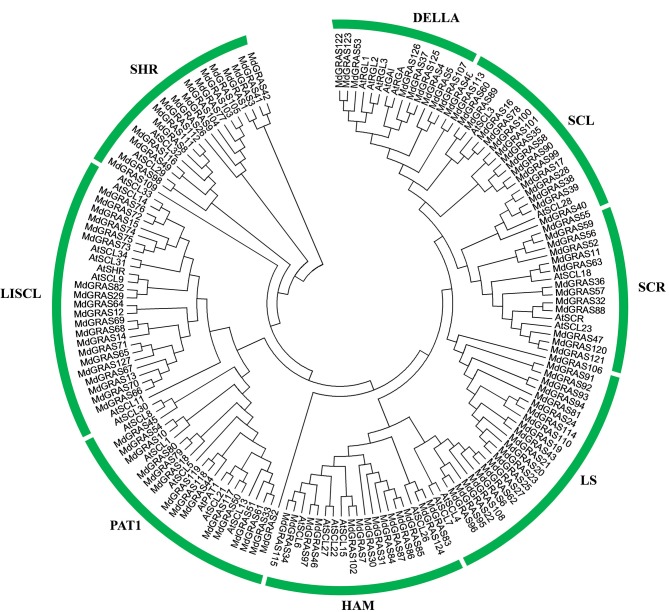
Since Arabidopsis is the most popular model species and the functions of some GRAS genes have been well characterized. Phylogenetic analysis of GRAS genes was investigated in apple and Arabidopsis. A total of 160 GRAS sequences were collected and analyzed. They were classified into eight distinct groups according to the phylogenetic tree: DELLA, SCL3, SCR, Ls, LISCL, PAT1, SHR, and HAM (Figure 2). 14 GRAS proteins were located in the DELLA subfamily, including five AtGRAS (AtGAI, AtRGA, AtRGL1, AtRGL2, and AtRGL3) and nine MdGRAS proteins. However, MdGRAS37 did not contain the integral DELLA and TVHYNP domains. MdGRAS37 was classified into the DELLA subfamily because of its high homology with the other proteins. To examine the gene structures, we analyzed the exon-intron structures of the 127 MdGRAS genes according to their genome sequences (Figure S2). Thirteen genes (MdGRAS100, MdGRAS101, MdGRAS25, MdGRAS7, MdGRAS102, MdGRAS2, MdGRAS33, MdGRAS51, MdGRAS80, MdGRAS45, MdGRAS29, MdGRAS1, and MdGRAS3) had two or more introns. Other genes had few or no introns.
Figure 2.
Phylogenetic tree showing the evolutionary relationships of GRAS proteins in apple and Arabidopsis. The phylogenetic tree was conducted using MEGA 5.10. The prefixes At and Md indicate Arabidopsis and apple, respectively.
Expansion patterns of the MdGRAS gene family
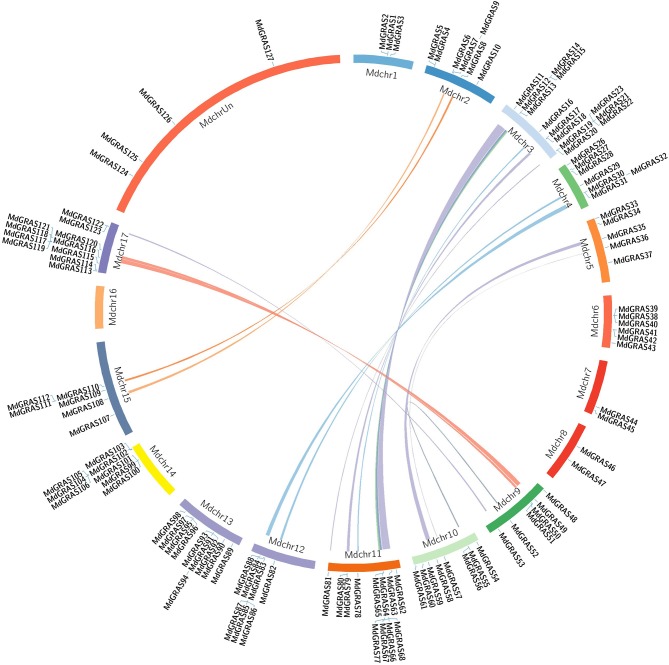
Segmental and tandem duplications provide information on the expansion of the gene family. Here, we used the Circos program to evaluate the segmental and tandem duplications of the MdGRAS genes. Tandem duplications were identified from adjacent homologs on a single chromosome, while segmental duplications were identified from homologs on different chromosomes. More than 15 pairs of MdGRAS genes, such as MdGRAS122/53, MdGRAS116/50, and MdGRAS115/49, were located in duplicated genomic regions (Figure 3). All were located in different chromosomes. Chromosomes 11 and 3 had the most duplication, which could partly explain the larger numbers of MdGRAS genes on chromosomes 11 and 3 (Figure S1). Chromosomes 1, 6, 7, 8, 13, and 14 did not contain any duplicated genes. In summary, segmental and tandem duplications among MdGRAS genes have contributed to the expansion of the gene family.
Figure 3.
Syntenic relationships of apple GRAS genes. Locations of MdGRAS genes on the apple chromosomes.
Comparative analysis of the GRAS gene families in different species
To further analyze the evolutionary relationships and expansion patterns of plant GRAS genes, we compared the published GRAS genes in different species (Table S2). The number of GRAS genes was the largest in apple and was close to that in Populus. However, the apple genome is twice the size of the Populus genome. The number of GRAS genes was also similar between Chinese cabbage (48) and P. mume (46), and between grape (52) and tomato (53). However, their genome sizes are also very different (Chinese cabbage: 485.0 Mb vs. P. mume: 280.0 Mb; grape: 487.0 Mb vs. tomato: 760.0 Mb). These results suggested that the number of plant GRAS genes was partly related to genome size.
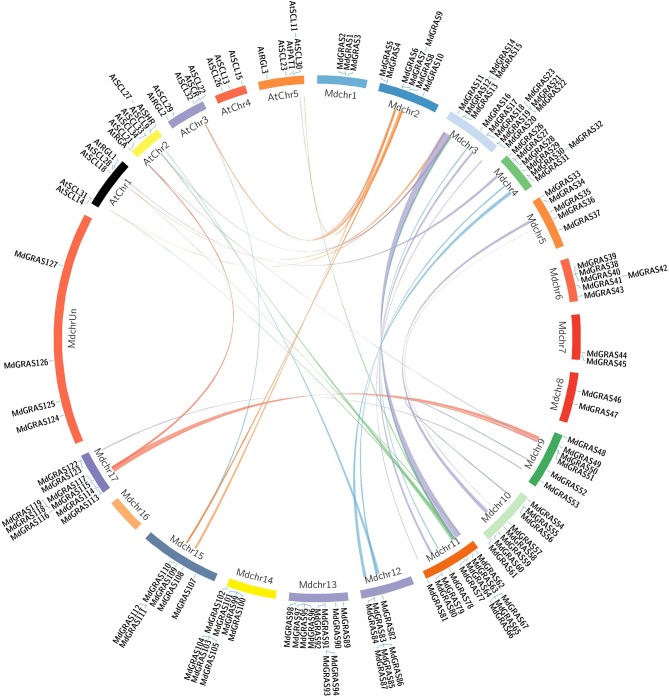
As Arabidopsis is a model plant species, the functions of the GRAS genes in Arabidopsis have been well researched. We generated a comparative GRAS synteny map between apple and Arabidopsis. Many pairs of syntenic orthologous genes were matched, such as AtSCL32-MdGRAS108, AtRGA-MdGRAS114, and AtPAT1-MdGRAS63 (Figure 4). These genes were detected on 14 chromosomes with duplicated regions, including 4 chromosomes in Arabidopsis and 10 chromosomes in apple.
Figure 4.
Syntenic relationships between apple and Arabidopsis GRAS genes. Colored curves represent apple and Arabidopsis syntenic gene regions.
Tissue-specific analysis of MdGRAS expression using ArrayExpress data
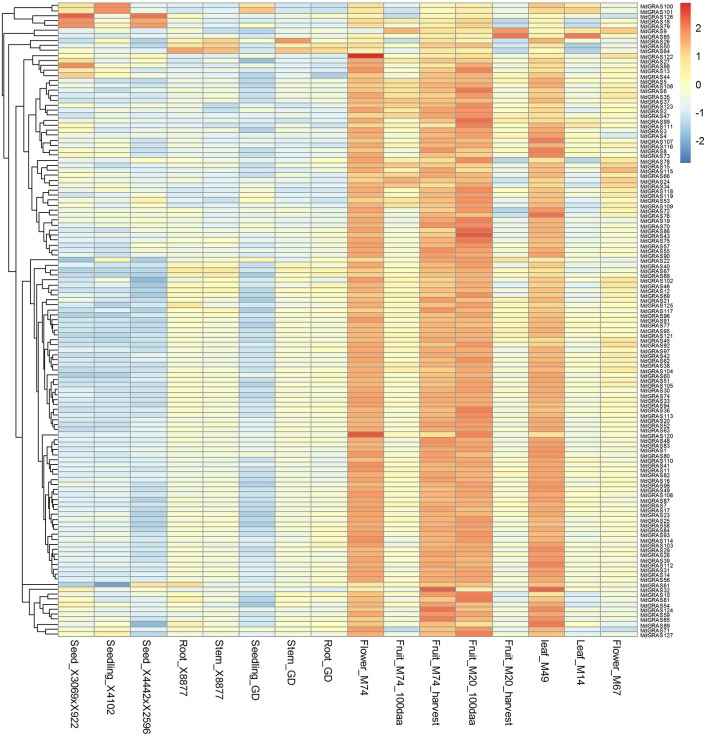
To elucidate the potential roles and functions of the MdGRAS genes in apple, we downloaded expression profile data for different tissues from the ArrayExpress database (E-GEOD-42873). Eight organs and samples with two biological replicates were represented in this expression array, including seedlings, roots, stems, flowers, fruit, and leaves (Figure 5). The GRAS genes showed different expression patterns among different apple varieties and tissues. In the “M74” apple genotype, most of the MdGRAS genes were expressed at higher levels in the flower than in the fruit. Moreover, the expression levels of the MdGRAS genes were higher in ripe fruit (at harvest) than in unripe fruit, indicating their important roles in fruit ripening. Most MdGRAS genes were expressed at lower levels in seedlings, except for MdGRAS100, MdGRAS101, MdGRAS126, MdGRAS18, MdGRAS79, and MdGRAS27. In “M14” leaves, MdGRAS85 showed the highest expression. However, other GRAS genes were hardly detected in leaves. Moreover, most GRAS showed lower levels in stems except MdGRAS26, MdGRAS50, and MdGRAS64.
Figure 5.
Heat map showing MdGRAS gene transcript levels in different tissues. Relative transcript levels are based on ArrayExpress data.
QRT-PCR analysis of the candidate flowering related MdGRAS genes in different tissues in “Nagafu No. 2”
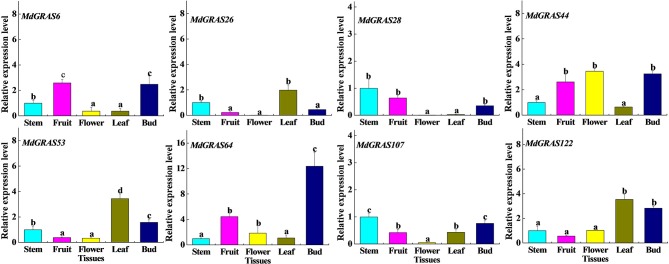
We selected eight higher expression genes (MdGRAS6, MdGRAS26, MdGRAS28, MdGRAS44, MdGRAS53, MdGRAS64, MdGRAS107, and MdGRAS122) from our transcriptome data for further analysis (Figure S4, Table S3). These eight genes were from different GRAS subfamilies, including the LS (MdGRAS6), SHR (MdGRAS26), SCL (MdGRAS28), PAT1 (MdGRAS44), DELLA (MdGRAS53, MdGRAS107, and MdGRAS122) and LISCL (MdGRAS64) subfamily. We employed qRT-PCR to investigate their expression patterns in different tissues (stems, leaves, flowers, fruits and buds). All candidate MdGRAS genes showed constitutive expression patterns in the five tissues, except that the levels of MdGRAS26 and MdGRAS28 were low in flowers (Figure 6). The transcription levels of MdGRAS6, MdGRAS26, MdGRAS44, MdGRAS53, MdGRAS64, MdGRAS107, and MdGRAS122 were high in buds and leaves, which indicated their important roles in mediating flower induction. Additionally, MdGRAS107 and MdGRAS28 were all showed lower levels than other candidate genes among the five tissues.
Figure 6.
Expression of candidate MdGRAS genes in different tissues. Each value represents the mean Âś standard error of three replicates. Means followed by small letters (a, b, and c) are significantly different at a level of 0.05.
QRT-PCR analysis of the candidate MdGRAS genes in different apple varieties
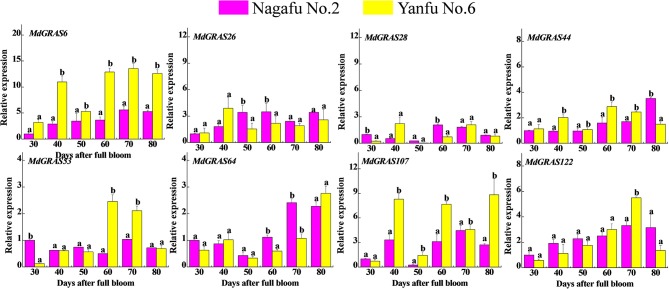
To examine the expression of the MdGRAS genes in two varieties with contrasting flowering rates, we selected the easy-flowering variety “Yanfu No. 6” and the difficult-flowering “Nagafu No. 2.” As shown in Table S4, “Yanfu No. 6” has a 1.5-fold higher flowering rate than “Nagafu No. 2.” We examined the expression patterns in these varieties using qRT-PCR (Figure 7). All candidate genes showed different expression patterns during flower induction. MdGRAS6 expression was higher in the buds of “Yanfu No. 6” during all stages. MdGRAS44 expression was higher in “Yanfu No. 6” except at 80 DAFB, while MdGRAS122 expression was higher in “Nagafu No. 2” except at 70 DAFB. MdGRAS26 expression was lower at the first stage in “Nagafu No. 2,” but was higher in all subsequent stages. The transcription level of MdGRAS107 was higher in “Yanfu No. 6” except at 30 DAFB.
Figure 7.
Expression of candidate MdGRAS genes in apple buds in the easy-flowering variety “Yanfu No. 6” and different-flowering “Nagafu No. 2.” Samples were collected at 30, 40, 50, 60, 70, and 80 days after full bloom (DAFB). Each value represents the mean ± standard error of three replicates. Means followed by small letters (a, b) are significantly different at a level of 0.05.
QRT-PCR analysis of the candidate MdGRAS genes in response to treatments affecting flowering
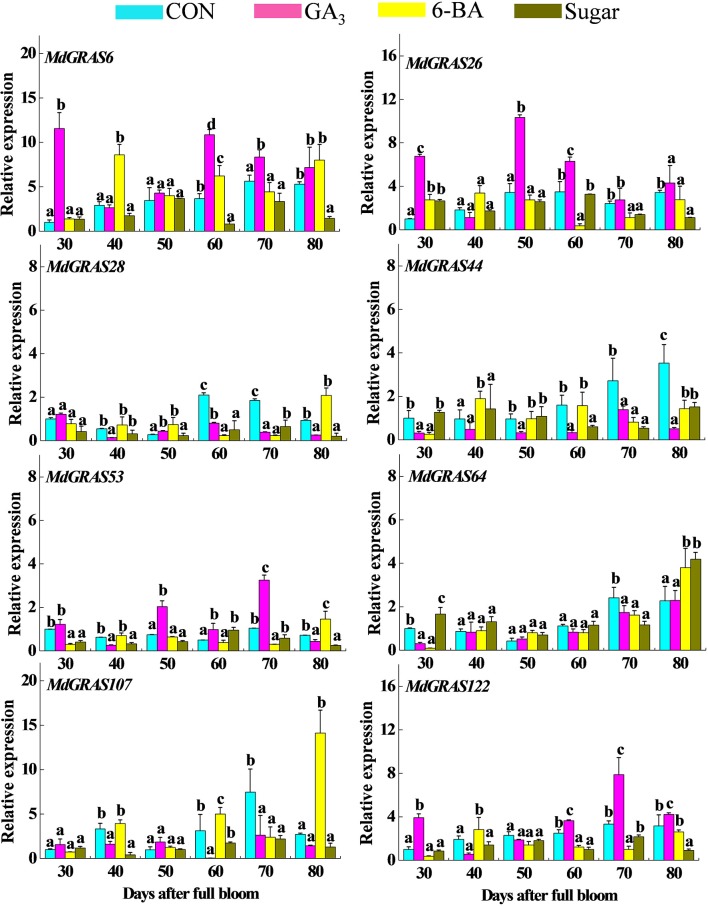
Previous studies have shown the exogenous treatments (GA, 6-BA, and sucrose) can alter flowering rates, among which GA always decreases the flowering rate, while 6-BA and sugar increase flowering rates (Xing et al., 2015; Li et al., 2016; Zhang et al., 2016). We used these kinds of treatments to further investigate the expression of the candidate MdGRAS genes during flowering. As shown in Table S4, exogenous GA3 treatment inhibited flowering rate while sugar and 6-BA promoted flowering. The genes displayed various expression patterns at different time points (Figure 8). Some of them were down-regulated, while others were up-regulated. MdGRAS6 was up-regulated after GA3 treatment except at 40 DAFB and after 6-BA treatment except at 70 DAFB, but was down-regulated after sugar treatment except at 30 DAFB. MdGRAS26 showed similar changes after GA3 treatment, and was up-regulated except at 40 DAFB. However, the level of MdGRAS26 was down-regulated after 6-BA and sugar treatment except at 30 DAFB. MdGRAS28 and MdGRAS44 were down-regulated after GA3 treatment except that MdGRAS28 was up-regulated at 30 and 50 DAFB. MdGRAS28 was down-regulated after sugar treatment at all time points. The expression of MdGRAS53 was promoted by exogenous GA3 except at 40 and 80 DAFB, but was inhibited by exogenous 6-BA and sugar at most time points. MdGRAS64 expression was up-regulated after sugar treatment, but was not consistent in response to GA and 6-BA. MdGRAS107 was down-regulated after GA3 treatment except at 30 and 50 DAFB, but was up-regulated by 6-BA except at 30 and 70 DAFB. MdGRAS107 was down-regulated after sugar treatment except at 30 DAFB. The transcription level of MdGRAS122 was lower in 6-BA treated buds than control except at 40 DAFB, and it was down-regulated after sugar treatment at all time points. In order to further confirm the results, we also investigated their expression patterns in GA-treated buds in 2014 and 6-BA treated buds in 2013 based on our previous research (Li et al., 2016; Zhang et al., 2016). Samples were collected in the early stage (ES), middle stage (MS), and later stage (LS) of flower induction. As shown Figures S5, S6, their expression patterns in 2014 and 2015 were similar with our results.
Figure 8.
Expression of candidate MdGRAS genes in apple buds treated with GA3, 6-BA, and sugar. Samples were collected at 30, 40, 50, 60, 70, and 80 days after full bloom (DAFB). Each value represents the mean ± standard error of three replicates. Means followed by small letters (a, b, and c) are significantly different at a level of 0.05.
Analysis related cis-elements in the candidate MdGRAS genes
To further analyze the potential roles of MdGRAS genes in response to various circumstances, a 1.5 kb promoter of candidate MdGRAS genes were used. The related cis-elements were identified as previous study (Fan et al., 2017). The candidate MdGRAS genes all shared large light-responsive boxes, followed by stress (Figure S7). Additionally, hormones-related cis-elements were also found in all the candidate MdGRAS genes, including MeJA, salicylic acid, gibberellin, auxin and ethylene. These identified motifs in the promoter of each gene indicated that MdGRAS may be regulated by various cis-elements within the promoter during growth.
Discussion
Genome-wide identification of GRAS genes has been reported in Arabidopsis, P. mume, Populus, rice, grape, castor bean, and Chinese cabbage (Tian et al., 2004; Song et al., 2014; Huang et al., 2015; Lu et al., 2015; Grimplet et al., 2016; Xu W. et al., 2016), but little in known in apple. In this study, we identified MdGRAS genes in the apple genome (Velasco et al., 2010), and analyzed their phylogenetic relationships, gene characteristics, gene structures, expansion history, and syntenic relationships. We also analyzed their tissue-specific expression patterns and their expression profiles in response to flower induction. Our comprehensive study provides a basic for further investigation of GRAS genes in apple, which can also have potential values in genetic improvement of flower induction in apple as well as some other relative species.
Phylogenesis of apple GRAS genes
We identified 127 putative MdGRAS genes in apple, a larger number than that reported for other plant species such as P. mume, Populus, rice, grape, and Chinese cabbage (Tian et al., 2004; Song et al., 2014; Huang et al., 2015; Lu et al., 2015; Grimplet et al., 2016; Xu W. et al., 2016). The larger number of GRAS genes in apple might be because its genome (881.3 Mb) is larger than those of P. mume (280.0 Mb), rice (372.0 Mb), grape (487.0 Mb), and Brassica rapa (283.3 Mb). The MdGRAS genes were located on almost all of the chromosomes except chromosome 16. The structures of most MdGRAS genes were similar, and most had few introns, similar to those in P. mume and Populus (Tian et al., 2004; Song et al., 2014). These findings indicated that the structures of GRAS genes are highly conserved among different plant species. A previous study identified six genes in the DELLA subfamily in apple, based on data in the EST database (Foster et al., 2007). In our study, nine putative genes belonging to the DELLA subfamily were identified from the apple genome (Xing et al., 2016) according to the phylogenetic tree (Figure 2). However, MdGRAS37, which was classified into the DELLA subfamily, lacked the conversed DELLA domain and other features of the DELLA subfamily (Figure S3), including gene length and structural domains. MdGRAS37 was probably placed in the DELLA clade because of its high sequence similarity with Arabidopsis sequences of the DELLA subfamily (Lu et al., 2015). The DELLA domain of MdGRAS37 may have gradually degenerated during the evolutionary process or its sequence may be partly similar with DELLA proteins. However, this needs to be further researched.
Expansion and synteny of GRAS genes
Gene duplications can be tandem, segmental, or arise from whole genome duplication. Duplicated genes are known to play important roles in plant growth and development.
To date, various kinds of genes encoding transcription factors have been identified, and most of them have shown gene duplications, including AP2, MADS, SPL, and DOF (Zahn et al., 2005; Moreno-Risueno et al., 2007; Shigyo et al., 2007; Kumar et al., 2016). Gene duplication is important for gene family expansion (Day et al., 2003; Cannon et al., 2004; Leister, 2004). The expansion and duplication of GRAS genes have also been reported in Arabidopsis, P. mume, and Chinese cabbage (Liu and Widmer, 2014; Song et al., 2014; Lu et al., 2015). It was reported that 85% of AtGRAS genes have undergone segmental duplications, including two tandemly duplicated genes (AtSCL33 and AtSCL34). Here, we used the Circos program to identify tandemly duplicated genes in the apple genome. Synteny analysis of the duplicated blocks in the apple genome showed that some MdGRAS genes were putative segmental duplicates (Figure 2). The present result was consistent with previous study in Arabidopsis, which GRAS also occurred gene duplication in apple genome. Plant GRAS genes were involved in various biological processes, and the expansion of the MdGRAS might contribute to their diversity functions in regulation plant growth and developemt. Additionally, it was reported that a genome-wide duplication (more than 50 million years ago) contributed to the expansion of the apple chromosome number, which led to a change to 17 chromosomes from nine chromosomes (Velasco et al., 2010). So the whole genome duplications promoted the expansion of GRAS genes in apple. This expanded by segmental duplication or whole genome duplications of GRAS genes in apple genome was also proved in grape and other species (Grimplet et al., 2016). Totally, the duplication and expansion of GRAS genes have played important roles in gene evolution, and contributed to their varied structures and functions.
To date, little functional research has been performed on rosaceous GRAS genes. Comparison with homologous GRAS genes of the model plant Arabidopsis can help us further analyze the MdGRAS genes. Comparisons of genome sequences among different species can be valuable for the reconstruction of individual gene families (Koonin, 2005). Such genome comparisons can be used to transfer annotations from a better-studied taxon (whose genome structure and functions have been elucidated) to a less-studied taxon (Lyons et al., 2008). Based on this, we inferred the functions of the less-studied MdGRAS genes from the better-understood AtGRAS genes. We used the Circos program to compare apple and Arabidopsis. Most GRAS genes were detected in syntenic genomic regions (Figure 4), allowing us to predict their functions. Currently, most AtGRAS genes are well understood, including AtSCL3 (Zhang et al., 2011), AtSHR (Levesque et al., 2006), AtSCL13 (Torres-Galea et al., 2006), and AtSLR (Aleman et al., 2008). Thus, we can analyze the potential functions of MdGRAS genes based on their homologs in Arabidopsis. However, functions predicted in this way needs to be experimentally verified.
Expression profiles of MdGRAS genes in different tissues
To reveal their potential functions in apple growth and development, we employed ArrayExpress data to analyze the expression of the MdGRAS genes in different tissues and organs. These potential investigations of each GRAS can be useful to access its possible function. Their various expression patterns were consistent with their diverse gene locations, lengths and sequences. As shown in Figure 5, the expression patterns of the GRAS genes differed among different tissues and varieties, which is consistent with other species (Tian et al., 2004; Song et al., 2014; Huang et al., 2015; Lu et al., 2015; Grimplet et al., 2016; Xu W. et al., 2016). For example, tomato SlGRAS18 and SlGRAS38 transcript levels were high at the breaker and red-ripening stages, and their expression was regulated by RIN (Fujisawa et al., 2012, 2013). In another study, transcripts of 14 SlGRAS genes were detected during fruit ripening (Huang et al., 2015). Our analyses of GRAS expression profiles confirmed that most of the MdGRAS genes had higher transcript levels in fruit than in other apple tissues, indicating their important roles in fruit ripening. MdGRAS9 and MdGRAS85 expression levels were extremely high in “M20” during the harvest stage. These results suggested that MdGRAS9 and MdGRAS85 play important roles during fruit development; this was consistent with previous study. In Arabidopsis, three AtGRAS genes (SCL6, SCL22, and SCL27) were shown to be involved in leaf development. In the apple genotype “M14,” MdGRAS85 transcripts were detected at high level in leaves, suggesting that this gene may share similar functions with AtSCL6, AtSCL22, and AtSCL27 in leaf development, which was consistent with their close evolutionary relationships to MdGRAS85 (Figure 2). However, MdGRAS85 did not show high level in leaves of “M49”; the different expressions of MdGRAS85 may be explained by their different genotypes of “M14” and “M49.” The high expression patterns of MdGRAS126, MdGRAS18, and MdGRAS79 in seeds were also consistent with PmGRAS20 and PmGRAS36 expression, and suggested that these genes may have important roles in seed development (Lu et al., 2015). These different expression patterns of GRAS indicated their various functions in apple development; further functional analysis needs to be confirmed.
We also analyzed the expression of the candidate MdGRAS genes in different tissues (stem, leaf, flower, fruit, and bud) in “Nagafu No. 2” with qRT-PCR. The MdGRAS genes all showed different expression patterns among the five tissues. Leaves and buds have been used to research flower induction in Arabidopsis and other plants (Galvao et al., 2012; Porri et al., 2012; Xing et al., 2015; Fan et al., 2016). The candidate genes MdGRAS6, MdGRAS26, MdGRAS44, MdGRAS53, MdGRAS64, and MdGRAS122 mostly showed high expression in buds and leaves, which proved their important roles in flower induction. A previous study showed that DELLA has important roles in regulating internode elongation (Bolle, 2004). We found that the expression of MdGRAS107, a DELLA subfamily member, was also higher expressed in the apple stem, which indicates it might also have functions in regulating internode elongation.
Characterization of MdGRAS expression during flower induction
To our best knowledge, the relationship between GRAS and flower induction was received less attention, apart from MdGRL2a. Flower induction is associated with several environmental and internal factors, including temperature, hormones and photosynthesis (Galvao et al., 2012; Porri et al., 2012; Xing et al., 2015, 2016). Hormones and sugar are of the most important factors affected apple flowering, the relationship about GRAS involved in hormones or sugar remains unknown. Up to now, GA was considered to be the most associated with flower induction; 6-BA and sugar were also involved in flower induction. And they were all well applied in orchard cultivation. Additionally, our previous studies showed that exogenous treatments can alter flowering rates; GA3 can inhibit flowering, while 6-BA and sugar can increase flowering rates (Xing et al., 2015; Li et al., 2016; Zhang et al., 2016). These exogenous treatments mainly alter the nutrition or hormone balance and influence the expression of related genes, which inhibits (GA) or promotes (sugar and 6-BA) flowering (Xing et al., 2015; Li et al., 2016; Zhang et al., 2016). Based on this, we investigated the candidate MdGRAS genes in response to these flowering-related treatments (Figure 8). With their multiple functions, the candidate MdGRAS genes showed different expression patterns after the treatments, and were down- or up-regulated at most time points. “Yanfu No. 6,” a bud mutant of “Nagafu No. 2,” has a higher proportion of spurs, shorter internodes and a higher flowering rate than “Nagafu No. 2” (Table S4). The differential expression of the candidate MdGRAS genes in these two varieties with contrasting flowering rates indicated their important roles in flower induction (Figure 7).
In recent years, although the roles of GRAS genes in regulating plant growth and development have become better understood, little is known about the roles of GRAS genes in regulating flower induction in fruit trees. Several studies have shown that GRAS genes are important in bud growth and development. Most DELLA subfamily gene regulate flower induction by the GA pathway, and it can influence the expression of some flowering control genes (Bolle, 2004; Galvao et al., 2012; Porri et al., 2012; Xing et al., 2014, 2015; Huang et al., 2015). However, genes from other subfamilies were also associated with flower induction. AtGRAS genes promoted the expression of FT and the della mutant always showed earlier flowering (Galvao et al., 2012). In Arabidopsis, DELLA proteins were shown to inhibit flower induction, and interact with SPL to control flower induction (Shigyo et al., 2007). Previous studies showed that the expression of one-fifth of the candidate genes was increased during flower development in P. mume, and DELLA gene expression was also higher in tomato buds, indicating their important roles in flower induction (Huang et al., 2015; Lu et al., 2015). In our study, we selected eight important MdGRAS genes from transcriptome data. They belong to different subfamilies, including LS (MdGRAS6), SHR (MdGRAS26), SCL (MdGRAS28), PAT1 (MdGRAS44), DELLA (MdGRAS53, MdGRAS107, and MdGRAS122) and LISCL (MdGRAS64). Our result suggested that these candidate MdGRAS genes from different subfamilies were important and they all have potential involvement in flower induction. Additionally, functions of their homologous genes have been described and verified in other species, which provided more evidence for their important response to flower induction (Fu et al., 2001; Hynes et al., 2003; Petty et al., 2003; Wang et al., 2010; Galvao et al., 2012). They can regulate flower induction directly and indirectly. In Arabidopsis, AtLAS and AtSHR were involved in shoot apical meristem. AtSCL6, AtSCL26, and AtSCL27 were highly expressed in flowers (Bolle, 2004); the Arabidopsis rga and gai mutant showed earlier flowering (Galvao et al., 2012). Additionally, GRAS genes in other species were all indicated their potential involvement in flowering induction, such as tomato (SlGRAS11 and SlGRAS18), castor beans (27568.m000253, 28650.m000187, 28650.m000190, and 28650.m000191), and grape (VviSHR1 and VviPAT6) (Huang et al., 2015; Grimplet et al., 2016; Xu W. et al., 2016). Our candidate MdGRAS genes were homologous with these well-known GRAS genes in other plants, which is in agreement with their important functions in flower induction. However, further functional research needs to be confirmed about these MdGRAS genes. Their different expression patterns during flower induction indicated they were involved in GA-mediated, 6-BA mediated or sugar-mediated flowering in apple trees. Previous transgenic Arabidopsis of MdRGL2a (one of the DELLA members in apple) showed delayed flowering phenotype, which proved their functions in regulating flowering. Additionally, the predication of cis-elements within their promoters indicated their potential roles in response to hormone mediated signal during apple growth and development (Figure S7). However, with the large number of GRAS gene in Arabidopsis and apple genome, we cannot capture more genes aiming at flower induction except the candidate eight MdGRAS. The other left MdGRAS genes need to be further confirmed. And more GRAS genes need to be functionally researched.
All in all, extensive cross-talk about GRAS genes has been identified in herbaceous and woody plants; extensive research effort has also been devoted to characterization GRAS genes in plant growth and development. However, the GRAS genes of woody economically important tree species have received less attention. In this study, we identified 127 putative GRAS genes from the apple genome, which were located on 16 chromosomes. We classified these genes and performed systematic phylogenetic, structural and synteny analyses. We also analyzed their expression in different tissues (stems, leaves, flowers, fruits and buds). The spatiotemporal expression patterns indicated they potentially have multiple functions in regulating plant growth and development. Additionally, some candidate MdGRAS genes were investigated under treatments with several flowering-related compounds, which indicated the candidate MdGRAS genes are involved in the complex flower induction process. These results may provide useful strategies for further improvement of flower induction in apple.
Author contributions
MH, SF, and DZ conceived and designed the experiment. SF, CG, and MZ performed the experiment. SF, HW, YL, and YS analyzed the data. MH and SF wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by China Apple Research System (CARS-28), National Natural Science Foundation of China (31672101), Science and Technology Innovative Engineering Project in Shaanxi province, China (2016KTZDNY01-10), China Postdoctoral Science Foundation (2014M56806), Science Foundation from the Northwest A&F University (2452015291), Yangling Subsidiary Center Project of the National Apple Improvement Center and Collaborative Innovation of the Center for Shaanxi Fruit Industry Development.
Glossary
Abbreviations
- DAFB
days after full blossom
- ES
early stage
- MS
middle stage
- LS
later stage
- GA3
gibberellic acid
- 6-BA
6-benzylaminopurine.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2017.00253/full#supplementary-material
Distribution of MdGRAS genes on the apple chromosomes.
MdGRAS gene structures. MdGRAS gene exon–intron composition. Red boxes and black lines represent exons and introns, respectively.
Alignment of DELLA subfamily members in apple and Arabidopsis.
Heat map showing the transcript levels of MdGRAS genes during flower induction. Transcript levels of GRAS genes in the buds of “Nagafu No. 2” and “Yanfu No. 6” were investigated during flower induction. Samples were collected three times at the early stage (ES), middle stage (MS) and later stages (LS) of flower induction.
Expression of candidate MdGRAS genes in GA-treated buds. Samples were collected three times at ES, MS and LS in 2014, which was consistent with 40, 50, and 70 DAFB in 2016. Each value represents the mean ± standard error of three replicates.
Expression of candidate MdGRAS genes in 6BA-treated buds. Samples were collected three times at ES, MS and LS in 2013, which was consistent with 30, 50, and 80 DAFB in 2016. Each value represents the mean ± standard error of three replicates.
Predicted cis-elements in the promoter regions of the candidate MdGRAS genes. The upstream 1.5 kb promoter sequences were analyzed.
Primer sequences of MdGRAS and reference genes in quantitative reverse transcription PCR.
Comparison of MdGRAS genes in different species.
Expression levels of MdGRAS genes in Nagafu No. 2 and Yanfu No. 6. Data were collected from the transcriptome.
Investigation of flowering rate in 2016.
References
- Ait-ali T., Rands C., Harberd N. P. (2003). Flexible control of plant architecture and yield via switchable expression of Arabidopsis gai. Plant Biotechnol. 1, 337–343. 10.1046/j.1467-7652.2003.00030.x [DOI] [PubMed] [Google Scholar]
- Aleman L., Kitamura J., Abdel-Mageed H., Lee J., Sun Y., Nakajima M., et al. (2008). Functional analysis of cotton orthologs of GA signal transduction factors GID1 and SLR1. Plant Mol. Biol. 68, 1–16. 10.1007/s11103-008-9347-z [DOI] [PubMed] [Google Scholar]
- Bolle C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218, 683–692. 10.1007/s00425-004-1203-z [DOI] [PubMed] [Google Scholar]
- Bolle C., Koncz C., Chua N. H. (2000). PAT1, a new member of the GRAS family, is involved in Phytochrome A signal transduction. Genes Dev. 14, 1269–1278. 10.1101/gad.14.10.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon S. B., Mitra A., Baumgarten A., Young N. D., May G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H. R., Zhang Z. R., Lv W., Xu J. N., Wang X. Y. (2015). Genome-wide characterization and analysis of F-box protein-encoding genes in the Malus domestica genome. Mol. Genet. Genomics 290, 1435–1446. 10.1007/s00438-015-1004-z [DOI] [PubMed] [Google Scholar]
- Czikkel B. E., Maxwell D. P. (2007). NtGRAS1, a novel stress-induced member of the GRAS family in tobacco, localizes to the nucleus. J. Plant Physiol. 164, 1220–1230. 10.1016/j.jplph.2006.07.010 [DOI] [PubMed] [Google Scholar]
- Dai C., Xue H. W. (2010). Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. Embo J. 29, 1916–1927. 10.1038/emboj.2010.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R. B., Shibuya N., Minami E. (2003). Identification and characterization of two new members of the GRAS gene family in rice responsive to N-acetylchitooligosaccharide elicitor. Biochim. Biophys. Acta 1625, 261–268. 10.1016/S0167-4781(02)00626-7 [DOI] [PubMed] [Google Scholar]
- Fan S., Zhang D., Lei C., Chen H. F., Xing L. B., Ma J. J., et al. (2016). Proteome Analyses Using iTRAQ Labeling Reveal Critical Mechanisms in Alternate Bearing Malus prunifolia. J. Proteome Res. 15, 3602–3616. 10.1021/acs.jproteome.6b00357 [DOI] [PubMed] [Google Scholar]
- Fan S., Zhang D., Xing L. B., Qi S. Y., Du L. S., Wu H. Q., et al. (2017). Phylogenetic analysis of IDD gene family and characterization of its expression in response to flower induction in Malus. Mol. Genet. Genomics. [Epub ahead of print]. 10.1007/s00438-017-1306-4 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Clements J., Eddy S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T., Kirk C., Jones W. T., Allan A. C., Espley R., Karunairetnam S., et al. (2007). Characterisation of the DELLA subfamily in apple (Malus x domestica Borkh.). Tree Genet. Genomes 3, 187–197. 10.1007/s11295-006-0047-z [DOI] [Google Scholar]
- Fu X. D., Sudhakar D., Peng J. R., Richards D. E., Christou P., Harberd N. P. (2001). Expression of arabidopsis GAI in transgenic rice represses multiple gibberellin responses. Plant Cell 13, 1791–1802. 10.1105/tpc.13.8.1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M., Nakano T., Shima Y., Ito Y. (2013). A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25, 371–386. 10.1105/tpc.112.108118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M., Shima Y., Higuchi N., Nakano T., Koyama Y., Kasumi T., et al. (2012). Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 235, 1107–1122. 10.1007/s00425-011-1561-2 [DOI] [PubMed] [Google Scholar]
- Galvao V. C., Horrer D., Kuttner F., Schmid M. (2012). Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139, 4072–4082. 10.1242/dev.080879 [DOI] [PubMed] [Google Scholar]
- Gambino G., Perrone I., Gribaudo I. (2008). A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 19, 520–525. 10.1002/pca.1078 [DOI] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schafer E., Muller D., Herrero R., Schmitz G., et al. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17, 1175–1187. 10.1101/gad.260703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J., Agudelo-Romero P., Teixeira R. T., Martinez-Zapater J. M., Fortes A. M. (2016). Structural and functional analysis of the GRAS gene family in grapevine indicates a role of GRAS proteins in the control of development and stress responses. Front. Plant Sci. 7:353. 10.3389/fpls.2016.00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitton B., Kelner J. J., Velasco R., Gardiner S. E., Chagne D., Costes E. (2012). Genetic control of biennial bearing in apple. J. Exp. Bot. 63, 131–149. 10.1093/jxb/err261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann A. B., Lombardo F., Miwa H., Perry J. A., Bunnewell S., Parniske M., et al. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142, 1739–1750. 10.1104/pp.106.089508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A. B., Downie J. A., Oldroyd G. E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21, 545–557. 10.1105/tpc.108.064501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Xian Z., Kang X., Tang N., Li Z. (2015). Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato. BMC Plant Biol. 15:209. 10.1186/s12870-015-0590-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes L. W., Peng J. R., Richards D. E., Harberd N. P. (2003). Transgenic expression of the Arabidopsis DELLA proteins GAI and gai confers altered gibberellin response in tobacco. Transgenic Res. 12, 707–714. 10.1023/B:Trag.0000005145.68017.6e [DOI] [PubMed] [Google Scholar]
- Jia F., Wu B., Li H., Huang J., Zheng C. (2013). Genome-wide identification and characterisation of F-box family in maize. Mol. Genet. Genomics 288, 559–577. 10.1007/s00438-013-0769-1 [DOI] [PubMed] [Google Scholar]
- Koizumi K., Hayashi T., Wu S., Gallagher K. L. (2012). The SHORT-ROOT protein acts as a mobile, dose-dependent signal in patterning the ground tissue. Proc. Natl. Acad. Sci. U.S.A. 109, 13010–13015. 10.1073/pnas.1205579109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. (2005). Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39, 309–338. 10.1146/annurev.genet.39.073003.114725 [DOI] [PubMed] [Google Scholar]
- Kumar G., Arya P., Gupta K., Randhawa V., Acharya V., Singh A. K. (2016). Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica). Sci. Rep. 6:20695. 10.1038/srep20695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. (2004). Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet. 20, 116–122. 10.1016/j.tig.2004.01.007 [DOI] [PubMed] [Google Scholar]
- Levesque M. P., Vernoux T., Busch W., Cui H. C., Wang J. Y., Blilou I., et al. (2006). Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 4, 739–752. 10.1371/journal.pbio.0040143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hou H. M., Li X. Q., Xiang J., Yin X. J., Gao H., et al. (2013). Genome-wide identification and analysis of the SBP-box family genes in apple (Malus x domestica Borkh.). Plant Physiol. Biochem. 70, 100–114. 10.1016/j.plaphy.2013.05.021 [DOI] [PubMed] [Google Scholar]
- Li Y. H., Zhou G. Y., Ma J. X., Jiang W. K., Jin L. G., Zhang Z. H., et al. (2014). De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 32, 1045–1052. 10.1038/nbt.2979 [DOI] [PubMed] [Google Scholar]
- Li Y. M., Zhang D., Xing L. B., Zhang S. W., Zhao C. P., Han M. Y. (2016). Effect of exogenous 6-benzylaminopurine (6-BA) on branch type, floral induction and initiation, and related gene expression in 'Fuji' apple (Malus domestica Borkh). Plant Growth Regul. 79, 65–70. 10.1007/s10725-015-0111-5 [DOI] [Google Scholar]
- Li Y., Wu B., Yu Y., Yang G., Wu C., Zheng C. (2011). Genome-wide analysis of the RING finger gene family in apple. Mol. Genet. Genomics 286, 81–94. 10.1007/s00438-011-0625-0 [DOI] [PubMed] [Google Scholar]
- Liu X. Y., Widmer A. (2014). Genome-wide comparative analysis of the GRAS gene family in populus, Arabidopsis and Rice. Plant Mol. Biol. Reporter 32, 1129–1145. 10.1007/s11105-014-0721-5 [DOI] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu J., Wang T., Xu Z., Sun L., Zhang Q. (2015). Genome-wide analysis of the GRAS gene family in Prunus mume. Mol. Genet. Genomics 290, 303–317. 10.1007/s00438-014-0918-1 [DOI] [PubMed] [Google Scholar]
- Lyons E., Pedersen B., Kane J., Alam M., Ming R., Tang H. B., et al. (2008). Finding and comparing syntenic regions among Arabidopsis and the Outgroups Papaya, Poplar, and Grape: CoGe with Rosids. Plant Physiol. 148, 1772–1781. 10.1104/pp.108.124867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno M. A., Martínez M., Vicente-Carbajosa J., Carbonero P. (2007). The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol. Genet. Genomics 277, 379–390. 10.1007/s00438-006-0186-9 [DOI] [PubMed] [Google Scholar]
- Park H. J., Jung W. Y., Lee S. S., Song J. H., Kwon S. Y., Kim H., et al. (2013). Use of heat stress responsive gene expression levels for early selection of heat tolerant cabbage (Brassica oleracea L.). Int. J. Mol. Sci. 14, 11871–11894. 10.3390/ijms140611871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty L. M., Harberd N. P., Carré I. A., Thomas B., Jackson S. D. (2003). Expression of the Arabidopsis gai gene under its own promoter causes a reduction in plant height in chrysanthemum by attenuation of the gibberellin response. Plant Sci. 164, 175–182. 10.1016/S0168-9452(02)00380-1 [DOI] [Google Scholar]
- Porri A., Torti S., Romera-Branchat M., Coupland G. (2012). Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139, 2198–2209. 10.1242/dev.077164 [DOI] [PubMed] [Google Scholar]
- Pysh L. D., Wysocka-Diller J. W., Camilleri C., Bouchez D., Benfey P. N. (1999). The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119. [DOI] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz C., Theres K. (1999). The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. U.S.A. 96, 290–295. 10.1073/pnas.96.1.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigyo M., Tabei N., Yoneyama T., Yanagisawa S. (2007). Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol. 48, 179–185. 10.1093/pcp/pcl044 [DOI] [PubMed] [Google Scholar]
- Song X. M., Liu T. K., Duan W. K., Ma Q. H., Ren J., Wang Z., et al. (2014). Genome-wide analysis of the GRAS gene family in Chinese cabbage (Brassica rapa ssp. pekinensis). Genomics 103, 135–146. 10.1016/j.ygeno.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Stuurman J., Jaggi F., Kuhlemeier C. (2002). Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 16, 2213–2218. 10.1101/gad.230702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Jones W. T., Harvey D., Edwards P. J., Pascal S. M., Kirk C., et al. (2010). N-terminal domains of DELLA proteins are intrinsically unstructured in the absence of interaction with GID1/gibberellic acid receptors. J. Biol. Chem. 285, 11557–11571. 10.1074/jbc.M109.027011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Jones W. T., Rikkerink E. H. (2012). GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem. J. 442, 1–12. 10.1042/BJ20111766 [DOI] [PubMed] [Google Scholar]
- Sun X., Xue B., Jones W. T., Rikkerink E., Dunker A. K., Uversky V. N. (2011). A functionally required unfoldome from the plant kingdom: intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 77, 205–223. 10.1007/s11103-011-9803-z [DOI] [PubMed] [Google Scholar]
- Tian C., Wan P., Sun S. H., Li J. Y., Chen M. S. (2004). Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 54, 519–532. 10.1023/B:Plan.0000038256.89809.57 [DOI] [PubMed] [Google Scholar]
- Torres-Galea P., Huang L. F., Chua N. H., Bolle C. (2006). The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome A responses. Mol. Genet. Genomics 276, 13–30. 10.1007/s00438-006-0123-y [DOI] [PubMed] [Google Scholar]
- Vandenbussche F., Fierro A. C., Wiedemann G., Reski R., Van Der Straeten D. (2007). Evolutionary conservation of plant gibberellin signalling pathway components. BMC Plant Biol. 7:65. 10.1186/1471-2229-7-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A., et al. (2010). The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 42, 833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Wang L., Mai Y. X., Zhang Y. C., Luo Q., Yang H. Q. (2010). MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol. Plant 3, 794–806. 10.1093/mp/ssq042 [DOI] [PubMed] [Google Scholar]
- Xing L. B., Zhang D., Li Y. M., Shen Y. W., Zhao C. P., Ma J. J., et al. (2015). Transcription profiles reveal sugar and hormone signaling pathways mediating flower induction in apple (Malus domestica Borkh.). Plant Cell Physiol. 56, 2052–2068. 10.1093/pcp/pcv124 [DOI] [PubMed] [Google Scholar]
- Xing L. B., Zhang D., Li Y. M., Zhao C. P., Zhang S. W., Shen Y. W., et al. (2014). Genome-wide identification of vegetative phase transition-associated microRNAs and target predictions using degradome sequencing in Malus hupehensis. BMC Genomics 15:1125. 10.1186/1471-2164-15-1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L. B., Zhang D., Zhao C. P., Li Y. M., Ma J. J., An N., et al. (2016). Shoot bending promotes flower bud formation by miRNA-mediated regulation in apple (Malus domestica Borkh.). Plant Biotechnol. J. 14, 749–770. 10.1111/pbi.12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. N., Xing S. S., Zhang Z. R., Chen X. S., Wang X. Y. (2016). Genome-wide identification and expression analysis of the tubby-like protein family in the Malus domestica Genome. Front. Plant Sci. 7:1693. 10.3389/fpls.2016.01693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Chen Z., Ahmed N., Han B., Cui Q., Liu A. (2016). Genome-Wide Identification, Evolutionary Analysis, and Stress Responses of the GRAS Gene Family in Castor Beans. Int. J. Mol. Sci. 17:1004. 10.3390/ijms17071004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Winter C. M., Wu M. F., Kanno Y., Yamaguchi A., Seo M., et al. (2014). Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344, 638–641. 10.1126/science.1250498 [DOI] [PubMed] [Google Scholar]
- Zahn L. M., Kong H. Z., Leebens-Mack J. H., Kim S., Soltis P. S., Landherr L. L., et al. (2005). The evolution of the SEPALLATA subfamily of MADS-Box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169, 2209–2223. 10.1534/genetics.104.037770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. P., Iyer L. M., Aravind L. (2012). Bacterial GRAS domain proteins throw new light on gibberellic acid response mechanisms. Bioinformatics 28, 2407–2411. 10.1093/bioinformatics/bts464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Zhang D., Fan S., Du L., Shen Y., Xing L., et al. (2016). Effect of exogenous GA3 and its inhibitor paclobutrazol on floral formation, endogenous hormones, and flowering-associated genes in ‘Fuji’ apple (Malus domestica Borkh.). Plant Physiol. Biochem. 107, 178–186. 10.1016/j.plaphy.2016.06.005 [DOI] [PubMed] [Google Scholar]
- Zhang Z. L., Ogawa M., Fleet C. M., Zentella R., Hu J., Heo J. O., et al. (2011). Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 2160–2165. 10.1073/pnas.1012232108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of MdGRAS genes on the apple chromosomes.
MdGRAS gene structures. MdGRAS gene exon–intron composition. Red boxes and black lines represent exons and introns, respectively.
Alignment of DELLA subfamily members in apple and Arabidopsis.
Heat map showing the transcript levels of MdGRAS genes during flower induction. Transcript levels of GRAS genes in the buds of “Nagafu No. 2” and “Yanfu No. 6” were investigated during flower induction. Samples were collected three times at the early stage (ES), middle stage (MS) and later stages (LS) of flower induction.
Expression of candidate MdGRAS genes in GA-treated buds. Samples were collected three times at ES, MS and LS in 2014, which was consistent with 40, 50, and 70 DAFB in 2016. Each value represents the mean ± standard error of three replicates.
Expression of candidate MdGRAS genes in 6BA-treated buds. Samples were collected three times at ES, MS and LS in 2013, which was consistent with 30, 50, and 80 DAFB in 2016. Each value represents the mean ± standard error of three replicates.
Predicted cis-elements in the promoter regions of the candidate MdGRAS genes. The upstream 1.5 kb promoter sequences were analyzed.
Primer sequences of MdGRAS and reference genes in quantitative reverse transcription PCR.
Comparison of MdGRAS genes in different species.
Expression levels of MdGRAS genes in Nagafu No. 2 and Yanfu No. 6. Data were collected from the transcriptome.
Investigation of flowering rate in 2016.