Abstract
Advances in molecular analyses based on high-throughput technologies can contribute to a more accurate classification of non–small cell lung cancer (NSCLC), as well as a better prediction of both the disease course and the efficacy of targeted therapies. Here we set out to analyze whether global gene expression profiling performed in a group of early-stage NSCLC patients can contribute to classifying tumor subtypes and predicting the disease prognosis. Gene expression profiling was performed with the use of the microarray technology in a training set of 108 NSCLC samples. Subsequently, the recorded findings were validated further in an independent cohort of 44 samples. We demonstrated that the specific gene patterns differed significantly between lung adenocarcinoma (AC) and squamous cell lung carcinoma (SCC) samples. Furthermore, we developed and validated a novel 53-gene signature distinguishing SCC from AC with 93% accuracy. Evaluation of the classifier performance in the validation set showed that our predictor classified the AC patients with 100% sensitivity and 88% specificity. We revealed that gene expression patterns observed in the early stages of NSCLC may help elucidate the histological distinctions of tumors through identification of different gene-mediated biological processes involved in the pathogenesis of histologically distinct tumors. However, we showed here that the gene expression profiles did not provide additional value in predicting the progression status of the early-stage NSCLC. Nevertheless, the gene expression signature analysis enabled us to perform a reliable subclassification of NSCLC tumors, and it can therefore become a useful diagnostic tool for a more accurate selection of patients for targeted therapies.
Introduction
Lung cancer is one of the most common causes of cancer-related deaths worldwide. Non–small cell lung cancer (NSCLC) accounts for 85% of all lung cancers and represents a heterogeneous group of malignancies comprised mainly of adenocarcinomas (ACs) and squamous cell carcinomas (SCCs) [1]. Recently, numerous novel targeted therapies have been established as treatment options for patients with nonresectable or metastatic NSCLC [2]. However, despite the significant therapeutic progress, novel targeted anticancer drugs used in distinct NSCLC subtypes presented different levels of efficacy. Notably, targeted lung cancer therapies directed against specific cellular alterations were found to be most successful in patients with nonsquamous tumors [3]. Diagnosis of lung cancer based on the histopathological analysis remains the gold standard. However, this method is seriously flawed due to several important limitations [4], [5], [6]. Therefore, recent advances in personalized targeted lung cancer therapies not only require an accurate histological classification of NSCLC but need to be extended by a more precise characterization of numerous molecular alterations [7], [8], [9]. Precise histopathological and molecular characterization of NSCLC plays a crucial role in the recruitment of patients for novel molecular targeted therapies [10].
Despite the fact that the majority of patients are diagnosed with locally advanced or metastatic disease, thus being eligible for only few therapeutic options [11], still about 15% of patients are candidates for surgical treatment. On the other hand, up to 30% of stage I patients develop recurrences within 5 years after surgical treatment [12]. Although the role of adjuvant chemotherapy in stage I remains controversial, some molecularly categorized high-risk patients may benefit from it. Therefore, it seems clear that there is a need to identify reliable predictors of relapse in order to improve the clinical management of early-stage NSCLC.
In recent years, there was a growing body of evidence that aberrations in the profiles of gene expression played a crucial role in carcinogenesis and progression of many human tumor types, including lung cancer [13]. On the other hand, tumor-specific molecular signatures can contribute to a more effective early detection of asymptomatic lung cancer and a better prediction of the disease course [14].
There are an increasing number of studies demonstrating that particular histotypes of NSCLC display distinct molecular characteristics [15]. Patterns of differential gene expression in lung AC and SCC may indicate the occurrence of different gene-related signaling pathways underlying the pathogenesis of these histologies. This feature has become a basis for numerous studies analyzing the use of specific genes as putative molecular markers defining both cancer type and origin [16], [17]. Indeed, we have recently demonstrated that certain molecular determinants are closely associated with some selected features of histological and genetic characteristics of lung cancer [18].
To date, the use of several combinations of gene expression profiles has not proved its clinical usefulness in prognostication of lung cancer [19]. However, advances in high-throughput next-generation sequencing and the microarray technology stimulate the research in molecular prognostic area and could become a more useful tool for the development of more accurate predictive markers of relapse following surgery [20], [21].
Therefore, the objective of the current study was to perform global gene expression profiling in a large and well-characterized group of patients with completely resected early-stage lung tumors in order to classify the NSCLC subtypes and predict the prognosis of the disease.
Materials and Methods
Patients and Materials
The study was approved by the Bioethical Committee of the Medical University of Bialystok. Informed consent was obtained from each patient. One hundred and fifty-two cases of surgically resected NSCLC were used for the purpose of the study. Inclusion criteria included: original diagnosis of lung AC, SCC, or large cell lung carcinoma (LCC) based on histologic evidence; completely resected tumor (free resection margins); stage I or stage II; a minimum of 3-year follow-up including monitoring for events of cancer recurrence and lung cancer–related death; availability of representative fresh-frozen tumor specimens (the material containing at least 50% of tumor cells for the RNA extraction); and no neoadjuvant chemotherapy. In the first phase of the research, we performed gene expression profiling of 108 NSCLC tumor samples (42 with AC, 56 with SCC, and 10 with LCC), generating a “training” data set (set 1). To confirm the microarray results, gene expression levels were evaluated on a subset of 44 samples (16 with AC, 25 with SCC, and 3 with LCC) as an independent “validation” data set (set 2). Patient subsets were recruited following a temporal order (108 patients were selected between the years 2003 and 2009 for the training set, and 44 patients were selected between 2009 and 2010 for the validation set). All the procedures connected with patient selection for the study, as well as the study design, were conducted according to the standard operating procedures, which were closely maintained throughout the time of the experiment. To evaluate differences in the gene expression profiles between different tumor subtypes, we focused on lung AC and SCC due to the relatively low numbers of LCC patients, especially in the validation set. With respect to clinical characteristics (age, gender, tumor histology, disease stage, and progression status), both groups were comparable (Table 1).
Table 1.
Patient Characteristics for the Training Set (n = 108) and the Validation Set (n = 44)
| Characteristic | Set 1, n = 108 | Set 2, n = 44 | All, n = 152 | P Values* | |
|---|---|---|---|---|---|
| Age (years) | Mean ± SD | 62.27 ± 8.36 | 64.78 ± 8.32 | 62.99 ± 8.39 | .095 |
| Median | 62.92 | 64.30 | 63.51 | ||
| Range | 39.83-78.08 | 46.3-78.8 | 39.83-78.8 | ||
| Gender | Female | 22 (20%) | 10 (23%) | 32 (21%) | .747 |
| Male | 86 (80%) | 34 (77%) | 120 (79%) | ||
| Histology | SCC | 56 (52%) | 25 (57%) | 81 (53%) | .813 |
| AC | 42 (39%) | 16 (36%) | 58 (38%) | ||
| LCC | 10 (9%) | 3 (7%) | 13 (9%) | ||
| Tumor stage | IA | 21 (19%) | 8 (18%) | 29 (19%) | .806 |
| IB | 30 (28%) | 11 (25%) | 41 (27%) | ||
| IIA | 24 (22%) | 8 (18%) | 32 (21%) | ||
| IIB | 33 (31%) | 17 (39%) | 50 (33%) | ||
| Progression at 3 years | Yes | 45 (42%) | 14 (32%) | 59 (39%) | .258 |
| No | 63 (58%) | 30 (68%) | 93 (61%) | ||
SD, standard deviation. Progression at 3 years: yes: recurrence and/or cancer-related death at 3 years; no: free from recurrence and/or cancer-related death at 3 years.
P values were calculated with Pearson's chi-squared test of independence; independent-samples t test for age.
Histologic Diagnosis
For all the samples used in the study, histologic diagnosis was based on medical records from the archival pathology files of the University Clinical Hospital in Bialystok and additionally validated: hematoxylin and eosin–stained slides of all of the cases were independently reevaluated by two experienced pathology experts (J.R. and L.C.). Histologic diagnosis was rendered according to the most recent WHO classification of tumors of the lung [22] and the IASLC/ATS/ERS International Multidisciplinary Lung Adenocarcinoma Classification [23]. In case of any disagreement with the original diagnosis, the slides were evaluated immunohistochemically (IHC) for the expression of thyroid transcription factor-1 (TTF-1) (immunohistochemical profile in ACs) and tumor protein p63 (p63) (squamous immunophenotype). Additionally, all tumor slices were reviewed to evaluate the amount of neoplastic cells for the RNA extraction.
RNA Extraction and Quality Control
Total RNA was isolated from fresh-frozen tumor samples using mirVana miRNA Isolation Kit (Ambion, Austin, TX) following the manufacturer's protocol. RNA quantity and quality were assessed using a UV/VIS spectrophotometer NanoDrop 2000c (Thermo Scientific, Wilmington, DE). The level of integrity required for the microarray analysis (RNA Integrity Number above 7) was determined for the extracted total RNA using Agilent RNA 6000 Nano Kit on apparatus Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA).
Microarray Analysis
All samples were analyzed on two-color Agilent Human Gene Expression v2 4x44K Microarray (Agilent Technologies, Santa Clara, CA). Tumor samples were hybridized against a lung cancer common reference pool that consisted of a pool of RNA derived from 108 of the NSCLC samples. Total RNA in the amount of 1000 ng from tumor sample and common reference sample was labeled with a Cyanine 3-pCp and a Cyanine 5-pCp fluorophores, respectively. Labeling reactions were performed using Agilent's Quick Amp Labeling Kit, two-color with the use of synthetic spike controls – RNA Spike-In Kit, two color, following the manufacturer's protocol (Agilent Technologies, Santa Clara, CA). Hybridization of the labeled RNA in the presence of spike controls was performed in SureHyb chambers (Agilent Technologies, Santa Clara, CA) for 17 hours at 65°C. We used the Spike-In solutions to help distinguish significant biological data from processing issues. Slides were washed using the Gene Expression Wash Buffer Kit (Agilent Technologies, Santa Clara, CA) following the manufacturer's instructions and scanned at 5-μm resolution using an Agilent G2505C DNA Microarray Scanner. The scanned slides were quantified using Feature Extraction 10.7.3 software (Agilent Technologies, Santa Clara, CA). Additionally, all the arrays were assessed using the QC Report generated by Agilent's software.
Statistics
All preprocessing steps were executed with the R-package “limma.” The background was corrected by fitting a convolution of normal and exponential distributions to the foreground intensities. Subsequently, two normalization steps were conducted: LOESS, a nonparametric regression method, was used for within-arrays normalization, and a quantile normalization was used for between-arrays normalization. We preprocessed the training data set (108 samples) and the independent validation data set (44 samples).
We used the R-package “limma” to assess the statistical significance of the differences in gene expression levels. Genes with adjusted P values due to multiple testing lower than .05 were taken as differentially expressed using the adjustment procedure of Benjamini-Hochberg [24]. Prediction analysis of microarray ranking (PAMR) and Gene Ontology (GO) analyses were done with R/Bioconductor packages “pamr” and “GOstats,” respectively. PAMR signature was selected with the shrunken parameter set to 3.5. For the overrepresentation analyses, Bonferroni correction was applied, and sets with the P value lower than .05 were called significant. For the survival analysis, a progression-free period of 3-year cutoff was arbitrarily chosen to discriminate between good and poor prognosis patients. The analysis of differential gene expression between recurrence-free and recurrence patients was adjusted for gender, histological subtype, and stage. For each gene, a linear model with the recurrence status after 3 years, gender, histological type, and stage as covariates was fitted. After fitting the models, the differences in gene expression were tested using the Student's t test.
Results
Patient Characteristics
A total of 152 patients were enrolled into the study. Clinical characteristics of the NSCLC patients whose samples were used in the training and validation sets are summarized in Table 1. No statistically significant differences were found in the main patient characteristics of these two sets. The training set included tumor tissues from 108 patients who underwent surgical resection between 2003 and 2009 in the University Clinical Hospital in Bialystok. Forty-four NSCLC cases resected between the years 2009 and 2010 were recruited for an independent validation group.
Gene Expression Profiling to Distinguish Lung AC from SCC Subtypes
A comprehensive evaluation of gene expression levels was focused on lung AC and SCC tissues using gene expression microarray analysis. LCC was not included in this part of the study as the number of samples was too low to obtain representative results.
Initially, we compared gene expression profiles in AC and SCC between the training group (set 1, n = 98) and the validation group (set 2, n = 41) in order to verify whether a consistent estimate of differential expression could be obtained in both sets. A strong association of gene expression profiles was observed between the training and validation sets (Supplementary Figure 1), showing that the analyzed groups of samples were suitable for both training and validation purposes.
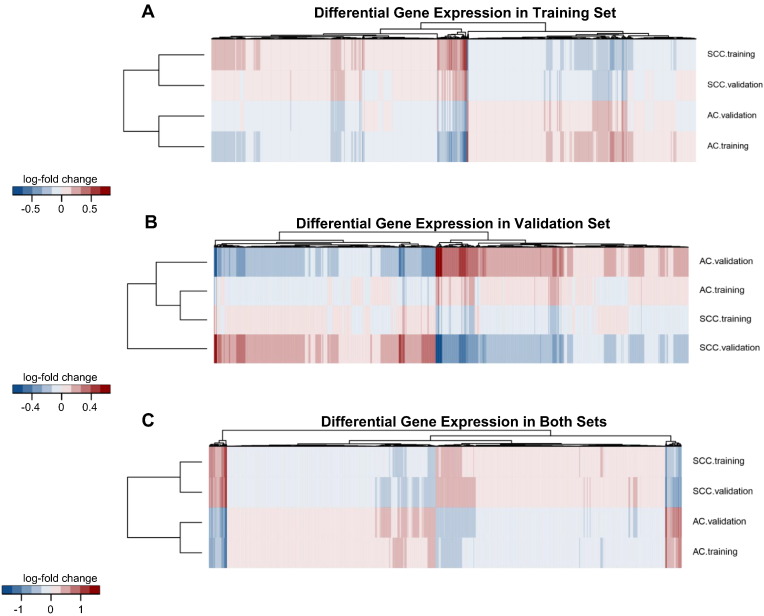
Next, we evaluated differences in gene expression levels between the two histological types of NSCLC. We demonstrated that gene expression profiles in lung AC and SCC differed significantly from each other in both the training and validation sets (Figure 1, A and B). The heat maps showed specific gene subclusters that were differentially expressed between the AC and SCC tumors and could therefore prove useful in the molecular classification of NSCLC subtypes. We identified 4752 genes whose expression levels differed significantly between the AC and SCC samples analyzed in the training set (adjusted P value < .05). More specifically, 2496 genes were upregulated in SCC and 2256 genes were upregulated in AC (Supplementary Materials, Appendix 1). In the analysis of the independent validation data set, a statistically significant difference (adjusted P value < .05) was found in the expression of 3504 genes (1780 genes were overexpressed in SCC, and 1724 were overexpressed in AC) (Supplementary Materials, Appendix 2). Moreover, we observed that the fold changes in the expression of 2549 genes were statistically significant (adjusted P value < .05) in both data sets (Figure 1C). The relative expression levels of 1334 genes were upregulated in SCC, whereas 1215 genes were upregulated in AC (Supplementary Materials, Appendix 3). However, it was quite surprising to note that the gene expression changes between AC and SCC that were found to be statistically significant only in the training data set were numerically not identical with the fold changes found in the validation set despite the fact that, in the vast majority, these gene expression changes had a consistent pattern (Figure 1A). This consistency was also observed in the validation data set. The majority of genes that were significantly differentially expressed between the AC and SCC samples in the validation set had the same direction of the expression change in the training set (Figure 1B). The names of the genes that were differentially expressed between the two histological NSCLC samples in the training, validation, and both sets are listed in Appendix 1, Appendix 2, Appendix 3.
Figure 1.
Hierarchical clustering of fold changes expression for the genes that displayed the statistical significance (adjusted P value < .05) for differential expression between AC and SCC samples in the training set (A), in the validation set (B), and in both sets (C). Columns correspond to individual genes, and rows represent AC or SCC tumors in appropriate group of the analyzed samples. For comparison of the gene expression between two data sets of samples, each heat map shows fold changes expression in both the training and validation sets. (A) The top and bottom rows represent expression data for genes significantly altered in AC samples compared with SCC samples (adjusted P value < .05) in the training set. Two middle rows correspond to expression levels of the same genes in the validation set. Despite the lack of statistically significant differences between AC and SCC in this set of samples, most of the genes have consistent sign with respect to training set. (B) The top and bottom rows displayed the differential expression pattern in SCC and AC samples (adjusted P value < .05) in the validation set. Two middle rows also reveal that the great majority of the genes in the training set have the same direction of change in expression compared with validation set. (C) Heat map of the statistically significant (adjusted P value < .05) results for differential gene expression profiles in AC and SCC samples in both sets. The scale represents the intensity of fold changes expression (log2 scale).
Histotypic Gene Signature to Distinguish SCC from AC
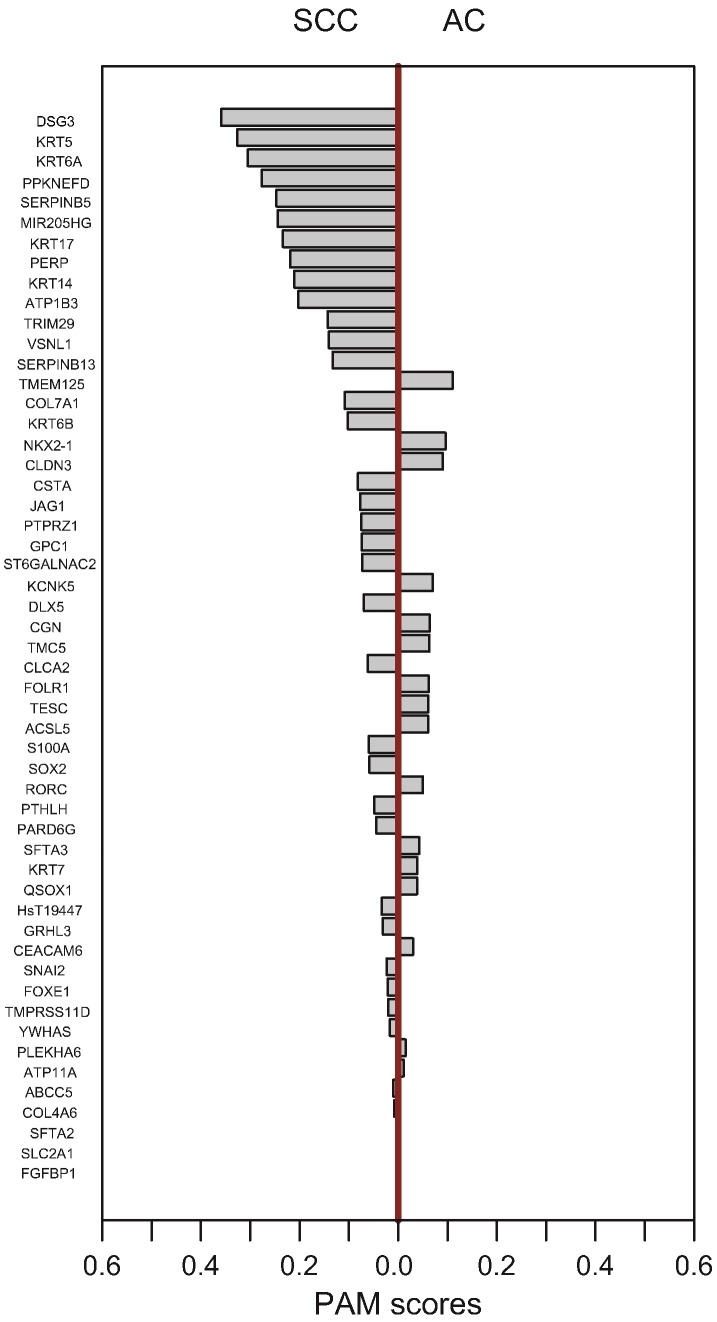
In order to identify a set of gene expression markers to best discriminate between the AC and SCC subtypes, we used the prediction analysis of microarray (PAM) method described in the study of Tibshirani and colleagues based on the nearest shrunken centroid classification algorithm [25]. The gene expression classifier was developed with a selected shrinkage threshold by training performed on the two classes in the training set. More specifically, the optimal number of genes in the predictor was chosen based on a visual analysis of the training errors and cross-validation results (Figure 2). On this basis, we set the optimal shrinkage threshold to 3.5 for the classification of SCC and AC tumors, minimizing misclassification errors in both the training and cross-validation confusion matrices. When 98 samples from the training set were included in the PAM analysis, we identified 53 gene signatures that accurately distinguished lung SCC from AC (Figure 3). Interestingly, some of the proteins encoded by the genes contained in our signature are already used as immunohistochemical markers in clinical diagnostic procedures and include DSG3 (desmoglein 3), NKX2-1 (TITF1), and high–molecular weight keratins (KRTs) such as KRT7, KRT17, KRT5, and KRT6 (keratin 6A, keratin 6B, keratin 6C). The list of genes constituting the histotypic signature and their corresponding values of PAM class scores in the training set are shown in Supplementary Materials, Appendix 4.
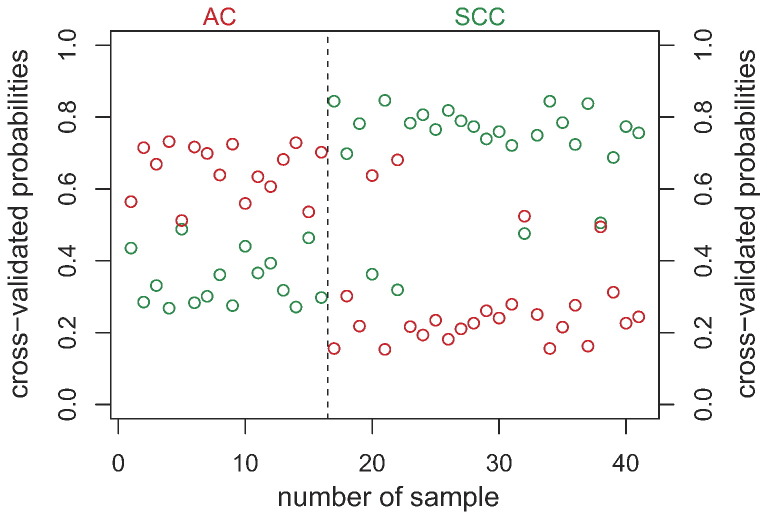
Figure 2.
The gene expression classifier construction using PAM algorithm with adopted shrinkage threshold by training performed on the two classes (AC and SCC histologies) in the training group. The optimal number of genes in the signature was selected based on the minimum number of misclassification errors using cross-validation procedure. The red color represents AC samples, and green color corresponds to SCC tumors.
Figure 3.
Gene expression–based signature for classification of SCC/AC subtypes using PAM method. The shrunken centroid algorithm was used to select 53 genes. The Y axis represents the individual genes according to their class scores, and the X axis displays the SCC and AC histologies.
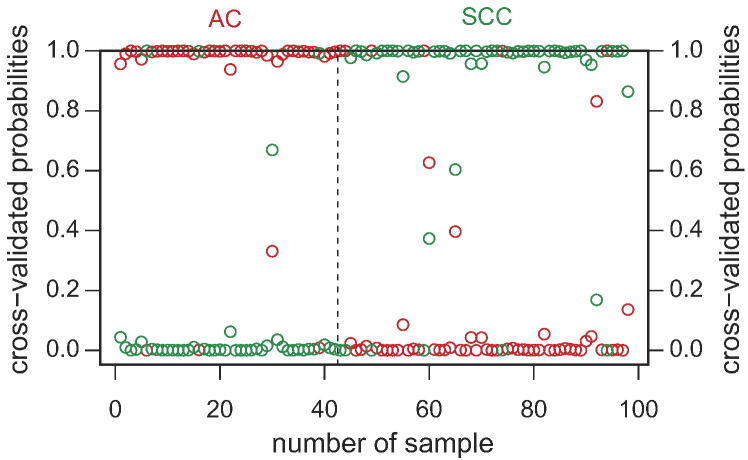
In the subsequent prediction analysis, our classifier was validated in the independent patient cohort (41 tumor samples) using the PAM algorithm with optimized parameters and a threshold established during the training set analysis. The PAM classification analysis showed that lung SCC and AC samples can be correctly classified into their respective groups (Figure 4). The predictor demonstrated a nearly perfect classification with only three SCC tumor samples assigned to the opposed group (93% accuracy). Our classifier was able to flawlessly classify all the AC samples (100% correct prediction for the AC set of samples).
Figure 4.
Classification of SCC/AC subtypes based on the 53-gene signature using PAM algorithm in the validation set. The red color represents AC samples, and the green color displays SCC tumors.
Performance evaluation of the predictive model using the classification results from the independent validation set showed that the 53-gene signature classified the AC patients with a sensitivity of 100% (16/16) and a specificity of 88% (22/25). Further examination of the classifier performance (Table 2) indicated that the 53-gene histotypic classifier was the most accurate with regard to the prediction of AC patients, with a negative predictive value (NPV) of 100% for AC. Classifier identification based on gene expression enabled us to obtain a reliable classification of NSCLC subtypes, and it can therefore constitute a useful diagnostic strategy for patient selection in targeted therapy.
Table 2.
Classification and Performance Evaluation of Predictive Model Using PAM Method in the Validation Cohort
| Classification, Numbers |
Classifier Performance % |
||||||
|---|---|---|---|---|---|---|---|
| Histology | PAM_AC | PAM_SCC | Sensitivity | Specificity | PPV | NPV | Accuracy |
| AC, n = 16 | 16 | 0 | 100 | 88 | 84.2 | 100 | 92.68 |
| SCC, n = 25 | 3 | 22 | |||||
PPV, positive predictive value.
GO Enrichment Analysis
In order to determine the differences between the two histological types of NSCLC at the level of biological processes, we performed a functional enrichment analysis using GO terms. We set out to search for significantly enriched GO terms in the differentially expressed genes (DEGs) from the two lung cancer subtypes in both the training and validation sets. The functional enrichment analysis indicated that the DEGs in the lung SCC group were significantly enriched in a number of biological processes. We identified at least several overrepresented terms in both the training and validation sets that are potentially involved in the pathogenesis of lung SCC such as nucleoplasm, mitotic cell cycle, poly(A) RNA binding, DNA repair, protein sumoylation, and epidermis development (Supplementary Materials, Appendix 5). On other hand, the GO enrichment analysis showed that many of the genes overexpressed in AC may be involved in critical biological processes affected by cancer, including extracellular exosome, plasma membrane, lysosome, and positive regulation of angiogenesis. Other highly ranked categories (adjusted P value < .05) included genes active in the immune response, regulation of the immune response, and the inflammatory response (Supplementary Materials, Appendix 6).
Next, we performed a functional enrichment analysis within the set of 53 histotypic genes constituting our signature in order to understand the crucial biological differences underlying the tumorigenesis of lung AC and SCC. The GO analysis of the 53 identified genes revealed that the histological profile was strongly enriched for genes associated with epidermis development, keratin filament, intermediate filament, structural constituent of cytoskeleton, and extracellular exosome. Significantly (adjusted P value < .05) overrepresented GO terms were identified only in the lung SCC group, most likely due to the relatively high number of genes overexpressed within the signature in SCC. A complete list of the GO categories associated with genes is reported in the Table 3 (detailed information can be found at Supplementary Materials, Appendix 7).
Table 3.
Functional GO Enrichment Analysis for Set of 53 Genes Included in the Histotypic Signature
| Biological Process | Genes Involved | Adj. P Values |
|---|---|---|
| Epidermis development | HsT19447, COL7A1, KRT5, KRT14, KRT17, PTHLH, GRHL3 | <.0001 |
| Keratin filament | KRT5, KRT6A, KRT6B, KRT14, PPKNEFD | <.0001 |
| Intermediate filament | KRT5, KRT6A, KRT14, KRT17, PPKNEFD | <.0001 |
| Structural constituent of cytoskeleton | KRT5, KRT6B, KRT14, KRT17 | <.0001 |
| Extracellular exosome | ATP1B3, CSTA, DSG3, YWHAS, GPC1, KRT5, KRT6A, KRT6B, KRT14, KRT17, SERPINB5, SERPINB13, SLC2A1, TMPRSS11D, PPKNEFD | <.0001 |
Significantly overrepresented GO terms in SCC are linked to genes.
Prognostic Significance of Gene Expression in NSCLC Patients
With regard to the potential role of the genes as prognostic factors in NSCLC, we examined the impact of gene expression levels in primary tumors on the progression status in NSCLC patients following surgical treatment. A comparison of patients with and without recurrence and/or cancer-related death within 3 years (at the 5% false discovery rate level) revealed no statistically significant genes in either the training or validation set, regardless of taking into account or neglecting the clinical factors.
Discussion
Several recent studies showed that global gene expression profiling could be exploited for both the histological and prognostic characterization of NSCLC [14], [26], [27]. In the current study, we used the microarray technology in order to discover gene expression differences among lung cancer histologies and disease progression status.
In the differential gene expression analysis between the samples derived from SCC and AC, we identified the specific gene patterns that clearly differed from each other in both the training and validation set. On this basis, using the training cohort, we developed the 53-gene classifier for identifying individual tumor subtypes. The validation procedure in the independent set of samples showed that our predictor distinguished SCC from AC with 93% accuracy. The gene expression–based signature for AC subtyping demonstrated a nearly perfect classification with 100% sensitivity and 88% specificity. To the best of our knowledge, this is currently one of the best predictive models showing such an excellent classification performance. Importantly, the 53-gene classifier was most accurate with regard to the prediction of AC histology (NPV: 100%). Our findings can thus prove to be of clinical importance as the targeted lung cancer therapies were found most successful in patients with lung AC.
Even though the diagnosis of lung cancer based on the histopathological analysis remains the gold standard, this method has some well-known difficulties and limitations. This is especially evident in the case of small tumor specimens and/or limited amount of cytological smears achieved from unresectable tumors [4], [6]. Diagnostic difficulties arise from the heterogeneity of the tumors, an insufficient number of cancer cells, poor NSCLC differentiation, and seizure of the tissue architecture [28]. Recent studies evaluating the degree of reproducibility of the histological classification performed in resected lung tumors have shown to be unreliable since one third of the cases was classified incorrectly [5]. Unfortunately, immunohistochemical staining (IHC) of lung tumors can improve the accuracy of classification only to a limited extent. To date, p63 has been recognized as the best single marker distinguishing AC from SCC (84% sensitivity and 85% specificity). A similar role can be played by a panel of four factors: p63, CK5/6, TTF1, and Napsin A [29]. IHC methods may be limited by the variability of the staining reactions caused by the heterogeneity of the tumor and some technical discrepancies, different sensitivities/specificities of individual markers, and the lack of standardization in the quantitative interpretation of the staining results [30]. Thus, there is an urgent need for the discovery of novel biomarkers to discriminate more precisely between SCC and AC.
Recently, several studies have attempted to identify the molecular factors which can become useful diagnostic markers allowing a more detailed stratification of NSCLC subtypes [31], [32], [33]. Girard and colleagues developed a combined 62-gene expression signature, including 42 genes for AC/SCC and 20 genes for nonmalignant/lung cancer discrimination [32]. Validation performed with the use of the TCGA and other public data sets resulted in high prediction accuracies (93%-95%). Nevertheless, our signature has a slight advantage over Girard's classifier. We obtained a 100% correct prediction for the AC set of samples. The authors also compared their results with six other signatures, indicating that 10% to 45% of genes were shared. However, the results of our study are in line with the data on the histotypic-associated genes that were identified by Girard and colleagues. Similarly to their findings, we confirmed that 17 genes including CLDN3, RORC, TESC, KCNK5, NKX2-1, ACSL5, SFTA2 (overexpressed in AC) and COL7A1, KRT6C, SERPINB5, VSNL1, DSG3, CLCA-2, KRT17, KRT6B, KRT6A, KRT5 (overexpressed in SCC) could be used to differentiate the two main histological types of lung cancer groups. Moreover, the estimated sign of the fold changes of all the genes identified in our study was completely consistent with the one observed by Girard and colleagues. Interestingly, some of the proteins encoded by the genes included in our and Girard's signature are already used as immunohistochemical markers in clinical diagnostic assays, such as DSG3 (desmoglein 3), NKX2-1 (TITF1), KRT7, KRT17, KRT5, and KRT6.
Hou and colleagues have presented a 75-gene signature for the classification of NSCLC subtypes, which was subsequently subjected to validation with the use of an external data set with the prediction accuracy at the level of 83% [33]. Similar results were obtained by Wilkerson and colleagues, who demonstrated a predictor comprising 51 unique genes [34]. They reported the prediction accuracy to stand at the level of 84%, as estimated by the Monte Carlo cross-validation procedure. It is worth noting that our group has identified and validated a gene signature associated with the histology of lung cancer, which allowed a faultless classification (NPV: 100%) of all the AC tumors. This finding seems to be very important since the efficacy of targeted lung cancer therapies appears limited to lung ACs harboring the oncogenic driver mutations [35], which then determine the further clinical approach. Precision in the classification of lung cancer subtypes should be considered of great importance especially in the case of AC tumors.
In the current study, we performed the functional GO enrichment analysis in order to understand better the critical biological differences underlying the tumorigenesis of lung AC and SCC. We showed that the DEGs in the lung SCC group were significantly enriched in a number of biological processes, such as the by Monte Carlo cross-validation procedure mitotic cell cycle, poly(A) RNA binding, DNA repair, protein sumoylation, and epidermis development. These overrepresented GO terms are potentially involved in the pathogenesis of lung SCC. On the other hand, we demonstrated that many of the genes overexpressed in AC may be involved in altering some of the crucial molecular processes affected by cancer, including the modification of extracellular exosome, plasma membrane, and lysosome and positive regulation of angiogenesis. Interestingly, other highly ranked categories in AC tumors include genes that are active in the processes associated with the immune response. Furthermore, we performed a GO analysis within the genes constituting our histology signature. We revealed the histological profile of the lung SCC to be strongly enriched for the genes linked to epidermis development, keratin filament, intermediate filament, structural constituent of cytoskeleton, and extracellular exosome. Our results showed clearly that the patterns of differential expression of genes in lung AC and SCC may indicate the occurrence of different gene-related biological processes underlying a different pathogenesis of these histologies.
Similar analyses aimed at identifying the differentially expressed genes have been conducted by Lu and colleagues [36]. The authors found that DEGs in an AC subset were not enriched in any specific pathways, whereas DEGs in their SCC samples were enriched in three pathways (Hsa04110, cell cycle; hsa03030, DNA replication; and hsa03430, mismatch repair). Additionally, these researchers constructed a global network of lung cancer with 341 genes and 1569 edges and appointed the top 5 genes, i.e., HSP90AA1, BCL2, CDK2, KIT, and HDAC2, that differentiate lung AC from SCC.
Daraselia and colleagues [37] used the microarray technology and described characteristic molecular networks and subtype-related differences between AC and SCC. They revealed that E2F, CTGF, and PDGF were significantly involved in lung cancer pathogenesis independently of the NSCLC subtype. These researchers showed that the cell cycle–related genes and DNA repair genes were upregulated mainly in SCC, while all the oncogenes in AC and the majority of oncogenes in SCC were downregulated.
In a more recent study, Liu and colleagues [38] defined a set of differentially expressed genes and 16 pathways for AC and SCC. The most significant genes participated in the following pathways: cell cycle (GSK3B, ATR, SKP2, CDK1, CDK2, SMC3, PLK1, CCND3), DNA replication (RFC2, PRIM2, MCM4, MCM5), spliceosome (PRPF19, SRSF2, THOC4), p53 signaling (CDK1, CDK2, GTSE1, ATR), adherent junction (IGF1R, TGFBR2, CTNND1), and tight junction (CKD4, CASK, MPP5). It has been noticed that pathways related to the immune response, metabolism, cell signal transduction, cell division, and proliferation had a higher prevalence in AC compared to SCC. These results support our findings regarding the importance of the processes related to the immune response in AC histology.
In our study, we showed that specific genes could be considered molecular drivers determining the histology of NSCLC. More specifically, the expression levels of SOX2 were significantly higher in lung SCC than in lung AC. According to the previous reports, SOX2 can control tumor initiation and cancer stem-cell functions in SCC [39]. We also found that the overexpression of claudin 3 gene was correlated with the histological diagnosis of AC. Moldvay and colleagues [40] revealed positive immunostaining for claudin 3 only in AC but not SCC cases. The authors suggested that claudin 3 may therefore constitute a diagnostic marker distinguishing ACs from SCCs. Another highly ranked gene in our signature was MIR205HG, which is the host gene for hsa-miR-205. Indeed, we have recently demonstrated that miR-205 is closely associated with the features of squamous histology of lung cancer [18]. Larzabal and colleagues [41] demonstrated for the first time a new intracellular signaling pathway involving two membrane-anchored proteins (ITGα5 and TMPRSS4) that cooperated in tumor growth promotion, metastasis, and migration through miR-205. Surprisingly, we observed that our histology signature also included the gene encoding the transmembrane protease (TMPRSS11D) from the same family of transmembrane serine proteases (TTSPs) as TMPRSS4 in Larzabal's studies. In line with their results, we also showed that both MIR205HG and TMPRSS11D were highly expressed in SCC compared to the AC samples. Our findings indicate that some of the top genes in the signature, such as CLDN3, TMPRSS11D, SERPINB5, and FGFBP1, could become good candidates for new immunostains in the pathology-based differential diagnosis of NSCLC subtypes.
With the hypothesis that the “intrinsic” molecular features of neoplastic cells may predict tumor progression [42], we set out to search for the prognostic significance of the gene expression profiles in NSCLC patients following surgical treatment. A comparison of patients with and without recurrence and/or cancer-related death within 3 years (at the 5% false discovery rate level) revealed no statistically significant genes in either the training or validation set. Altogether, the majority of previously published papers used the overall survival or the relapse-free survival as the main end-point measurements, both being quite insensitive to the prognostic test development [43]. Therefore, in the current study, we set out to perform a correlation analysis between the mRNA expression levels and the progression status of our NSCLC patients.
Although several studies have demonstrated a correlation between the mRNA expression levels and the disease recurrence, they found no repeatable gene expression patterns [44], [45], [46]. The problems with consistency could be linked with several limitations of the experimental design and such technical reasons as the type of technology used (microarray versus next-generation sequencing versus real-time polymerase chain reaction), type of tested material (fresh-frozen versus FFPE tissues), and methods of statistical analysis. Despite these methodological concerns, such biological parameters as histological subtypes, differential treatments, and varying disease stages play crucial roles in regulating differences in the gene expression levels [47]. Notably, our results indicated that the analysis of gene expression profiles in the early stages of NSCLC—AC, SCC, and LCC—does not bear a potential to predict relapse after surgery. On the other hand, Chang and colleagues [44] showed a significant association between the mRNA expression levels and survival only in a group of AC patients. These investigators found a 21-gene signature in the HMGB1/RAGE signaling pathway and a 31-gene signature in the clathrin-coated vesicle cycle pathway which were significantly associated with the prognosis of lung AC patients. Lu and colleagues [45] identified a reproducible gene expression signature in lung AC. Not only did the 16-gene signature reproduce the gene signatures presented in literature, but it also was able to identify a set of predictive genes for AC. However, the authors demonstrated that the above-mentioned genes could not serve as prognostic markers in patients with other NSCLC subtypes.
Roepman and colleagues [46] developed and validated a 72-gene classifier for early-stage NSCLC with a high and low risk of disease recurrence. This method was shown to be statistically significant for both SCC and AC histology. The comparison of their data with seven other publications demonstrated that the accuracy of all the classifiers stood at a similar level, ranging from 70% to 80%. Nevertheless, different gene sets were identified as prognostic, and the profiles of only 5 out of 327 genes overlapped between the two studies [48]. However, the bottleneck of this approach seems to be the validation procedure related to the complex classifier models and the initial revalue of the classifier efficiency.
In summary, we developed and validated here a novel 53-gene expression-based predictor distinguishing SCC from AC with nearly perfect accuracy. To the best of our knowledge, this is currently one of the best predictive models demonstrating such a high classification performance. Our results indicated that gene expression profiles in the early stages of NSCLC may help elucidate the histological distinctions of NSCLC tumors via identification of different gene-mediated biological processes involved in the pathogenesis of histologically distinct tumors. Moreover, we demonstrated that the analysis of gene expression profiles does not bear the capacity to predict the progression status of the early-stage NSCLC. Altogether, molecular tests based on the gene expression allow a reliable subclassification of NSCLC tumors and can thereby constitute a useful diagnostic tool for more effective patient recruitment for targeted therapies.
The following are the supplementary data related to this article.
Correlation analysis showed a strong association of gene expression profiles between the training and validation sets, demonstrating that analyzed groups of samples are suitable for training and validation purposes.
The differentially expressed genes (DEGs) between the AC and SCC tumors in the training set.
The differentially expressed genes (DEGs) between the AC and SCC tumors in the validation set.
The differentially expressed genes (DEGs) between the AC and SCC tumors in both the training and validation sets.
The list of genes constituting the histotypic signature and their corresponding values of PAM class scores in the training set.
Significantly overrepresented GO terms in the differentially expressed genes (DEGs) from the SCC, in both the training and validation sets.
Significantly enriched GO terms in the differentially expressed genes (DEGs) from the AC, both in the training and validation sets.
Functional GO enrichment analysis for set of 53 genes included in the histotypic signature. The list of the GO categories in SCC associated with genes.
Conflicts of Interest
The authors disclose no potential conflicts of interest.
Acknowledgement
We thank Prof. Lech Chyczewski from the Department of Medical Pathomorphology, Medical University of Bialystok, for his professional help in the histopathological analysis. We gratefully acknowledge Dr. Jakub Mieczkowski for the expert statistical analysis.
Footnotes
Funding: This work was supported by the National Science Centre [grant no. NN401 3785 39], Leading National Research Centre (KNOW) [Cancer/Mutagenesis research area]. Equipment used in this study was purchased by the Medical University of Bialystok as part of the OP DEP 2007-2013, Priority Axis I.3, Contract No. POPW.01.03.00-20-022/09.
References
- 1.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 2.Manegold C. Treatment algorithm in 2014 for advanced non–small cell lung cancer: therapy selection by tumour histology and molecular biology. Adv Med Sci. 2014;59:308–313. doi: 10.1016/j.advms.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Chan BA, Hughes BG. Targeted therapy for non–small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res. 2015;4:36–54. doi: 10.3978/j.issn.2218-6751.2014.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorda M, Gomez-Fernandez C, Garcia M, Mousavi F, Walker G, Mejias A, Fernandez-Castro G, Ganjei-Azar P. P63 differentiates subtypes of nonsmall cell carcinomas of lung in cytologic samples: implications in treatment selection. Cancer Cytopathol. 2009;117:46–50. doi: 10.1002/cncy.20015. [DOI] [PubMed] [Google Scholar]
- 5.Stang A, Pohlabeln H, Muller KM, Jahn I, Giersiepen K, Jockel KH. Diagnostic agreement in the histopathological evaluation of lung cancer tissue in a population-based case-control study. Lung Cancer. 2006;52:29–36. doi: 10.1016/j.lungcan.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Khayyata S, Yun S, Pasha T, Jian B, McGrath C, Yu G, Gupta P, Baloch Z. Value of P63 and CK5/6 in distinguishing squamous cell carcinoma from adenocarcinoma in lung fine-needle aspiration specimens. Diagn Cytopathol. 2009;37:178–183. doi: 10.1002/dc.20975. [DOI] [PubMed] [Google Scholar]
- 7.Matsuo N, Azuma K, Sakai K, Hattori S, Kawahara A, Ishii H, Tokito T, Kinoshita T, Yamada K, Nishio K. Association of EGFR exon 19 deletion and EGFR-TKI treatment duration with frequency of T790M mutation in EGFR-mutant lung cancer patients. Sci Rep. 2016;6:36458. doi: 10.1038/srep36458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 9.Charkiewicz R, Niklińska W, Zalewski G, Charkiewicz A, Kozlowski M, Sulewska A, Chyczewski L. New monoallelic combination of KRAS gene mutations in codons 12 and 13 in the lung adenocarcinoma. Adv Med Sci. 2013;58:83–89. doi: 10.2478/v10039-012-0080-0. [DOI] [PubMed] [Google Scholar]
- 10.Greenhalgh J, Dwan K, Boland A, Bates V, Vecchio F, Dundar Y, Jain P, Green JA. First-line treatment of advanced epidermal growth factor receptor (EGFR) mutation positive non-squamous non–small cell lung cancer. Cochrane Database Syst Rev. 2016;5 doi: 10.1002/14651858.CD010383.pub2. [DOI] [PubMed] [Google Scholar]
- 11.Al-Farsi A, Ellis PM. Treatment paradigms for patients with metastatic non–small cell lung cancer, squamous lung cancer: first, second, and third-line. Front Oncol. 2014;4:157. doi: 10.3389/fonc.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uramoto H, Tanaka F. Recurrence after surgery in patients with NSCLC. Transl Lung Cancer Res. 2014;3:242–249. doi: 10.3978/j.issn.2218-6751.2013.12.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grigoroiu M, Tagett R, Draghici S, Dima S, Nastase A, Florea R, Sorop A, Ilie V, Bacalbasa N, Tica V. Gene-expression profiling in non–small cell lung cancer with invasion of mediastinal lymph nodes for prognosis evaluation. Cancer Genomics Proteomics. 2015;12:231–242. [PubMed] [Google Scholar]
- 14.Välk K, Vooder T, Kolde R, Reintam MA, Petzold C, Vilo J, Metspalu A. Gene expression profiles of non–small cell lung cancer: survival prediction and new biomarkers. Oncology. 2010;79:283–292. doi: 10.1159/000322116. [DOI] [PubMed] [Google Scholar]
- 15.Pikor LA, Ramnarine VR, Lam S, Lam WL. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer. 2013;82:179–189. doi: 10.1016/j.lungcan.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 16.Rose-James A, Tt S. Molecular markers with predictive and prognostic relevance in lung cancer. Lung Cancer Int. 2012;2012:729532. doi: 10.1155/2012/729532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su Y, Pan L. Identification of logic relationships between genes and subtypes of non–small cell lung cancer. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charkiewicz R, Pilz L, Sulewska A, Kozlowski M, Niklinska W, Moniuszko M, Reszec J, Manegold C, Niklinski J. Validation for histology-driven diagnosis in non–small cell lung cancer using hsa-miR-205 and hsa-miR-21 expression by two different normalization strategies. Int J Cancer. 2016;138:689–697. doi: 10.1002/ijc.29816. [DOI] [PubMed] [Google Scholar]
- 19.Massuti B, Sanchez JM, Hernando-Trancho F, Karachaliou N, Rosell R. Are we ready to use biomarkers for staging, prognosis and treatment selection in early-stage non–small-cell lung cancer? Transl Lung Cancer Res. 2013;2:208–221. doi: 10.3978/j.issn.2218-6751.2013.03.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang HL, Wu YC, Su LJ, Huang YJ, Charoenkwan P, Chen WL, Lee HC, Chu WC, Ho SY. Discovery of prognostic biomarkers for predicting lung cancer metastasis using microarray and survival data. BMC Bioinformatics. 2015;16:54. doi: 10.1186/s12859-015-0463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma J, Mannoor K, Gao L, Tan A, Guarnera MA, Zhan M, Shetty A, Stass SA, Xing L, Jiang F. Characterization of microRNA transcriptome in lung cancer by next-generation deep sequencing. Mol Oncol. 2014;8:1208–1219. doi: 10.1016/j.molonc.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, editors. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon; IARC Press: 2004. [10 pp.] [Google Scholar]
- 23.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 25.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R, Fárez-Vidal ME. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129:355–364. doi: 10.1002/ijc.25704. [DOI] [PubMed] [Google Scholar]
- 27.Der SD, Sykes J, Pintilie M, Zhu CQ, Strumpf D, Liu N, Jurisica I, Shepherd FA, Tsao MS. Validation of a histology-independent prognostic gene signature for early-stage, non–small-cell lung cancer including stage IA patients. J Thorac Oncol. 2014;9:59–64. doi: 10.1097/JTO.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 28.Field RW, Smith BJ, Platz CE, Robinson RA, Neuberger JS, Brus CP, Lynch CF. Lung cancer histologic type in the surveillance, epidemiology, and end results registry versus independent review. J Natl Cancer Inst. 2004;96:1105–1107. doi: 10.1093/jnci/djh189. [DOI] [PubMed] [Google Scholar]
- 29.Terry J, Leung S, Laskin J, Leslie KO, Gown AM, Ionescu DN. Optimal immunohistochemical markers for distinguishing lung adenocarcinomas from squamous cell carcinomas in small tumor samples. Am J Surg Pathol. 2010;34:1805–1811. doi: 10.1097/PAS.0b013e3181f7dae3. [DOI] [PubMed] [Google Scholar]
- 30.Sinna EA, Ezzat N, Sherif GM. Role of thyroid transcription factor-1 and P63 immunocytochemistry in cytologic typing of non–small cell lung carcinomas. J Egypt Natl Canc Inst. 2013;25:209–218. doi: 10.1016/j.jnci.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, Ashkenazi K, Gefen N, Izraeli S, Rechavi G, Pass H. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non–small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 32.Girard L, Rodriguez-Canales J, Behrens C, Thompson DM, Botros IW, Tang H, Xie Y, Rekhtman N, Travis WD, Wistuba II. An expression signature as an aid to the histologic classification of non–small cell lung cancer. Clin Cancer Res. 2016;22:4880–4889. doi: 10.1158/1078-0432.CCR-15-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC. Gene expression-based classification of non–small cell lung carcinomas and survival prediction. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkerson MD, Schallheim JM, Hayes DN, Roberts PJ, Bastien RR, Mullins M, Yin X, Miller CR, Thorne LB, Geiersbach KB. Prediction of lung cancer histological types by RT-qPCR gene expression in FFPE specimens. J Mol Diagn. 2013;15:485–497. doi: 10.1016/j.jmoldx.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farhat FS, Houhou W. Targeted therapies in non–small cell lung carcinoma: what have we achieved so far? Ther Adv Med Oncol. 2013;5:249–270. doi: 10.1177/1758834013492001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu C, Chen H, Shan Z, Yang L. Identification of differentially expressed genes between lung adenocarcinoma and lung squamous cell carcinoma by gene expression profiling. Mol Med Rep. 2016;14:1483–1490. doi: 10.3892/mmr.2016.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daraselia N, Wang Y, Budoff A, Lituev A, Potapova O, Vansant G, Monforte J, Mazo I, Ossovskaya VS. Molecular signature and pathway analysis of human primary squamous and adenocarcinoma lung cancers. Am J Cancer Res. 2012;2:93–103. [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Yang XY, Shi WJ. Identifying differentially expressed genes and pathways in two types of non–small cell lung cancer: adenocarcinoma and squamous cell carcinoma. Genet Mol Res. 2014;13:95–102. doi: 10.4238/2014.January.8.8. [DOI] [PubMed] [Google Scholar]
- 39.Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–250. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 40.Moldvay J, Jäckel M, Páska C, Soltész I, Schaff Z, Kiss A. Distinct claudin expression profile in histologic subtypes of lung cancer. Lung Cancer. 2007;57:159–167. doi: 10.1016/j.lungcan.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 41.Larzabal L, de Aberasturi AL, Redrado M, Rueda P, Rodriguez MJ, Bodegas ME, Montuenga LM, Calvo A. TMPRSS4 regulates levels of integrin α5 in NSCLC through miR-205 activity to promote metastasis. Br J Cancer. 2014;110:764–774. doi: 10.1038/bjc.2013.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weigelt B, Peterse JL, van't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 43.Shen J, Jiang F. Applications of MicroRNAs in the diagnosis and prognosis of lung cancer. Expert Opin Med Diagn. 2012;6:197–207. doi: 10.1517/17530059.2012.672970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang YH, Chen CM, Chen HY, Yang PC. Pathway-based gene signatures predicting clinical outcome of lung adenocarcinoma. Sci Rep. 2015;5:10979. doi: 10.1038/srep10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu TP, Chuang EY, Chen JJ. Identification of reproducible gene expression signatures in lung adenocarcinoma. BMC Bioinformatics. 2013;14:371. doi: 10.1186/1471-2105-14-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roepman P, Jassem J, Smit EF, Muley T, Niklinski J, van de Velde T, Witteveen AT, Rzyman W, Floore A, Burgers S. An immune response enriched 72-gene prognostic profile for early-stage non–small-cell lung cancer. Clin Cancer Res. 2009;15:284–290. doi: 10.1158/1078-0432.CCR-08-1258. [DOI] [PubMed] [Google Scholar]
- 47.Beane J, Spira A, Lenburg ME. Clinical impact of high-throughput gene expression studies in lung cancer. J Thorac Oncol. 2009;4:109–118. doi: 10.1097/JTO.0b013e31819151f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation analysis showed a strong association of gene expression profiles between the training and validation sets, demonstrating that analyzed groups of samples are suitable for training and validation purposes.
The differentially expressed genes (DEGs) between the AC and SCC tumors in the training set.
The differentially expressed genes (DEGs) between the AC and SCC tumors in the validation set.
The differentially expressed genes (DEGs) between the AC and SCC tumors in both the training and validation sets.
The list of genes constituting the histotypic signature and their corresponding values of PAM class scores in the training set.
Significantly overrepresented GO terms in the differentially expressed genes (DEGs) from the SCC, in both the training and validation sets.
Significantly enriched GO terms in the differentially expressed genes (DEGs) from the AC, both in the training and validation sets.
Functional GO enrichment analysis for set of 53 genes included in the histotypic signature. The list of the GO categories in SCC associated with genes.