Abstract
The cypress family (Cupressaceae) possesses highly rearranged plastomes that lack a pair of large inverted repeats typically found in land plants. A few cypress species have been reported to contain isomeric plastomes, but whether the existence of isomeric plastomes is ubiquitous in the family remains to be investigated with a broader taxon sampling. In this study, we sequenced the complete plastomes of ten species in Cupressoideae, the largest cypress subfamily. Cupressoideae showed relatively accelerated rates of substitutions at both nonsynonymous and synonymous sites as compared with other subfamilies of Cupressaceae. Our PCR and read mapping analyses together suggested the existence of isomeric plastomes in eight of the ten sequenced Cupressoideae species. The isomeric plastomes were also detected in 176 individuals from nine wild populations of four Cupressoideae species. Within Calocedrus macrolepis, we discovered a new type of isomeric plastomes that was likely derived from homologous recombination mediated by an 11-bp repeat. We conclude that isomeric plastomes are commonly present in Cupressoideae, thereby contributing to increased plastomic complexity.
Keywords: Cupressaceae, Cupressoideae, isomeric plastomes, plastomic complexity, repeat
Introduction
Inversions of DNA fragments are one of the major mechanisms increasing plastomic complexity (Palmer 1991; Raubeson and Jansen 2005; Jansen and Ruhlman 2012; Wu and Chaw 2016). Land plant plastomes typically possess large inverted repeats (hereafter called typical IRs) that contain four ribosomal RNA genes in each copy (Wang et al. 2008; Wicke et al. 2011). The typical IRs are capable of triggering intra-plastomic homologous recombination to generate two isomeric plastomes in equal abundance (Palmer 1983; Martin et al. 2013). In addition, short repeats were considered to cause plastomic re-organization on the basis of a positive correlation between dispersed repeats and rearrangements (Lee et al. 2007; Guisinger et al. 2011; Weng et al. 2014).
Interestingly, the plastomes of conifers do not contain a pair of typical IRs, but they have evolved specific IRs that are associated with plastomic re-organizations. For example, two specific IRs are responsible for the generation of four different plastomic organizations among Pinaceae genera (Tsumura et al. 2000; Wu et al. 2011; Sudianto et al. 2016). In cupressophytes, Cephalotaxus oliveri (Cephalotaxaceae; Yi et al. 2013) and four Juniperus species (junipers of the cypress family, Cupressaceae; Guo et al. 2014) contain isomeric plastomes, which are associated with a ∼36-kb inversion mediated by a “trnQ-IR” sequence. Unlike the isomeric plastomes mediated by the typical IRs, the isomeric plastomes of cupressophytes differ in abundance, with major and minor ones within plastids (Guo et al. 2014).
The cypress family (Cupressaceae) diverged from other cupressophytes during the Late Permian and Early Triassic (Mao et al. 2012; Yang et al. 2012). The family includes ∼149 tree species in 29 genera (Christenhusz and Byng 2016) scattered over all continents except Antarctica. They are trees having the widest geographical range among living gymnosperms (Farjon 2005; Mabberley 2008; Mao et al. 2012; Yang et al. 2012; Wang and Ran 2014). Molecular phylogenetic studies have revealed seven cypress subfamilies, among which Cupressoideae is the largest, containing ∼100 species in 13 genera (Gadek et al. 2000; Mabberley 2008; Mao et al. 2012; Yang et al. 2012; Wang and Ran 2014).
In this study, we assembled the complete plastomes of ten species representing nine genera of Cupressoideae. Including other available Cupressaceous plastomes, our sampling comprised 17 species from 11 genera of Cupressoideae and five species from the four other Cupressaceous subfamilies. This allowed for comparisons of nucleotide substitution rates among Cupressaceous subfamilies. We also investigated three questions in isomeric plastomes. First, do isomeric plastomes commonly exist in Cupressoideae? Second, are there other types of isomeric plastomes not previously reported? Third, in terms of abundance, do the relative ratios of isomeric plastomes shift frequently among species or populations?
Materials and Methods
Plant Materials
To sequence complete plastomes, young and fresh leaves were harvested from ten Cupressoideae species, including Calocedrus formosana, Calocedrus macrolepis, Chamaecyparis lawsoniana, Fokienia hodginsii, Hesperocyparis glabra, Juniperus formosana, Platycladus orientalis, Thuja standishii, Thujopsis dolabrata, and Xanthocyparis vietnamensis (supplementary table S1, Supplementary Material online). To investigate isomeric plastomes at population levels, we chose Calocedrus formosana, Calocedrus macrolepis, Cupressus duclouxiana, Juniperus gaussenii, and Juniperus squamata (supplementary table S1, Supplementary Material online) because their wild populations were accessible. In total, we gathered 186 individuals from 11 populations. The vouchers were deposited in the herbarium at Kunming Institute of Botany, Chinese Academy of Sciences (KUN).
DNA Extraction
For each of the ten sequenced species, we used ∼50 g of young and fresh leaves to isolate plastid DNA using an improved extraction method (Zhang et al. 2011) based on the methods of Gong et al. (1994) and Jansen et al. (2005). The purified DNA concentration and quality were determined by doing agarose gel electrophoresis. For PCR assays, total genomic DNA was extracted from the examined plants by using the CTAB method (Doyle and Doyle 1987).
Plastome Sequencing, Assembly and Annotation
For each of the ten sequenced species, 5 µg of purified DNA (including nonplastid DNA) was sheared into 500-bp fragments to construct a paired-end library according to the manufacturer’s manual (Illumina Inc., San Diego). DNA sequencing was performed on the Illumina HiSeq 2000 at the Beijing Genomics Institute (Shenzhen, Guangdong, China). Approximately four million 90-bp paired-end reads were obtained from each species. After quality trimming by using the NGS QC Toolkit (Patel and Jain 2012) with the default parameters, we used CLC Genomics Workbench v7.0.3 (CLC Inc, Arhus, Denmark) for de novo assembly with the default parameters (word size = 64). To validate the plastome assembly, the paired-end reads were mapped onto the assembled plastomes by using Bowtie v2.2.3 (Langmead and Salzberg 2012) with the default preset options (–sensitive-local, -D 15 -R 2 -N 0 -L 20 -i S,1,0.75) in –local mode.
Plastomes were annotated with Dual Organellar GenoMe Annotator (DOGMA; Wyman et al. 2004). The boundaries of predicted genes were confirmed manually by aligning them with their orthologs from other published gymnosperms plastomes. Transfer RNA (tRNA) genes were further determined by using tRNAscan-SE (Schattner et al. 2005). Physical maps of the plastomes were generated by using Circos v0.66 (Krzywinski et al. 2009).
Phylogenetic Analyses
The 73 common protein-coding genes (supplementary tables S2 and S3, Supplementary Material online) were extracted by using a custom Perl script. The codon-based alignment for each gene involved use of MUSCLE (Edgar 2004) implemented in MEGA v6.06 (Tamura et al. 2013). Concatenation of the 73 aligned genes yielded a matrix of 52,929 bp. We used jModelTest v2.1.6 (Darriba et al. 2012) to determine the best substitution model for the matrix. A maximum likelihood (ML) tree inferred from the matrix were estimated by using RAxML v8.2.4 (Stamatakis 2014) with a GTRGAMMAI model. Supports for the tree nodes were assessed by rapid bootstrapping analysis of 1,000 replicates.
Analysis of Rearrangement Distance
To estimate locally collinear blocks (LCBs) among the examined plastomes, we performed whole genome alignment by using progressiveMauve implemented in Mauve v2.3.1 (Darling et al. 2010) with the default parameters. Before the alignment, the stop codon of psbA was specified as the starting point for all compared plastomes. On the basis of the yielded matrix of LCBs (supplementary table S4, Supplementary Material online), we used Grappa v2.0 (Bader et al. 2001; Moret et al. 2002) with the default parameters to estimate the pairwise breakpoint distance (the number of nonconserved adjacencies between genomes) as well as the pairwise inversion distance (the minimum number of inversions required for transformation of one genome to the other).
Exploration of Isomeric Plastomes
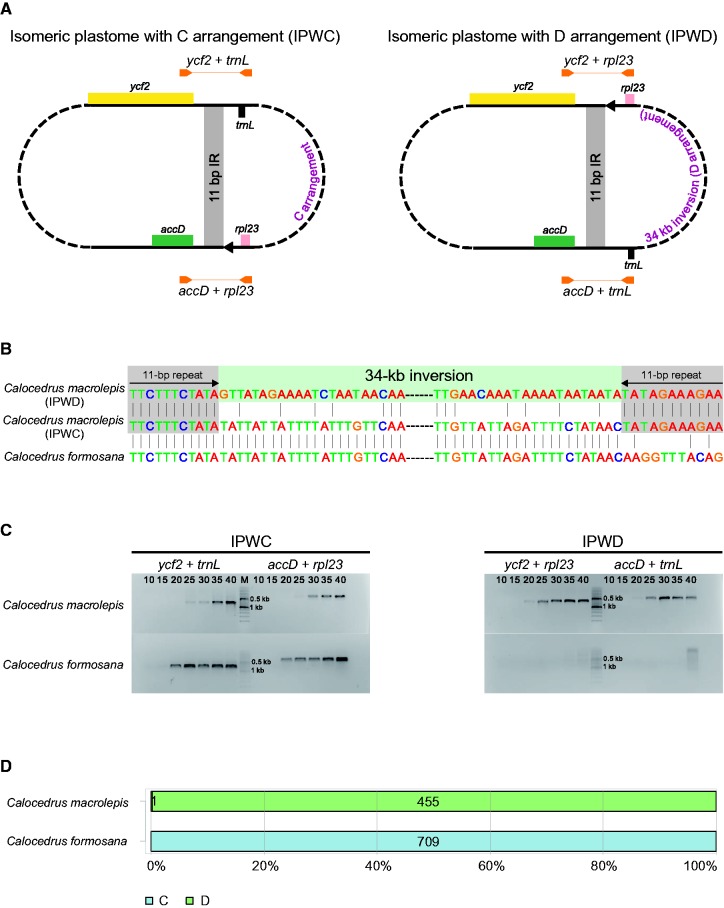
Previously, Guo et al. (2014) found that the plastomes within some Juniperus species differ in the orientation of a 36-kb fragment, and they subsequently designated these plastomes as isomeric plastomes with an A or B arrangement (also see supplementary fig. S4, Supplementary Material online). In Cal. macrolepis, we designated the plastomes that differ in the orientation of a 34-kb fragment as the isomeric plastomes with a C or D arrangement (fig. 1). We adopted two approaches to explore the presence of isomeric plastomes within species (i.e., isomeric plastomes with the A, B, C, or D arrangement). First, semi-quantitative PCR involved use of specific primer pairs for amplifying the unique regions of the isomeric plastomes. The isomeric plastome with the A arrangement could be amplified by a combination of chlB + psbK and rps4 + trnL; and the B, C, and D arrangements could be amplified by a combination of chlB + rps4 and psbK + trnL, ycf2 + trnL and rpl23 + accD, and ycf2 + rpl23 and trnL + accD, respectively. The primers used for PCRs are listed in supplementary table S5, Supplementary Material online. PCR reactions were performed in a 25 µl mixture, including 1 µl total DNA (20 ng/µl), 0.5 µl each of the forward and reverse primers (10 µmol/l), 13 µl Tiangen 2 × Taq PCR MasterMix, and 10 µl ddH2O. After an initial denaturation at 94 °C for 4 min, PCR reactions were conducted for 10, 15, 20, 25, 30, 35 and 40 cycles, respectively. Each cycle included denaturation at 94 °C for 50 s, annealing at 55 °C for 50 s, and elongation at 72 °C for 1 min 30 s. This PCR procedure was also used for detecting isomeric plastomes in population samples with 20, 25, 30, and 35 cycles, respectively.
Fig. 1.—
Detection of isomeric plastomes with C (IPWC) or D (IPWD) arrangement in Calocedrus macrolepis. (A) PCR primers (orange arrows) designed for detection of IPWC and IPWD. Grey areas link two copies of 11-bp inverted repeats (IRs). (B) Alignment showing the sequences of 11-bp IRs in Cal. macrolepis but not in Cal. formosana. (C) Semi-quantitative PCR demonstrating the co-existence of IPWC and IPWD in Cal. macrolepis but not in Cal. formosana. Primers for detection of isomeric plastomes are indicated, and 10–40 are PCR cycles. (D) Results of read mapping analyses. Light blue and green bars are paired-end reads for supporting IPWC and IPWD, respectively. Numbers within bars are reads that span a 11-bp IR copy.
Second, Illumina paired-end reads were mapped to the regions specific to each of the isomeric plastomes by using Bowtie v2.2.3 with the default settings. Paired-end reads that spanned trnQ-IR or 11-bp IR were collected if sequence identity was > 90%. SAMTools v0.1.19 (Li et al. 2009) was used to view and check the mapping scenario.
Estimation of Nucleotide Substitution Rates
The codeml program of PAML v4.8 (Yang 2007) was used to estimate pairwise substitution rates between Cunninghamia lanceolata and other cupressaceous species under the F3 × 4 codon model and four categories of the Gamma distribution (Goldman and Yang 1994). We used the Wilcoxon rank-sum test to assess the significant difference of the estimated substitution rates between Cupressoideae and other subfamilies (including Sequoioideae, Taiwanioideae, Taxodioideae). Gene groups were categorized according to Guisinger et al. (2008). All statistical tests involved use of R v3.2.2 (R Development Core Team 2015).
Results
Characteristics of the Ten Newly Sequenced Plastomes
The ten deciphered Cupressoideae plastomes range from 127,064 to 130,505 bp (table 1). Their plastome maps are depicted in supplementary figure S1, Supplementary Material online. The assemblies are well supported by high sequencing coverage ranging from 100.24× to 264.29× (supplementary fig. S2, Supplementary Material online). These ten plastomes are similar in gene content (table 1). However, a few differences were observed: 1) the gene trnP-GGG is absent from Th. standishii, Thu. dolabrata and J. formosana; 2) the gene trnT-GGU is absent from Ch. lawsoniana; 3) duplication of trnP-UGG is unique in J. formosana; 4) specific duplication of trnI-GAU is found in Ch. lawsoniana; 5) H. glabra, Th. standishii and X. vietnamensis contain a pseudo-rps16 gene; and 6) one of the duplicated trnI-CAU genes is pseudogenized in J. formosana.
Table 1.
Summary of Sequencing Outputs and Brief Features of the 10 Cupressoideae Plastomes Sequenced in This Study
| Species | No. of reads | Mapped reads (no./%) | Mapped bases (bp) | Insert size (median) | Coverage (mean) | Plastome size (bp) | GC content (%) | Gene (protein, rRNA, tRNA) |
|---|---|---|---|---|---|---|---|---|
| Juniperus formosana | 4,889,566 | 376,628/7.7 | 33,849,014 | 458 | 264.29 | 128,073 | 35.21 | 119 (83, 4, 32) |
| Hesperocyparis glabra | 4,795,638 | 226,383/4.72 | 20,246,334 | 472 | 159.34 | 127,064 | 35.12 | 119 (82, 4, 33) |
| Xanthocyparis vietnamensis | 4,592,018 | 506,992/11.04 | 45,531,575 | 476 | 357.17 | 127,479 | 35.71 | 119 (82, 4, 33) |
| Platycladus orientalis | 4,823,022 | 170,893/3.54 | 15,330,344 | 450 | 120.6 | 127,113 | 35.73 | 120 (83, 4, 33) |
| Calocedrus formosana | 4,291,862 | 202,305/4.71 | 18,187,106 | 467 | 142.79 | 127,370 | 34.83 | 120 (83, 4, 33) |
| Calocedrus macrolepis | 4,749,080 | 141,912/2.99 | 12,746,278 | 443 | 100.24 | 127,157 | 34.5 | 120 (83, 4, 33) |
| Fokienia hodginsii | 3,543,258 | 191,565/5.41 | 17,182,343 | 476 | 133.41 | 128,789 | 35.02 | 120 (83, 4, 33) |
| Chamaecyparis lawsoniana | 4,712,988 | 213,911/4.54 | 19,094,825 | 474 | 150.28 | 127,064 | 35.49 | 120 (83, 4, 33) |
| Thuja standishii | 4,871,026 | 237,093/4.87 | 21,174,422 | 455 | 162.25 | 130,505 | 35.38 | 118 (82, 4, 32) |
| Thujopsis dolabrata | 4,086,050 | 377,197/9.23 | 33,114,932 | 484 | 258.12 | 128,291 | 35.02 | 119 (83, 4, 32) |
Species are arranged by their divergence in the phylogenetic tree shown in figure 2.
Intra-Generic Variation of Plastomic Organizations in Calocedrus
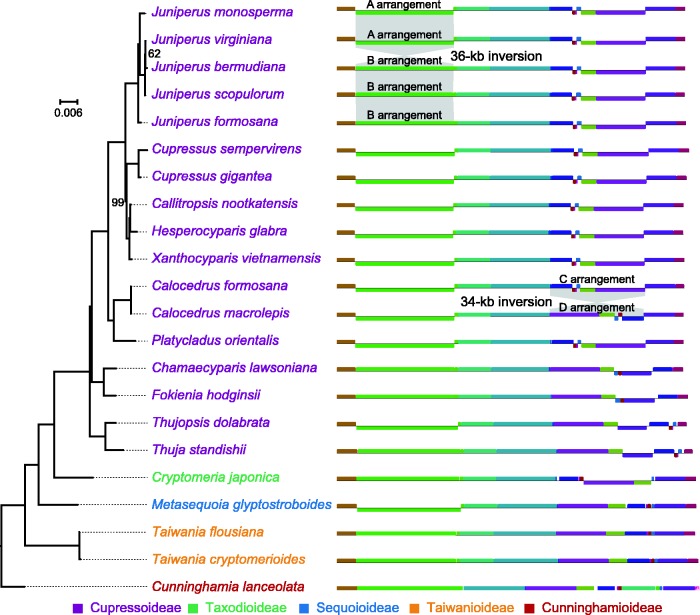
Comparisons of the 22 sampled Cupressaceous plastomes yielded 15 LCBs (supplementary table S4, Supplementary Material online), and their relative orders are depicted in the right panel of figure 2. The pairwise breakpoint and inversion distances range from 0 to 14 (the number of nonconserved adjacencies) and from 0 to 13 (the minimum number of inversions), respectively (supplementary fig. S3, Supplementary Material online). These data indicate that the Cupressaceous plastomes experienced numerous rearrangements during evolution. We observed variation in the LCB order within congeneric species (fig. 2). For example, Juniperus contains A and B arrangements that differs in the orientation of a 36-kb fragment, in agreements with the finding that the A and B arrangements are the suspected major isomers among the Juniper species (Guo et al. 2014). Moreover, we found that Cal. formosana and Cal. macrolepis are distinguishable by the orientation of a 34-kb fragment because the former and the latter contain the C and D arrangements, respectively.
Fig. 2.—
Comparison of plastomic organization among 22 Cupressaceae species. A maximum likelihood (ML) tree inferred from 73 plastid genes is shown in the left side. Bootstrapping supports are indicated when they are <100%. Plastomic maps are depicted with locally collinear blocks (LCBs) coded by different colors in the right side. LCBs below the horizontal line are opposite in orientations as compared to their counterparts in Cunninghamia lanceolata. The grey area highlights the intra-generic variation in LCBs with an 36-kb inversion resulting in formation of A and B arrangements in Juniperus, and an 34-kb inversion generating C and D arrangements in Calocedrus.
trnQ-IR Associated Isomeric Plastomes in Cupressoideae
Each of the ten newly sequenced plastomes contains a trnQ-IR sequence ranging from 194 to 297 bp (supplementary table S6, Supplementary Material online). This trnQ-IR is capable of triggering a 36-kb inversion, resulting in the coexistence of an isomeric plastome with the A arrangement (hereafter abbreviated as IPWA) and that with the B arrangement (IPWB) within Juniperous species (Guo et al. 2014; also see supplementary fig. S4A, Supplementary Material online). Our semi-quantitative PCR results suggest that eight of the ten sequenced species contain both IPWA and IPWB (supplementary fig. S4B, Supplementary Material online). Our read mapping analyses also show specific reads that support the coexistence of IPWA and IPWB within each of the species (supplementary fig. S4C, Supplementary Material online). Furthermore, the coexistence of IPWA and IPWB was also detected within each of the 85 individuals sampled from four populations of the three examined species, Cup. duclouxiana, J. gaussenii and J. squamata (supplementary fig. S5A, Supplementary Material online). However, neither PCR nor read mapping analyses detected the coexistence of IPWB and IPWA in Cal. macrolepis and Th. standishii.
Although IPWA and IPWB coexist in each of the eight species, they differ in relative ratios. IPWA is the major plastome in Cal. formosana, Cup. duclouxiana, H. glabra, J. gaussenii, J. squamata, P. orientalis, Thu. dolabrata and X. vietnamensis, but it is the minor one (in other words, IPWB is major) in Cha. lawsoniana, F. hodginsii and J. formosana (supplementary figs. S4 and S5A, Supplementary Material online). The recombinant frequency of trnQ-IRs was estimated in a range from 0.0012 (number of reads for IPWA/that for IPWB = 1/801 in supplementary fig. S4, Supplementary Material online) inversions per plastome in J. formosana to 0.0377 (12/318) in H. glabra. Together, these data clearly suggest that all of our assemblies are the major isomeric plastome.
Detection of a New Type of Plastomic Isomer in Calocedrus Macrolepis
As mentioned previously, Cal. formosana and Cal. macrolepis differ in the orientation of a 34-kb fragment flanked by rpl23 and trnL-CAA. We, therefore, designated the 34-kb fragments in Cal. formosana and Cal. macrolepis as the C and D arrangements, respectively (fig. 2). We conclude that this 34-kb fragment is inverted uniquely in Cal. macrolepis because its counterparts in other Cupressoideae species are identical in the orientation (fig. 2). In addition, we detected a pair of 11-bp inverted repeats specifically residing at the boundaries of the 34-kb fragment in Cal. macrolepis (fig. 1A and B).
Our semi-quantitative PCR and read mapping assays revealed that Cal. macrolepis contain two isomeric plastomes with the C (IPWC) and D (IPWD) arrangements, respectively, but the coexistence of IPWC and IPWD was not detected in Cal. formosana (fig. 1B–D). The PCR and read mapping results also indicate that IPWD is the major isomeric plastome in Cal. macrolepis (fig. 1C and D). Moreover, PCR assays of 91 Cal. macrolepis individuals also support IPWD as the major isomeric plastome across all five sampled populations (supplementary fig. S5B, Supplementary Material online). In contrast, IPWD was not detected in all of the ten individuals sampled from two populations of Cal. formosana (supplementary fig. S5C, Supplementary Material online).
Accelerated Nucleotide Substitution Rates in Cupressoideae
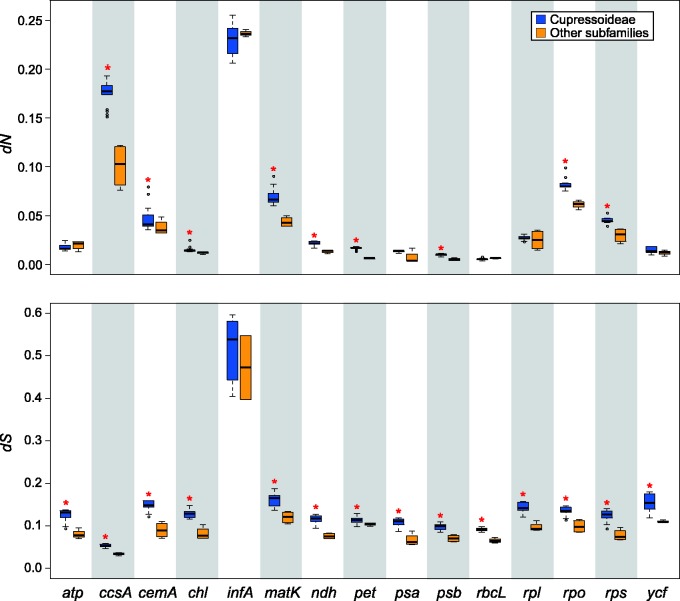
The ML tree inferred from the combined 73 plastid genes reveals five major clades: Cunninghamoideae, Taiwanioideae, Sequoioideae, Taxodioideae, and Cupressoideae (fig. 2). The pairwise substitution rates between Cupressoideae and other subfamilies are compared in figure 3. The estimated substitution rates differ greatly among gene groups. For example, the nonsynonymous (dN) substitution rates of Cupressoideae vary from a mean of 0.0052 in rbcL to 0.2298 in infA, with the former being 44-fold slower than the latter (fig. 3). Such variation among gene groups was also observed in the synonymous (dS) substitution rates of Cupressoideae with a range from a mean of 0.0536 in ccsA to 0.5126 in infA. As compared with other subfamilies, the Cupressoideae show significantly accelerated dN substitution rates: nine of the 15 gene groups are significantly faster in Cupressoideae than other subfamilies. In terms of dS sites, the Cupressoideae are significantly accelerated in all gene groups except infA. In short, these data suggest a plastome-wide acceleration of substitution rates in Cupressoideae, remarkably at the dS sites.
Fig. 3.—
Boxplots for comparisons of nucleotide substitution rates between Cupressoideae and other Cupressaceae subfamilies. (A) Nonsynonymous (dN) and (B) synonymous (dS) substitution rates estimated from 15 gene categories. Thick lines within boxes are medians, and outliers are shown as circles. Red asterisks denote that the substitution rates are significantly (P < 0.05) higher in Cupressoideae than other Cupressaceous subfamilies.
Discussion
Ubiquitous Existence of Isomeric Plastomes in Cupressoideae
The trnQ-IR associated isomeric plastomes were previously discovered in Cephalotaxus (Cephalotaxaceae; Yi et al. 2013) and Juniperus species (Cupressaceae; Guo et al. 2014). Such trnQ-IR associated isomeric plastomes were also detected in eight of our ten newly elucidated Cupressoideae species (supplementary fig. S4, Supplementary Material online). Nevertheless, we detected no coexistence of IPWA and IPWB in Cal. macrolepis, although its two trnQ-IR copies are 99.19% identical in their sequences. The two trnQ-IR copies of Th. standishii have the lowest sequence identity (88.14%) among the studied species (supplementary table S6, Supplementary Material online), which likely explains its absence of IPWA. With the evidence from Juniperus (Guo et al. 2014) and our experimental results (supplementary figs. S4 and S5A, Supplementary Material online), the coexistence of IPWA and IPWB has been discovered in nine Cupressoideae genera. This suggests that the existence of isomeric plastomes might be a common feature in Cupressoideae.
Additionally, we discovered that IPWA is the major isomeric plastome in the 85 individuals of the three examined species (Cup. duclouxiana, J. gaussenii and J. squamata), despite being sampled from different populations (supplementary fig. S5A, Supplementary Material online). Moreover, the 91 individuals from five populations of Cal. macrolepis are consistent in retention of IPWD as the major isomeric plastome (supplementary fig. S5B, Supplementary Material online). Collectively, these data suggest that the isomeric plastomes have existed for a long time and the major isomeric plastomes have been fixed within each of these species.
Shifts of the abundance between IPWA and IPWB were found among congeneric species (Guo et al. 2014) and among genera (supplementary fig. S4, Supplementary Material online). This finding implies that the existence of isomeric plastomes and shift in their abundance together contribute to the plastome complexity in Cupressoideae. However, the evolutionary mechanisms that underlie the shift of the abundance between isomeric plastomes are unclear and require further investigation in the future.
Large Inversions Mediated by Short Inverted Repeats
In plastomes, small repeats (usually <100 bp) have been associated with inversions of small fragments. In the plastome of Chusquea, a genus of bamboos, stems (<12 bp) of a hairpin structure are able to induce minute inversions (Kelchner and Wendel 1996). Investigations of the plastomes of Panax, ginseng of the flowering family Araliaceae, revealed an association between 4 and 16 bp inversions and repeats of smaller than 25 bp (Kim and Lee 2004). Kim et al. (2005) found that in angiosperm plastomes, inversions of small fragments are able to be mediated by short repeats of 11–24 bp. In this study, we discovered a specific plastomic inversion of a 34-kb fragment in Cal. macrolepis, which is likely mediated by an 11-bp IR (fig. 1 and supplementary fig. S5B, Supplementary Material online).
Double-strand breaks (DSBs) are one of the most severe forms of DNA damage (Kwon et al. 2010). Meanwhile, homologous recombination (HR) is an effective mechanism that repairs DSBs (Marechal and Brisson 2010). DSBs followed by HR account for the frequent inversion of large or small single-copy regions involving the typical IRs in plastomes (Palmer 1983; Martin et al. 2013). The 34-kb inversion, which resulted in the D arrangement, might be a consequence of HR involving the 11-bp IR unique to Cal. macrolepis (fig. 1). We detected no indels between IPWC and IPWD (fig. 1B). This suggests that the IPWD may not be derived from nonhomologous end-joining nor microhomology-mediated end joining, as both mechanisms would cause short (1–4 bp) to long (3.1–4.6 kb) indels (Kwon et al. 2010; Marechal and Brisson 2010; Massouh et al. 2016). Inversions of large fragments that are not associated with the typical IRs have been repeatedly found in plastomes. For instances, Martin et al. (2014) reported that in the plastomes of the legume family, a 36-kb inversion was associated with a short IR of 29 bp in length. A 45-kb plastomic inversion between two subspecies of Medicago truncatula was likely mediated by ≥20 bp IRs (Gurdon and Maliga 2014). In terms of IRs that are able to mediate inversions of large fragments, the 11-bp IR we detected in Cal. macrolepis is by far the shortest.
We used PCR to detect the presence of isomeric plastomes in different populations of Cal. macrolepis (supplementary fig. S5B, Supplementary Material online). Hsu et al. (2016) noted that amplification of plastid DNA could be contaminated by nuclear plastid DNA (the so-called nupt) when using total DNA as PCR templates. We cannot rule out the possibility that our amplicons of IPWC were contaminants from nupts in Cal. macrolepis. In addition, mitochondrial plastid DNA (mtpt) is common in many seed plants (Stern and Lonsdale 1982; Wang et al. 2007; Goremykin et al. 2009; Alverson et al. 2010). Since the nuclear and mitochondrial genome sequences of Cal. macrolepis are not available, whether our PCR assays were contaminated by nupts and mtpts is unknown. However, we have provided evidence of NGS reads to support the presence of IPWC in Cal. macrolepis (fig. 1B and D). Moreover, nupts were experimentally demonstrated to be eliminated quickly from nuclear genomes (Sheppard and Timmis 2009). The coexistence of IPWC and IPWD was demonstrated by our PCR assays in the 91 individuals sampled from five populations of Cal. macrolepis (supplementary fig. S5B, Supplementary Material online), suggesting that IPWC and IPWD might have co-existed prior to the diversification of these populations.
Plastome-Wide Acceleration of Nucleotide Substitution Rates in Cupressoideae
In Cupressoideae plastomes, nine of the 15 examined gene groups have accelerated dN substitution rates, whereas all gene groups except infA are accelerated at dS sites (fig. 3). It is of great interested to understand why the dN and dS substitution rates are co-accelerated in Cupressoideae plastomes. Substitutions at dS sites are evolutionarily neutral and largely dependent on mutation rates (Kimura 1983), whereas those at dN sites might be affected by multiple mechanisms, including mutation rates, population size, and selection pressure (Bromham et al. 2015). In the plastomes of Cupressoideae, the co-accelerated substitution rates at both dN and dS sites are likely being affected by mutation rates.
Multiple mechanisms have been proposed to account for genome-wide accelerations of substitution rates. Relatively shorter generation times of herbs than those of trees and shrubs were proposed to be associated with their higher substitution rates (Smith and Donoghue 2008). Population subdivisions followed by speciation events generally cause accelerated substitution rates genome-wide (Pagel et al. 2006; Venditti and Pagel 2010). Indeed, a correlation between substitution rates and species diversity were found in flowering plants (Barraclough et al. 1996; Barraclough and Savolainen 2001; Duchene and Bromham 2013). On the other hand, high error rates of DNA repair systems might be responsible for accelerations of plastomic substitution rates in Geraniaceae (Guisinger et al. 2011).
In this study, we calculated pairwise substitution rates, which make it difficult to determine when rate acceleration occurred in the history of Cupressoideae. In addition, our sampled taxa are largely biased toward samples of Cupressoideae species, which might not be able to illustrate an entire picture of plastomic substitution rates in Cupressaceae. Therefore, more data are required to assess the mechanisms underlying the accelerated substitution rates in Cupressoideae.
Conclusion
We found that the existence of isomeric plastomes is ubiquitous in Cupressoideae. In addition to trnQ-IR associated isomeric plastomes, a new type of isomeric plastomes was detected within Cal. macrolepis, which is likely associated with the specific 11-bp IR. We also provided solid evidence showing shift of the abundance between isomeric plastomes among genera and among species (e.g., Cal. formosana and Cal. macrolepis). However, such shift of the abundance is absent at the population level, indicating that the major isomeric plastomes have been fixed within species. As a result, we suggest that the existence of isomeric plastomes and shift in their abundance together contribute to the plastome complexity within Cupressoideae.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Yu-Chung Chiang from National Sun Yat-Sen University for help with collecting plant materials; and Zhi-Rong Zhang, Si-Yun Chen, Jun-Bo Yang and Hong-Tao Li at the laboratory of the Germplasm Bank of Wild Species, Kunming Institute of Botany for technical assistance. We gratefully acknowledge anonymous referees for helpful comments and valuable suggestions. This work was financially supported by the National Key Basic Research Program of China (grant no. 2014CB954100-01) and the Talent Project of Yunnan Province (project no. 2011CI042) and in part by a research grant from the Ministry of Science and Technology, Taiwan (MOST 103-2621-B-001-007-MY3). This study was facilitated by the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences.
Literature Cited
- Alverson AJ, et al. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 27:1436–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader DA, Moret BME, Yan M.. 2001. A linear-time algorithm for computing inversion distance between signed permutations with an experimental study. J Comput Biol. 8:483–491. [DOI] [PubMed] [Google Scholar]
- Barraclough TG, Harvey PH, Nee S.. 1996. Rate of rbcL gene sequence evolution and species diversification in flowering plants (angiosperms). Proc R Soc Lond B. 263:589–591. [Google Scholar]
- Barraclough TG, Savolainen V.. 2001. Evolutionary rates and species diversity in flowering plants. Evolution 55:677–683. [DOI] [PubMed] [Google Scholar]
- Bromham L, Hua X, Lanfear R, Cowman PF.. 2015. Exploring the relationships between mutation rates, life history, genome size, environment, and species richness in flowering plants. Am Nat. 185:507–524. [DOI] [PubMed] [Google Scholar]
- Christenhusz MJM, Byng JW.. 2016. The number of known plants species in the world and its annual increase. Phytotaxa 261:201–217. [Google Scholar]
- Darling AE, Mau B, Perna NT.. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772–772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Duchene D, Bromham L.. 2013. Rates of molecular evolution and diversification in plants: chloroplast substitution rates correlate with species-richness in the Proteaceae. BMC Evol Biol. 13:65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjon A. 2005. A monograph of Cupressaceae and Sciadopitys. Richmond, Surrey: Royal Botanic Gardens, Kew. [Google Scholar]
- Gadek PA, Alpers DL, Heslewood MM, Quinn CJ.. 2000. Relationships within Cupressaceae sensu lato: a combined morphological and molecular approach. Am J Bot. 87:1044–1057. [PubMed] [Google Scholar]
- Goldman N, Yang Z.. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 11:725–736. [DOI] [PubMed] [Google Scholar]
- Gong XS, Zeng FH, Yan LF.. 1994. An efficient method for the purification of chloroplast DNA from higher plants. J Wuhan Bot Res. 12:277–280. [Google Scholar]
- Goremykin VV, Salamini F, Velasco R, Viola R.. 2009. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol. 26:99–110. [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JNV, Boore JL, Jansen RK.. 2008. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc Natl Acad Sci U S A. 105:18424–18429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK.. 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 28:583–600. [DOI] [PubMed] [Google Scholar]
- Guo WH, et al. 2014. Predominant and substoichiometric isomers of the plastid genome coexist within Juniperus plants and have shifted multiple times during cupressophyte evolution. Genome Biol Evol. 6:580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon C, Maliga P.. 2014. Two distinct plastid genome configurations and unprecedented intraspecies length variation in the accD coding region in Medicago truncatula. DNA Res. 21:417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CY, Wu CS, Chaw SM.. 2016. Birth of four chimeric plastid gene clusters in Japanese umbrella pine. Genome Biol Evol. 8:1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, et al. 2005. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 395:348–384. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Ruhlman TA.. 2012. Plastid genomes of seed plants In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Dordrecht: Springer; p. 103–126. [Google Scholar]
- Kelchner SA, Wendel JF.. 1996. Hairpins create minute inversions in non-coding regions of chloroplast DNA. Curr Genet. 30:259–262. [DOI] [PubMed] [Google Scholar]
- Kim KJ, Choi KS, Jansen RK.. 2005. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol Biol Evol. 22:1783–1792. [DOI] [PubMed] [Google Scholar]
- Kim KJ, Lee HL.. 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11:247–261. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T, Huq E, Herrin DL.. 2010. Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc Natl Acad Sci U S A. 107:13954–13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HL, Jansen RK, Chumley TW, Kim KJ.. 2007. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol. 24:1161–1180. [DOI] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabberley DJ. 2008. Mabberley’s plant-book: a portable dictionary of plants, their classification, and uses. Cambridge: Cambridge University Press. [Google Scholar]
- Mao KS, et al. 2012. Distribution of living Cupressaceae reflects the breakup of Pangea. Proc Natl Acad Sci U S A. 109:7793–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A, Brisson N.. 2010. Recombination and the maintenance of plant organelle genome stability. New Phytol. 186:299–317. [DOI] [PubMed] [Google Scholar]
- Martin G, Baurens FC, Cardi C, Aury JM, D’Hont A.. 2013. The complete chloroplast genome of banana (Musa acuminata, Zingiberales): insight into plastid monocotyledon evolution. PLoS One 8:e67350.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GE, et al. 2014. The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann Bot. 113:1197–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massouh A, et al. 2016. Spontaneous chloroplast mutants mostly occur by replication slippage and show a biased pattern in the plastome of Oenothera. Plant Cell 28:911–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moret BME, Tang JJ, Wang LS, Warnow T.. 2002. Steps toward accurate reconstructions of phylogenies from gene-order data. J Comput Syst Sci. 65:508–525. [Google Scholar]
- Pagel M, Venditti C, Meade A.. 2006. Large punctuational contribution of speciation to evolutionary divergence at the molecular level. Science 314:119–121. [DOI] [PubMed] [Google Scholar]
- Palmer JD. 1983. Chloroplast DNA exists in two orientations. Nature 301:92–93. [Google Scholar]
- Palmer JD. 1991. Plastid chromosomes: structure and evolution In: Hermann RG, editor. The molecular biology of plastids: cell culture and somatic cell genetics of plants. Vienna: Springer; p. 5–53. [Google Scholar]
- Patel RK, Jain M.. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raubeson LA, Jansen RK.. 2005. Chloroplast genomes of plants In: Henry RJ, editor. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. Cambridge: CABI; p. 45–68. [Google Scholar]
- Schattner P, Brooks AN, Lowe TM.. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard AE, Timmis JN.. 2009. Instability of plastid DNA in the nuclear genome. PLoS Genet. 5:e1000323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Donoghue MJ.. 2008. Rates of molecular evolution are linked to life history in flowering plants. Science 322:86–89. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Lonsdale DM.. 1982. Mitochondrial and chloroplast genomes of maize have a 12-kilobase DNA-sequence in common. Nature 299:698–702. [DOI] [PubMed] [Google Scholar]
- Sudianto E, Wu CS, Lin CP, Chaw SM.. 2016. Revisiting the plastid phylogenomics of Pinaceae with two complete plastomes of Pseudolarix and Tsuga. Genome Biol Evol. 8:1804–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura Y, Suyama Y, Yoshimura K.. 2000. Chloroplast DNA inversion polymorphism in populations of Abies and Tsuga. Mol Biol Evol. 17:1302–1312. [DOI] [PubMed] [Google Scholar]
- Venditti C, Pagel M.. 2010. Speciation as an active force in promoting genetic evolution. Trends Ecol Evol. 25:14–20. [DOI] [PubMed] [Google Scholar]
- Wang D, et al. 2007. Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 MYA. Mol Biol Evol. 24:2040–2048. [DOI] [PubMed] [Google Scholar]
- Wang RJ, et al. 2008. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 8:36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Ran JH.. 2014. Evolution and biogeography of gymnosperms. Mol Phylogen Evol. 75:24–40. [DOI] [PubMed] [Google Scholar]
- Weng ML, Blazier JC, Govindu M, Jansen RK.. 2014. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol. 31:645–659. [DOI] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Muller KF, Quandt D.. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76:273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Chaw SM.. 2016. Large-scale comparative analysis reveals the mechanisms driving plastomic compaction, reduction, and inversions in conifers II (cupressophytes). Genome Biol Evol. 8:3740–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Lin CP, Hsu CY, Wang RJ, Chaw SM.. 2011. Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 3:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yang ZH. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang ZY, Ran JH, Wang XQ.. 2012. Three genome-based phylogeny of Cupressaceae s.l.: further evidence for the evolution of gymnosperms and Southern Hemisphere biogeography. Mol Phylogen Evol. 64:452–470. [DOI] [PubMed] [Google Scholar]
- Yi X, Gao L, Wang B, Su YJ, Wang T.. 2013. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): evolutionary comparison of Cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol Evol. 5:688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Ma PF, Li DZ.. 2011. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS One 6:e20596.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.