Abstract
Objective
MicroRNAs (miRs) play important regulatory roles in lipid metabolism. ApoB, as the only essential scaffolding protein in the assembly of very low density lipoproteins, is a target to treat hyperlipidemia and atherosclerosis. We aimed to find out miRs that reduce apoB expression.
Approach and Results
Bioinformatics analyses predicted that hsa-miR-548p can interact with apoB mRNA. MiR-548p or Control miR was transfected in human and mouse liver cells to test its role in regulating apoB secretion and mRNA expression levels. Site-directed mutagenesis was used to identify the interacting site of miR-548p in human apoB 3′-untranslated region. Fatty acid oxidation and lipid syntheses were examined in miR-548p overexpressing cells to investigate its function in lipid metabolism. We observed that miR-548p significantly reduces apoB secretion from human hepatoma cells and primary hepatocytes. Mechanistic studies showed that miR-548p interacts with the 3′-untranslated region of human apoB mRNA to enhance posttranscriptional degradation. Bioinformatics algorithms suggested two potential binding sites of miR-548p on human apoB mRNA. Site-directed mutagenesis studies revealed that miR-548p targets site I involving both seed and supplementary sequences. MiR-548p had no effect on fatty acid oxidation but significantly decreased lipid synthesis in human hepatoma cells by reducing HMGCR and ACSL4 enzymes involved in cholesterol and fatty acid synthesis. In summary, miR-548p reduces lipoprotein production and lipid synthesis by reducing expression of different genes in human liver cells.
Conclusions
These studies suggest that miR-548p regulates apoB secretion by targeting mRNA. It is likely that it could be useful in treating atherosclerosis, hyperlipidemia and hepatosteatosis.
Keywords: ApoB, lipoproteins, lipid synthesis, cholesterol, microRNA, MTP
Subject codes: Lipids and cholesterol, Metabolism
Introduction
MicroRNAs (miRs) are small endogenous non-coding RNAs. Since 1993, when the first miR was identified and cloned from C. elegans (1), the regulatory functions of miRs have been studied extensively in different species (2). Further, significant attempts have been made to test potential use of miRs as therapeutic agents. MiR-122, the most abundant miR expressed in the liver, binds to the 5′-UTR of the Hepatitis C virus genome to positively affect RNA stability and replication (3). Therefore, inhibition of endogenous miR-122 shows efficacy in treating chronic Hepatitis C infection. And, a locked nucleic acid-modified anti-miR-122, miraversin, is in Phase II clinical trials (4–6). MiR-34, which is induced by a tumor suppressor p53, reduces the expression of multiple genes in apoptosis and cell cycle arrest to suppress tumor genesis (7, 8). MRX34, a double-stranded miR-34 mimic has been shown to enhance cancer cell death and inhibit metastasis in a Phase I clinical trial (9). These promising results from preclinical and clinical studies encourage us to seek for miRs that can potentially treat cardiovascular diseases that remain the major cause of morbidity and mortality in western world (10).
Atherosclerosis, the major cause of cardiovascular diseases, occurs due to excess lipid accumulation on the inner wall of arteries (11). Atherosclerotic lesions in major arteries narrow their diameters and reduce blood circulation (12). Several risk factors of atherosclerosis have been identified such as hyperlipidemia, hypertension, smoking, lack of exercise and family history. Hyperlipidemia refers to increases in plasma lipids and lipoproteins. High plasma concentrations of low-density lipoproteins (LDL) and their modified forms, such as oxidized-LDL, contribute to atherosclerosis. And, lowering their concentrations is associated with reduced incidence of cardiovascular disease (13, 14).
Plasma lipid levels can be lowered by increasing their catabolism or decreasing production. Statins and PCSK9 inhibitors reduce plasma LDL by increasing their clearance via upregulating hepatic LDL receptors (15, 16). Hence, these drugs are not useful in the treatment of homozygous familial hypercholesterolemia (HoFH) subjects with null LDL receptor mutations (17, 18). Hypercholesterolemia in these patients can be lowered using drugs that decrease apolipoproteins B (apoB) or microsomal triglyceride transfer protein (MTP); two critical proteins required for the assembly and secretion of very low-density lipoprotein (VLDL) (17, 19, 20). During VLDL assembly, newly translated apoB peptides are lipidated in the endoplasmic reticulum by MTP (19–21). ApoB siRNAs and antisense oligonucleotides (ASOs) reduce lipoprotein production and plasma cholesterol in both rodents and non-human primates (22–25). Mipomersen, a specific apoB ASO, has been approved to treat HoFH (17, 19). Mipomersen reduces plasma cholesterol but is associated with lipid accumulation in the liver (17, 19, 26–29). Similarly, small molecule inhibitors and ASO mediated reductions in MTP lower plasma cholesterol but are also associated with increases in hepatic lipids (19, 20, 30–32). Therefore, novel therapeutic agents that reduce VLDL production while avoiding hepatosteatosis might be useful treatment modalities.
Few miRs have been reported to regulate metabolism of apoB-containing lipoproteins (33, 34). Inhibition of miR-122 results in significant reductions in plasma cholesterol, apoB and triglyceride (35–37). Further, miR-122 knockout mice have lower plasma cholesterol and triglyceride due to reductions in both LDL and HDL. These mice have higher hepatic lipids and plasma alanine aminotransferase (ALT) and alkaline phosphatase and develop hepatocellular carcinoma with age (38, 39). MiR-34a expression reduces lipoprotein production and causes hepatosteatosis (40). Mir34a−/− mice have high plasma triglyceride and cholesterol (40). Recently, we showed that miR-30c reduces MTP expression and lowers plasma cholesterol and atherosclerosis without causing steatosis (41, 42). Mechanistic studies indicated that miR-30c reduces lipoprotein production and lipid synthesis. Moreover, miR-33a/b was identified for its role in down regulating ABCA1 in macrophage to inhibit cholesterol efflux and HDL biosynthesis (43–45). Here, we investigated whether there are miRs that target apoB to regulate lipoprotein production and identified a novel human miR-548p, which targets apoB mRNA, decreases apoB secretion and reduces hepatic lipid synthesis in human hepatoma cells.
Materials and methods
Materials and Methods are available in the on-line Data Supplement.
Results
MiR-548p lowers apoB secretion from human liver cells
Using TargetScan 5.2, we observed that human apoB mRNA has a target site for hsa-miR-548p (Supplementary Fig I) that is conserved in primates, but not in other organisms including dog, mouse, rat, chicken, and frog (Fig 1A). To test whether hsa-miR-548p affects human apoB secretion, different indicated concentrations of control (Ctrl) miR, miR-548p, and anti-548p were transfected in Huh-7 cells (Fig 1B). MiR-548p dose-dependently reduced media and cellular apoB without changing apoAI levels (Fig 1B). Anti-548p, an inhibitor of miR-548p, increased both media and cellular apoB without affecting apoAI secretion indicating the role of endogenous miR-548p in apoB regulation (Fig 1B). Further studies showed that miR-548p reduces media and cellular apoB in a time dependent manner (Fig 1C). The maximum reduction of 67% was observed after 72 hours. Anti-548p increased media and cellular apoB levels without affecting apoAI (Fig 1C). In addition, we observed the same effect of miR-548p on apoB in another human hepatoma cell line, HepG2 (Fig 1D). Forced expression of miR-548p reduced media and cellular apoB, but had no effect on apoAI (Fig 1D). On the other hand, anti-548p increased media and cellular apoB. We also tested the effect of miR-548p on apoB and apoAI secretion in human primary hepatocytes compared to negative control Ctrl. MiR-30c was used as a positive control of transfection efficiency, as it is known to target MTP and reduce apoB secretion (41, 42). MiR-548p significantly reduced apoB secretion from the primary hepatocytes without affecting apoAI (Fig 1E). Thus, miR-548p reduces cellular and media apoB in human liver cells without altering apoAI levels.
Figure 1. MiR-548p decreases cellular and secreted apoB from human liver cells without affecting apoAI secretion.
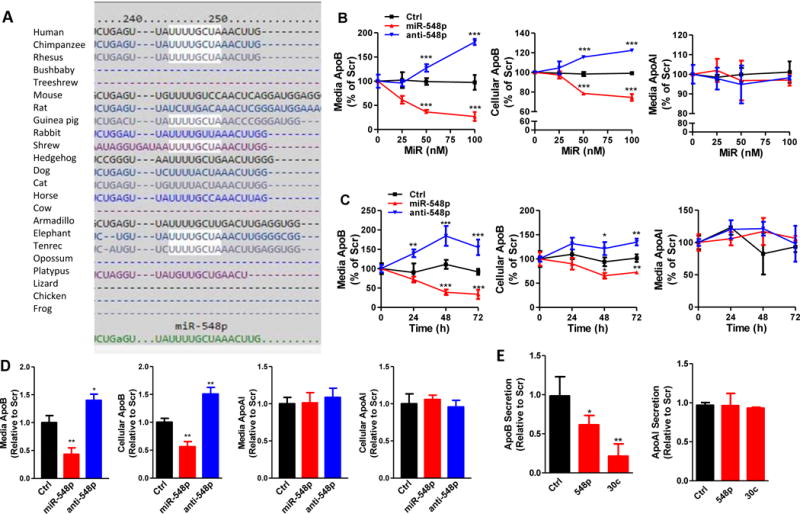
A. TargetScan 6.2 was used to predict putative miRs that could target apoB 3′-UTR. The miR-548p seed sequence interacting site in apoB mRNA is well conserved in primates, including human, chimpanzee and rhesus monkeys, but not in mouse and rat. B. Different amounts of Ctrl, miR-548p, and anti-548p (100 nM) were reverse transfected in Huh-7 cells. Cellular apoB, as well as media apoB and apoAI were quantified using ELISA 48 hours after transfection. The data are representative of two independent experiments. C. Time course experiments in Ctrl, miR-548p, and anti-548p (100 nM) transfected Huh-7 cells. Media and cell lysates were collected at indicate time points to measure secreted and cellular apoB and apoAI. D. MiR-548p (100 nM) decreased apoB secretion in HepG2 cells without affecting apoAI. ApoB and apoAI in cells and media were measured 48 hours after transfection. The data are representative of two independent experiments. E. MiR-548p and miR-30c (100 nM) decreased apoB secretion in human primary hepatocytes without affecting apoAI. * p<0.05, **p<0.01, ***p<0.001. Significance was calculated by two-way ANOVA and student’s t-test.
MiR-548p reduces apoB mRNA levels in human, but not in mouse, liver cells
To begin understand how miR-548p regulates apoB secretion, we quantified apoB, MTP and apoAI mRNA levels in miR-548p, miR-30c and Ctrl transfected human primary hepatocytes. MiR-548p significantly reduced apoB mRNA levels without affecting MTP or apoAI, while miR-30c only reduced MTP mRNA levels (Fig 2A). In Huh-7 cells, miR-548p reduced and anti-548p increased apoB mRNA levels. MiR-548p and anti-548 had no effect on MTP and apoAI mRNA levels (Fig 2B). These studies indicate that miR-548p reduces steady state apoB mRNA levels in liver cells.
Figure 2. MiR-548p reduces apoB mRNA levels in human primary hepatocytes and Huh7 cells.
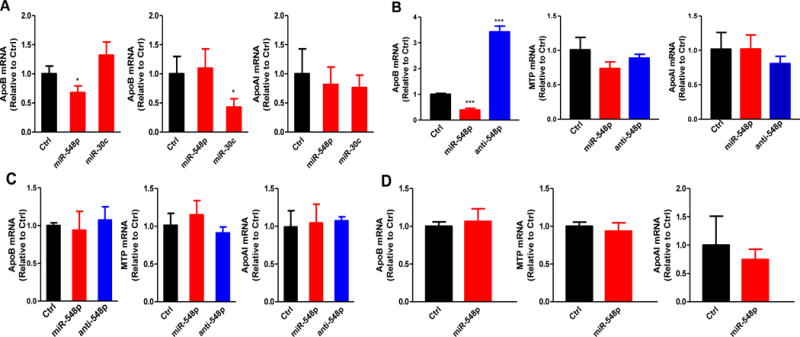
A. In the human primary hepatocytes, miR-548p (100 nM) reduced apoB mRNA levels, miR-30c (100 nM) reduced MTP mRNA levels. B. MiR-548p and anti-miR-548p (100 nM) decreased and increased apoB mRNA levels repectively in Huh-7 cells. The data are representative of two independent experiments. C. MiR-548p or anti-miR-548p (100 nM) has no effect on mouse apoB in mouse primary hepatocytes. D. MiR-548p (100 nM) has no effect on mouse apoB in AML12 cells. The data are representative of two independent experiments. The mRNA expression levels of apoB, MTP or apoAI were quantified after 48 h of transfection. Relative mRNA levels were normalized to 18S. * p<0.05, **p<0.01, ***p<0.001. Significance was calculated by student’s t-test.
Since mouse is the most widely used animal model to study physiological effects of genetic manipulations and drug treatments, we transfected miR-548p in mouse primary hepatocytes (Fig 2C) and in mouse hepatoma AML12 cells (Fig 2D). MiR-548p and anti-548p had no effect on mouse apoB, MTP or apoAI. This is consistent with the absence of miR-548p interacting site in the 3′-UTR of mouse apoB (Fig 1A). Thus, miR-548p reduces apoB mRNA levels in human liver cells.
MiR-548p interacts with the 3′-UTR of apoB to induce posttranscriptional degradation
MiRs, in general, reduce mRNA levels by interacting with 3′-untranslated region (3′-UTR) of mRNAs and inducing post-transcriptional degradation (2, 46). Therefore, we hypothesize that miR-548p may induce post-transcriptional degradation of apoB mRNA. ApoB mRNA disappeared faster in miR-548p expressing cells than in Ctrl transfected cells treated with actinomycin D to inhibit transcription (Fig 3A). Hence, miR-548p promotes post-transcriptional degradation of apoB mRNA.
Figure 3. MiR-548p interacts with 3′-UTR of apoB mRNA to promote posttranscriptional degradation.
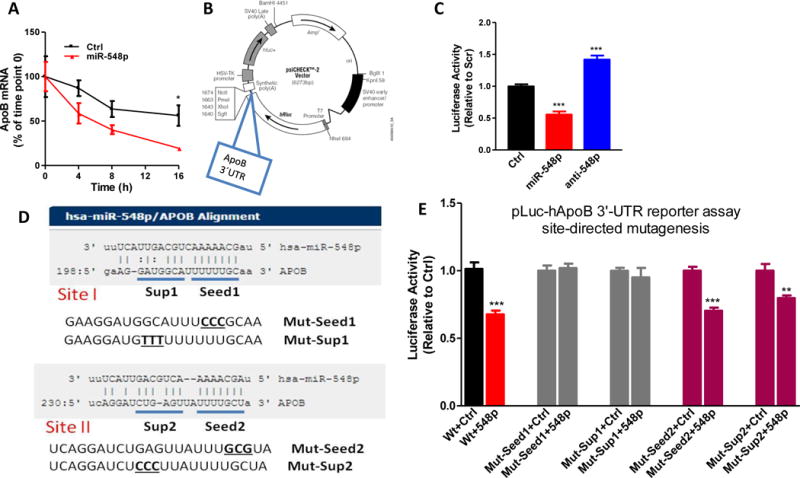
A. Huh-7 cells were transfected with Ctrl or miR-548p (100 nM). After 24 hours, cells were incubated with actinomycin D (1 μg/ml, from 1mg/ml stock). ApoB mRNA levels at different time points were normlaized using 18S as internal control. The apoB mRNA levels relative to time point 0 were plotted. B. ApoB 3′-UTR was amplified from genomic DNA by PCR and cloned into psiCHECK2 vector at the end of Renilla luciferase cDNA. C. Plasmids (1.5 μg) containing human apoB 3′-UTR were transfected in Huh-7 cells using transfection reagent Turbofect (Life Technologies). After 24 h, Ctrl, miR-548p and anti-548p (100 nM) were introduced into cells. After 48 h, dual luciferase activities were assayed. The data are representative of two independent experiments. D. Two potential targeting sites (Site I and Site II) were predicted by miRanda. ApoB 3′-UTR has complementary bases that could interact with both seed and supplementary sequences. Every seed and supplementary sequence was mutated using site-directed mutagenesis. E. Luciferase reporter plasmids (1.5 μg) containing wild type or different mutant human apoB 3′-UTR were transfected in Huh7 cells. Cells were then transfected with Scr or miR-548p (100 nM). Luciferase activity was measured after 24 h. The data are representative of two independent experiments. p<0.05, **p<0.01, ***p<0.001. Significance was calculated by two-way ANOVA and student’s t-test.
To identify interactions between miR-548p and apoB mRNA, we cloned wild type 3′-untranslated region (3′-UTR) of apoB mRNA at the end of luciferase cDNA in psiCHECK plasmid (Fig 3B) and expressed in Huh-7 cells. Compared to the Ctrl, miR-548p reduced and anti-548p increased luciferase activity in cells transfected with luciferase containing wild type apoB 3′-UTR (Fig 3C). These studies suggest that miR-548p interacts with 3′-UTR of apoB.
Both seed and supplementary sites in miR-548p interact with the 3′-UTR of apoB mRNA at site II
Attempts were then made to identify miR-548p interacting site in the apoB mRNA. Miranda predicted that human apoB 3′-UTR contains two potential target sites for miR-548p (Fig 3D). Both sites contain perfect complementary binding sites for miR-548p seed sequence and also contain possible interaction sites involving supplementary sequences. To determine if miR-548p interacts with these sequences, we mutated these sites to perturb potential base-paring (Fig 3D). Mutations in either seed or supplementary sequences in Site I resulted in loss of miR-548p effect on luciferase activity (Fig 3E). However, mutations of Site II seed and supplementary sequences did not affect luciferase activity (Fig 3E). These studies suggest that miR-548p interacts with the 3′-UTR of apoB by interacting with both seed and supplementary sequences at Site I.
MiR-548p decreases hepatic lipid synthesis
Inhibition of apoB secretion is usually associated with cellular lipid accumulation (47). Therefore, we asked whether miR-548p would increase cellular lipids. No significant differences were observed in total cellular triglyceride and cholesterol levels in cells transfected with Ctrl and miR-548p (Fig 4A–B). Thus, miR-548p expression does not cause lipid accumulation in Huh-7 cells.
Figure 4. MiR-548p decreases lipid synthesis and the expression of genes involved in cholesterol and fatty acid synthesis.
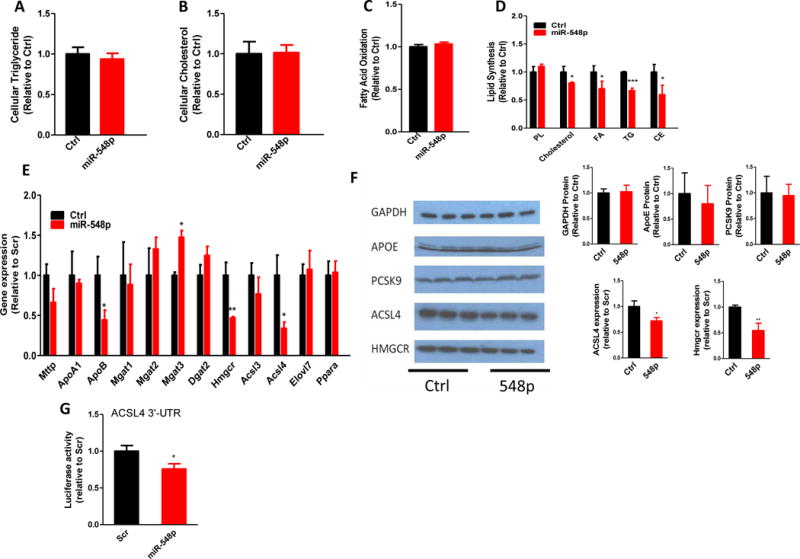
(A–B) Overexpression of miR-548p had no effect on cellular lipid levels. Cellular triglyceride and cholesterol were extracted and quantified in Ctrl or miR-548p (100 nM) transfected Huh-7 cells. (C–D) MiR-548p (100 nM) reduced cholesterol and triglyceride synthesis without affecting fatty acid oxidation in Huh-7 cells. PL, phospholipid; FA, fatty acid; TG, triglyceride; CE cholesteryl ester. The data are representative of three independent experiments. (E) Genes in lipoprotein assembly, triglyceride, cholesterol, and fatty acid biosynthesis were quantified using quantitative RT-PCR. The data are representative of three independent experiments. (F) Western blots of GAPDH, ACSL4, and HMGCR in Ctrl or miR-548p (100 nM) transfected Huh-7 cells (left). Medium from transfected cells was used to immunoprecipitate apoE and PCSK9 and then subjected to western blotting (left). Specific protein bands were quantified using ImageJ and plotted as bar graphs (right). (G) Luciferase reporter plasmids (1.5 μg) containing human ACSL4 or HMBCR 3′-UTRs were transfected in Huh-7 cells. Cells were then distributed in different wells and transfected with Ctrl or miR-548p (100 nM). Luciferase activity was measured after 24 h. p<0.05, **p<0.01, ***p<0.001. Significance was calculated by student’s t-test. The data are representative of two independent experiments.
To explain reasons for the absence of lipid accumulation in miR-548p expressing cells, we examined the effects of miR-548p on fatty acid oxidation and lipid synthesis (Fig 4C–D). Fatty acid oxidation was unaffected (Fig 4C). However, miR-548p significantly decreased cholesterol, fatty acid, and triglyceride synthesis by 20%, 30%, and 33%, respectively, but had no effect on phospholipid synthesis (Fig 4D). These studies indicate that miR-548p decreases lipid synthesis.
To explain how miR-548p decreases cholesterol and fatty acid synthesis, we measured mRNA levels of genes involved in lipid synthesis that are predicted to contain potential target sites of miR-548p (Fig 4E). HMGCR and ACSL4 mRNA levels were significantly reduced in miR-548p expressing cells. However, mRNA levels of other enzymes involved in triglyceride and fatty acid biosynthesis, as well as fatty acid oxidation were not affected (Fig 4E). Further, cellular proteins levels of these enzymes were significantly reduced (Fig 4F) in miR-548p expressing cells while GAPDH protein levels were unaffected. Further, media apoE and PCSK9 levels were unaffected by miR-548p expression. HMGCR is a rate-limiting enzyme in cholesterol synthesis (48). ACSL4 plays a role in fatty acid and triglyceride synthesis (49, 50). Thus, miR-548p may target HMGCR and ACSL4 to reduce lipid synthesis.
To determine whether miR-548p directly regulates ACSL4, we utilized luciferase constructs containing the 3′-UTRs of ACSL4. MiR-548p overexpression significantly decreased the luciferase activity when it was expressed with theACSL4 3′-UTR indicating a direct interaction between ACSL4 3′-UTR and miR-548p (Fig 1G).
Discussion
Here we report identification of a novel miR, miR-548p, that targets apoB mRNA and reduces intracellular and media apoB levels. MiR-548p interacts with the 3′-UTR of apoB mRNA involving both seed and supplementary sequences at site I. These interactions induce post-transcriptional degradation of mRNA and reduce secretion of apoB from human liver cells. In addition, we observed that miR-548p significantly reduces lipid synthesis. This is most likely due to diminished expression of ACSL4 and HMGCR, two enzymes involved in fatty acid and cholesterol biosynthesis. Thus, miR-548p reduces lipid synthesis and lipoprotein production via independent mechanisms targeting different mRNAs.
The importance of interactions between apoB mRNA and miR-548p in the regulation of apoB secretion is supported by several experiments. Bioinformatics analyses predict conserved interacting sites in primates. Cloning of 3′-UTR of apoB mRNA at the end of luciferase renders luciferase susceptible to miR-548p dependent regulation. Mutagenesis of 3′-UTR sequence abolishes the regulation by miR-548p. Mouse apoB is not regulated by miR-548p most likely due to the absence of miR-548p interacting site.
According to miRanda, human apoB 3′-UTR contains two targeting sites for miR-548p involving perfect seed sequence Watson-Crick base pair interactions. In contrast, TargetScan predicts only one site that corresponds to the site II predicted by miRanda. Site-directed mutagenesis studies revealed that only one predicted site, site I, contributes to the regulation of apoB mRNA (Fig 3). This is consistent with the general understanding that the first miR interacting site after the stop codon is important for physiological activity. However, this interaction starts with the residue 3 of the miR-548p. Usually, residue 2 of miR is involved in the first H-bonding with mRNAs. In fact, the predicted binding of miR-548p at site II starts with the second residue. It is likely that this site is not available for miR-548p as the binding start site is ~ 12 nucleotides away from the end of the first binding site. Thus, if the site I is occupied by a RISC complex then the site II may not be accessible for additional interactions. At this site I, both seed and supplementary site interactions appear important for miR-548p to regulate apoB mRNA levels as mutagenesis of residues only involved in these supplementary site interactions abolish response to miR-548p. A previous study about the interaction of miR-30c and MTP has highlighted the importance of supplementary site interactions (51). Therefore, it is likely that miR-548p interacts with the first possible interaction site available after the stop codon and these interactions involve both seed and supplementary sequences.
MiR-548p is a member of a large primate specific family (52). Most of the miRs are conserved in different species (42, 53, 54). The conservation allows an easy way to evaluate physiological effects of overexpression or loss-of-function of specific miRs in model organisms before human studies. Nevertheless, many human or primate specific miRs have been identified that play important roles in physiological and pathological pathways, such as cellular differentiation and cancer development (55–57). It is likely that primate miRs have evolved to regulate species specific phenotypes. Primates carry most of the cholesterol in LDL, whereas mice carry it in HDL. Consistent with this, 3′-UTR of primate apoB mRNA contains miR-548p targets sites that carry cholesterol mainly in LDL (Fig 1A). And, this interacting site is not present in animals that carry cholesterol mainly in HDL, such as mouse. Thus, miR-548p might have evolved to regulate plasma LDL levels.
MiR-548p is predicted to be transcribed from an independent gene (MIR548P) that is present on the opposite strand of the ST8 alpha-N-acetyl-neuraminide alpha-2, 8-sialyltransferase 4 (ST8SIA4) gene (Supplementary Fig IIA) that encodes an enzyme required for the synthesis of polysialic acid (58). MiRs are known to be transcribed as pri-miR, undergo cleavage and transported across the nucleus as pre-miR. Not only the mature miR, but also the pre-miR548p is conserved in primates (Supplementary Fig IIB). It has been reported that miR-548p is present in several human tissues including blood, brain, liver, lung etc. (Supplementary Fig III).
To our knowledge, miR-548p is the first miR that has been shown to target apoB mRNA and decrease apoB secretion from human hepatoma cells. MTP and apoB are the two critical proteins in apoB-containing lipoprotein assembly and secretion. Inhibition of MTP causes hepatosteatosis and elevates plasma AST and ALT activities. In the absence of MTP, lipids are not transferred from endoplasmic reticulum (ER) membrane to ER lumen. Therefore, the newly synthesized lipids, mainly triglyceride, are stored in cytosolic lipid droplets. However, when apoB expression is reduced, lipids can still be transferred to ER lumen by MTP. Lipid accumulation in the ER lumen has been shown to activate autophagy, which engulfs and hydrolyzes lipids in lysosomes (25). Fatty acids generated from lipolysis are then oxidized in mitochondria. Therefore, in mice treated with apoB ASOs, hepatic triglyceride levels increase soon after injection (first 3 weeks) and later resolve due to higher fatty acid oxidation (25). Hence, targeting apoB might be an appropriate way to reduce VLDL production. In this study, we show that miR-548p reduces apoB mRNA to decrease apoB protein secretion from human hepatocytes, and Huh-7 and HepG2cells (Fig 1) and also lowers hepatic cholesterol and fatty acid synthesis by suppressing the expression of HMGCR and ACSL4 (Fig 4). Thus, it is possible that early steatosis seen with apoB ASO can be avoided by miR-548p overexpression, and may be evaluated in the future in primates.
With this report, we have now identified two miRs that appear to have similar physiological regulatory effects. MiR-30c reduces lipoprotein production by targeting MTP and lipid synthesis by targeting ELOVL5 and LPGAT3 (41, 42). MiR-548p reduces lipoprotein production by targeting apoB and lipid synthesis by lowering ACSL4 and HMGCR. To our knowledge, this is the first report describing two different miRs affecting the same physiological processes by targeting different genes. It is likely that nature has evolved multiple miRs to regulate similar pathways due to their physiological importance. Alternatively, the evolution of different miRs may represent redundant mechanisms for their regulation.
From genetic and therapeutic studies, it is well known that separate inhibition of lipoprotein synthesis and lipid synthesis results in different diseases. For example, defects in lipoprotein production result in hypolipidemia that is associated with accumulation of lipids in tissues (19). On the other hand, defects in lipid synthesis are associated with lipodystrophy (59). In contrast, combined reduction of both these metabolic pathways appears to avoid pathologies associated with defects in individual pathways. Therefore, targeting of multiple related pathways might be more beneficial than the classical approach of targeting individual proteins/pathways.
Although these studies advocate for the identification and use of miRs that affect multiple pathways with beneficial outcomes, these approaches need extensive evaluation. A major concern about miR therapies is unanticipated effects. Hence, a rigorous evaluation of all anticipated and unanticipated effects are warranted before advocating their use in humans.
In short, these studies have identified miR-548p that reduces expression of apoB, HMGCR and ACSL4 to diminish lipoprotein production and lipid synthesis. Due to its ability of controlling these two pathways, similar to miR-30c, it might reduce plasma cholesterol and atherosclerosis without causing steatosis. However, this can only be evaluated in primates. Such studies will provide strong evidence for further clinical investigations in humans.
Supplementary Material
Highlights.
Hsa-miR-548p reduces apoB-containing lipoprotein secretion in human liver cells.
MiR-548p interacts with apoB 3′-UTR involving both seed and supplementary sequences at Site I.
MiR-548p decreases lipid synthesis in human hepatoma cells by suppressing the expression levels of ACSL4 and HMGCR.
MiR-548p does not alter apoB expression levels in mouse liver cells.
Acknowledgments
None.
Sources of Funding: This work was supported in part by AHA pre-doctoral fellowship to LZ; and VA Merit Award to MMH.
Abbreviations used
- ApoB
apolipoprotein B
- ASO
antisense oligonucleotides
- Ctrl
control
- FBS
fetal bovine serum
- HDL
high density lipoproteins
- HoFH
homozygous familial hypercholesterolemia
- LDL
low density lipoproteins
- miR
microRNA
- MTP
microsomal triglyceride transfer protein
- UTR
untranslated region
- VLDL
very low density lipoproteins
Footnotes
Disclosures: The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thibault PA, Huys A, Amador-Canizares Y, Gailius JE, Pinel DE, Wilson JA. Regulation of Hepatitis C Virus Genome Replication by Xrn1 and MicroRNA-122 Binding to Individual Sites in the 5′ Untranslated Region. J Virol. 2015;89:6294–6311. doi: 10.1128/JVI.03631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014;42:609–621. doi: 10.1093/nar/gkt852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 6.Qiu Z, Dai Y. Roadmap of miR-122-related clinical application from bench to bedside. Expert Opin Investig Drugs. 2014;23:347–355. doi: 10.1517/13543784.2014.867327. [DOI] [PubMed] [Google Scholar]
- 7.Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gulla A, Tagliaferri P, Tassone P, Caraglia M. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 9.Agostini M, Knight RA. miR-34: from bench to bedside. Oncotarget. 2014;5:872–81. doi: 10.18632/oncotarget.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendis S, Davis S, Norrving B. Organizational update: the world health organization global status report on noncommunicable diseases 2014; one more landmark step in the combat against stroke and vascular disease. Stroke. 2015;46:e121–122. doi: 10.1161/STROKEAHA.115.008097. [DOI] [PubMed] [Google Scholar]
- 11.Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 12.Singh RB, Mengi SA, Xu YJ, Arneja AS, Dhalla NS. Pathogenesis of atherosclerosis: A multifactorial process. Exp Clin Cardiol. 2002;7:40–53. [PMC free article] [PubMed] [Google Scholar]
- 13.Gotto AM., Jr Jeremiah Metzger Lecture: cholesterol, inflammation and atherosclerotic cardiovascular disease: is it all LDL? Trans Am Clin Climatol Assoc. 2011;122:256–289. [PMC free article] [PubMed] [Google Scholar]
- 14.Holvoet P, Harris TB, Tracy RP, Verhamme P, Newman AB, Rubin SM, Simonsick EM, Colbert LH, Kritchevsky SB. Association of high coronary heart disease risk status with circulating oxidized LDL in the well-functioning elderly: findings from the Health, Aging, and Body Composition study. Arterioscler Thromb Vasc Biol. 2003;23:1444–1448. doi: 10.1161/01.ATV.0000080379.05071.22. [DOI] [PubMed] [Google Scholar]
- 15.Choumerianou DM, Dedoussis GV. Familial hypercholesterolemia and response to statin therapy according to LDLR genetic background. Clin Chem Lab Med. 2005;43:793–801. doi: 10.1515/CCLM.2005.134. [DOI] [PubMed] [Google Scholar]
- 16.Alkindi M, Siminovitch KA, Gupta M, Genest J. Monoclonal Antibodies for the Treatment of Hypercholesterolemia: Targeting PCSK9. Can J Cardiol. 2016;32:1552–1560. doi: 10.1016/j.cjca.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Rader DJ, Kastelein JJ. Lomitapide and mipomersen: two first-in-class drugs for reducing low-density lipoprotein cholesterol in patients with homozygous familial hypercholesterolemia. Circulation. 2014;129:1022–1032. doi: 10.1161/CIRCULATIONAHA.113.001292. [DOI] [PubMed] [Google Scholar]
- 18.Raal FJ, Honarpour N, Blom DJ, Hovingh GK, Xu F, Scott R, Wasserman SM, Stein EA, Investigators T Inhibition of PCSK9 with evolocumab in homozygous familial hypercholesterolaemia (TESLA Part B): a randomised, double-blind, placebo-controlled trial. Lancet. 2015;385:341–350. doi: 10.1016/S0140-6736(14)61374-X. [DOI] [PubMed] [Google Scholar]
- 19.Walsh MT, Hussain MM. Targeting microsomal triglyceride transfer protein and lipoprotein assembly to treat homozygous familial hypercholesterolemia. Crit Rev Clin Lab Sci. 2016;54:26–48. doi: 10.1080/10408363.2016.1221883. [DOI] [PubMed] [Google Scholar]
- 20.Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- 21.Davidson NO, Shelness GS. APOLIPOPROTEIN B: mRNA editing, lipoprotein assembly, and presecretory degradation. Annu Rev Nutr. 2000;20:169–93. doi: 10.1146/annurev.nutr.20.1.169. [DOI] [PubMed] [Google Scholar]
- 22.Zimmermann TS, Lee AC, Akinc A, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama T, Butler JS, Sehgal A, et al. Harnessing a physiologic mechanism for siRNA delivery with mimetic lipoprotein particles. Mol Ther. 2012;20:1582–1589. doi: 10.1038/mt.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tadin-Strapps M, Peterson LB, Cumiskey AM, et al. siRNA-induced liver ApoB knockdown lowers serum LDL-cholesterol in a mouse model with human-like serum lipids. J Lipid Res. 2011;52:1084–1097. doi: 10.1194/jlr.M012872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conlon DM, Thomas T, Fedotova T, Hernandez-Ono A, Di Paolo G, Chan RB, Ruggles K, Gibeley S, Liu J, Ginsberg HN. Inhibition of apolipoprotein B synthesis stimulates endoplasmic reticulum autophagy that prevents steatosis. J Clin Invest. 2016;126:3852–3867. doi: 10.1172/JCI86028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gebhard C, Huard G, Kritikou EA, Tardif JC. Apolipoprotein B antisense inhibition–update on mipomersen. Curr Pharm Des. 2013;19:3132–3142. doi: 10.2174/13816128113199990312. [DOI] [PubMed] [Google Scholar]
- 27.Bell DA, Hooper AJ, Burnett JR. Mipomersen, an antisense apolipoprotein B synthesis inhibitor. Expert Opin Investig Drugs. 2011;20:265–272. doi: 10.1517/13543784.2011.547471. [DOI] [PubMed] [Google Scholar]
- 28.Crooke ST, Geary RS. Clinical pharmacological properties of mipomersen (Kynamro), a second generation antisense inhibitor of apolipoprotein B. Br J Clin Pharmacol. 2013;76:269–276. doi: 10.1111/j.1365-2125.2012.04469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashemi N, Odze RD, McGowan MP, Santos RD, Stroes ES, Cohen DE. Liver histology during Mipomersen therapy for severe hypercholesterolemia. J Clin Lipidol. 2014;8:606–611. doi: 10.1016/j.jacl.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letteron P, Sutton A, Mansouri A, Fromenty B, Pessayre D. Inhibition of microsomal triglyceride transfer protein: another mechanism for drug-induced steatosis in mice. Hepatology. 2003;38:133–140. doi: 10.1053/jhep.2003.50309. [DOI] [PubMed] [Google Scholar]
- 31.Joy TR, Hegele RA. Microsomal triglyceride transfer protein inhibition-friend or foe? Nat Clin Pract Cardiovasc Med. 2008;5:506–508. doi: 10.1038/ncpcardio1251. [DOI] [PubMed] [Google Scholar]
- 32.Hussain MM, Bakillah A. New approaches to target microsomal triglyceride transfer protein. Curr Opin Lipidol. 2008;19:572–578. doi: 10.1097/MOL.0b013e328312707c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou L, Irani S, Sirwi A, Hussain MM. MicroRNAs regulating apolipoprotein B-containing lipoprotein production. Biochim Biophys Acta. 2016;1861:2062–2068. doi: 10.1016/j.bbalip.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 34.Irani S, Hussain MM. Role of microRNA-30c in lipid metabolism, adipogenesis, cardiac remodeling and cancer. Curr Opin Lipidol. 2015;26:139–146. doi: 10.1097/MOL.0000000000000162. [DOI] [PubMed] [Google Scholar]
- 35.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 37.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai WC, Hsu SD, Hsu CS, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu SH, Wang B, Kota J, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, Zalzala M, Xu J, Li Y, Yin L, Zhang Y. A metabolic stress-inducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nat Commun. 2015;6:7466. doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irani S, Pan X, Peck BC, Iqbal J, Sethupathy P, Hussain MM. MicroRNA-30c Mimic Mitigates Hypercholesterolemia and Atherosclerosis in Mice. J Biol Chem. 2016;291:18397–18409. doi: 10.1074/jbc.M116.728451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernandez-Hernando C, Suarez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Curr Opin Lipidol. 2011;22:86–92. doi: 10.1097/MOL.0b013e3283428d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Izaurralde E. GENE REGULATION. Breakers and blockers-miRNAs at work. Science. 2015;349:380–382. doi: 10.1126/science.1260969. [DOI] [PubMed] [Google Scholar]
- 47.Koornneef A, Maczuga P, van Logtenstein R, Borel F, Blits B, Ritsema T, van Deventer S, Petry H, Konstantinova P. Apolipoprotein B knockdown by AAV-delivered shRNA lowers plasma cholesterol in mice. Mol Ther. 2011;19:731–740. doi: 10.1038/mt.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharpe LJ, Brown AJ. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) J Biol Chem. 2013;288:18707–18715. doi: 10.1074/jbc.R113.479808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooke M, Orlando U, Maloberti P, Podesta EJ, Cornejo Maciel F. Tyrosine phosphatase SHP2 regulates the expression of acyl-CoA synthetase ACSL4. J Lipid Res. 2011;52:1936–1948. doi: 10.1194/jlr.M015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Golej DL, Askari B, Kramer F, Barnhart S, Vivekanandan-Giri A, Pennathur S, Bornfeldt KE. Long-chain acyl-CoA synthetase 4 modulates prostaglandin E(2) release from human arterial smooth muscle cells. J Lipid Res. 2011;52:782–793. doi: 10.1194/jlr.M013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soh J, Hussain MM. Supplementary site interactions are critical for the regulation of microsomal triglyceride transfer protein by microRNA-30c. Nutr Metab (Lond) 2013;10:56. doi: 10.1186/1743-7075-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS One. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ouimet M, Ediriweera HN, Gundra UM, et al. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J Clin Invest. 2015;125:4334–4348. doi: 10.1172/JCI81676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu SH, Delgado ER, Otero PA, Teng KY, Kutay H, Meehan KM, Moroney JB, Monga JK, Hand NJ, Friedman JR, Ghoshal K, Duncan AW. MicroRNA-122 regulates polyploidization in the murine liver. Hepatology. 2016;64:599–615. doi: 10.1002/hep.28573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu HY, He L, Fominykh K, Yan Z, Guo S, Zhang X, Taylor MS, Tang L, Li J, Liu J, Wang W, Yu H, Khaitovich P. Evolution of the human-specific microRNA miR-941. Nat Commun. 2012;3:1145. doi: 10.1038/ncomms2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu HY, Guo S, Xi J, Yan Z, Fu N, Zhang X, Menzel C, Liang H, Yang H, Zhao M, Zeng R, Chen W, Pääbo S, Khaitovich P. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet. 2011;7:e1002327. doi: 10.1371/journal.pgen.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu B, Ying X, Wang J, et al. Identification of a tumor-suppressive human-specific microRNA within the FHIT tumor-suppressor gene. Cancer Res. 2014;74:2283–2294. doi: 10.1158/0008-5472.CAN-13-3279. [DOI] [PubMed] [Google Scholar]
- 58.Sato C, Kitajima K. Impact of structural aberrancy of polysialic acid and its synthetic enzyme ST8SIA2 in schizophrenia. Front Cell Neurosci. 2013;7:61. doi: 10.3389/fncel.2013.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simha V, Garg A. Lipodystrophy: lessons in lipid and energy metabolism. Curr Opin Lipidol. 2006;17:162–169. doi: 10.1097/01.mol.0000217898.52197.18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


