Abstract
Hepatocellular carcinoma (HCC) is one of the most deadly cancers that still lacks effective treatments. Dysregulation of kinase signaling has frequently been reported to contribute to HCC. In this study, we used bioinformatic approaches to identify kinases that regulate gene expression changes in human HCCs and two murine HCC models. We identified a role for calcium/calmodulin-dependent protein kinases II gamma isoform (CAMK2γ) in hepatocarcinogenesis. CAMK2γ−/− mice displayed severely enhanced chemical-induced hepatocarcinogenesis compared with wild-type controls. Mechanistically, CAMK2γ deletion potentiates hepatic activation of mechanistic target of rapamycin complex 1 (mTORC1), which results in hyperproliferation of hepatocytes. Inhibition of mTORC1 by rapamycin effectively attenuats the compensatory proliferation of hepatocytes in CAMK2γ−/− livers. We further demonstrated that CAMK2γ suppressed growth factor- or insulin-induced mTORC1 activation by inhibiting IRS1/AKT signaling. Taken together, our results reveal a novel mechanism by which CAMK2γ antagonizes mTORC1 activation during hepatocarcinogenesis.
Keywords: Hepatocellular Carcinoma, Kinase, CAMK2γ, mTORC1, Rapamycin, AKT
Introduction
Liver cancers, including the hepatocellular carcinoma (HCC), are the second most common cause of cancer deaths worldwide (World Cancer Report 2014, WHO). Patients with liver cancer have poor prognosis due to recurrence and the lack of effective treatments.1, 2 The development of liver cancer is associated with various risk factors.3 Chronic infection by virus, certain types of parasites in the liver, abuse of alcohol, or steatohepatitis can all cause long-term hepatocyte necrosis and inflammation, followed by formation of post-necrotic cirrhosis of the liver.3 Insults from xenobiotics or chemicals such as Aflatoxin and Arsenic not only cause hepatocyte death, but also directly induce oncogenic mutations at tumor suppressor genes such as p53.1, 4 A common pathological consequence of these risk factors is continuous hepatocyte death accompanied by compensatory hepatocyte proliferation, which promotes hepatocyte transformation and malignant cell expansion.5 However, the underlying molecular mechanisms controlling hepatocarcinogenesis are still largely unclear.
Kinase regulation plays important roles during hepatocarcinogenesis. Adult hepatocytes are normally in a quiescent state. Upon carcinogen stimuli, pro-inflammatory cytokines, for instance IL-6 and TNFα, are synthesized and released from the Kupffer cells to activate kinase pathways such as JAK/STAT and IKK/IκBα in hepatocytes, which in turn prime hepatocytes out of quiescence and drive them into cell cycling.5 Growth factors and hormones subsequently stimulate the proliferation of the transformed hepatocytes by interacting with the receptor-tyrosine kinase (RTK) family. Several kinase inhibitors are under active development to treat liver cancer. For example, Sorafenib, which targets both the Raf/MEK/ERK pathway and receptor tyrosine kinases VEGFR/PDGFR in tumor cells, can prolong the survival of liver cancer patients by about 3 months through inhibiting cancer cell proliferation and angiogenesis.6 Population studies show that metformin decreases cancer risk by activating AMPK to suppress mTORC1, which induces apoptosis of malignant liver cells.7, 8
Current approaches to systemically predict kinase activities include Mass Spectrometry (MS)-based phosphor-proteomic analysis and substrate peptide based phosphor-kinase arrays.9–13 Both approaches have their limitations when applying to in vivo studies. MS-based phosphor-proteomic analysis is limited by abundance of a specific phosphorylation peptide, efficiency of phosphor-specific peptide enrichment, as well as sensitivity of MS instruments.9–11 Peptide based kinase arrays, on the other hand, can only detect a limited number of kinases with known substrate motifs, and the results are highly influenced by specificity of the substrate peptide towards different kinases.11–13 Moreover, activity of kinases is regulated by transiently induced cellular chemical and mechanical signals. Differential subcellular localization or kinase/adaptor/scaffold protein interaction could spatially regulate kinase activity or accessibility to specific substrates, which cannot be reflected by the peptide-based kinase arrays.
Taking advantage of the rapidly expanding transcriptome and kinase function studies, new bioinformatics algorithms have recently been developed to predict activation of potential upstream transcription factors and associated kinases.14–16 These bioinformatics tools use gene expression profiles as input to identify differentially expressed genes and can further examine transcription factor or kinase correlated to gene expression signatures. They can then provide candidate gene lists for further analyses. Among these tools, Expression2Kinases (X2K) and Ingenuity Pathway Analysis (IPA) have successfully been used to predict the change of kinase activity in various human diseases and animal models.17–20
mTORC1 is a protein complex that senses nutrient/energy/redox status and regulates protein synthesis.21 mTORC1 activities are essential for liver cell proliferation as the S6 knockout mice almost fail to display liver regeneration after 70% partial hepatectomy.22 Noteworthy, up to 50% of liver cancer patients show up-regulation of mTORC1 activities.7 However, regulation of mTORC1 during hepatocarcinogenesis remains elusive.
Calcium/calmodulin-dependent protein kinases II (CAMK2s) have been extensively studied in central neural systems due to its key roles in learning and memory.23 The γ and δ isoforms of CAMK2s are key mediators of different upstream signaling pathways, including oxidative and ER stresses, non-canonical Wnt signaling, and GPCRs.24–28 In this study, we used novel bioinformatic approaches to analyze gene expression profiling from human HCCs and two HCC animal models. Among the identified list of kinases, CAMK2γ stood out to play a critical role during hepatocarcinogenesis by antagonizing mTORC1 activation.
Results
Identification of new kinases in hepatocarcinogenesis
To identify novel kinases involved in liver hepatocarcinogenesis, we analyzed genome-wide mRNA profiling datasets of both animal HCC models and human HCC specimens by computational approaches, including the ChIP-X Enrichment Analysis (ChEA) and Kinase Enrichment Analysis (KEA).29, 30 Two well-characterized HCC rodent models, the chemical carcinogen diethylnitrosamine (DEN)-treated mice and the spontaneously tumorigenic FXR−/− mice,31–34 were selected for these analyses. Both models recapitulate the human HCC pathological progression.31, 35 In addition, a dataset of human HCC specimens was also included,36 in which the gene expression of HCC and surrounding nontumorous liver tissue was compared. Differentially expressed genes were called for each dataset using limma 37 (see Materials and Methods for details) and these genes were further analyzed for potential upstream transcription factors (TFs) with ChIP Enrichment Analysis (ChEA).29 ChEA utilizes a database of TF binding sites from chromatin immunoprecipitation studies (ChIP-seq and ChIP-chip) to determine the over-representation of TF targets for the given set of genes compared to the whole genome. The top-ranked TFs reported by ChEA from each dataset are shown in Fig. 1A (Table. S1). Seven TFs (HNF4A, PPARG, MITF, CREM, FLI1, KDM5B and RUNX1) were identified in the top 10 in at least two of the datasets, indicating that they are involved in gene regulation in the development of HCC. These seven TFs were further used to predict the upstream kinases that contribute to liver tumorigenesis with kinase enrichment analysis (KEA).30 KEA utilizes an underlying kinase-substrate interaction database to identify the kinases that phosphorylate given proteins/TFs.38–40 KEA showed that p38α (MAPK14), GSK3α/β, CAMK2γ/δ, ERK2 (MAPK1), AMPKα2 (PRKAA2), Ribosomal s6 kinase (RPS6KA1, RSK1), Casein kinase 1α (CSNK1α1), and TGFBR2 were the top-ranked kinases that most likely to be involved in HCC. Ingenuity Pathway Analysis was then used to characterize connections among these kinases. ERK2 and p38α appear to constitute the center of the liver cancer kinase network and affect each other. Interestingly, CAMK2γ is known to activate both kinases (Fig. S1A).
Figure 1. Gene profiling and pathway analyses of kinases involved in liver carcinogenesis.
(A) Microarray data from mouse models and clinical HCC specimens were analyzed by ChIP-X Enrichment Analysis (ChEA) and Kinase Enrichment Analysis (KEA). The top hits of ChEA and KEA were listed for each dataset. (B) X2K analyses on the same datasets. Top 50 kinases in each dataset were selected and compared among the 3 data sets. Four kinases, CAMK2γ, GSK3β, MAPK1 (ERK2), and RPS6Kα1 (RSK1), were enriched in both KEA and X2K analyses, and are highlighted in the figure.
To account for the fact that kinases may be involved in hepatocarcinogenesis without directly phosphorylating TFs, we further analyzed the three datasets using Expression2Kinases (X2K) to include the kinases whose substrates are not TFs (Fig. 1B).15 X2K analysis unitizes known protein-protein interactions to construct a network that connects identified TFs and predict upstream regulators.17–20 For each dataset, the top 10 TFs from ChEA (Fig. 1A) were further used to generate regulation networks based on the known protein-protein interactions of these TFs (Fig. S1B–D). All the proteins identified in these networks were then used to identify the upstream kinases using KEA in each dataset. Because the X2K analyses gave rise to many kinase candidates, we only selected the top 50 kinases enriched from each dataset (Fig. 1B). The kinases that emerged in all 3 datasets as common upstream kinases are listed in Fig. 1B. CAMK2γ was significantly enriched (p-value < 0.05, Fisher’s exact test) in all the three datasets. Taken together with the Ingenuity Pathway Analysis (IPA) results (Fig. S1A), these results suggest that CAMK2γ is located upstream of the kinase cascades and plays an important role in hepatocarcinogenesis. Consistent with the bioinformatics study, the phosphorylation of liver CAMK2γ, which indicates its activation, was induced by a genotoxic stress via DEN treatment (Fig. S1E).
Deletion of CAMK2γ promoted DEN-induced hepatocarcinogenesis in mice
Our bioinformatics analyses suggest that CAMK2γ may be involved in hepatocarcinogenesis.41 Previous studies from our lab also revealed an oncogenic role of CAMK2γ in human chronic myeloid leukemia.42 We therefore hypothesized that CAMK2γ might promote HCC development. To address our hypothesis, we generated two chemical-induced HCC animal models to determine the roles of CAMK2γ in hepatocarcinogenesis. For the first model, 2-weeks-old male CAMK2γ−/− mice and the wild-type littermates were injected with one single dose of 25 mg/kg DEN. This regimen is sufficient to induce hepatocarcinogenesis in wild-type mice after 8–10 months (Fig. 2A). To our surprise, while wild-type mice only showed small tumors or neoplastic foci in liver, deletion of CAMK2γ resulted in a significant increase in hepatocarcinogenesis (Fig. 2B). Deletion of CAMK2γ did not change liver size, but significantly increased the tumor number and volume. To confirm these observations, we induced HCC using another protocol that DEN was injected and followed by a tumor promotor (TCPOBOP) as described previously.43 This model better represents the liver carcinogenesis caused or enhanced by xenobiotic insults. Four weeks old male mice were treated with one single dose of DEN and thereafter TCPOBOP injection every two weeks for 8 times. More significant increase of liver carcinogenesis in CAMK2γ−/− mice was observed in this model (Fig. 2C). The CAMK2γ−/− mice exhibited more than 3-fold increase of both liver tumor sizes and tumor volumes compared with their wild-type littermates (Fig. 2D).
Figure 2. Enhanced DEN-induced hepatocarcinogenesis in CAMK2γ−/− mice.
(A) A DEN-induced liver cancer model was compared in CAMK2γ−/− and wild-type (WT) mice. Representative images of liver tumors in the WT and CAMK2γ−/− mice after DEN injection. (B) Statistical analyses of liver size, tumor number, and tumor size in the mice. HCC was induced by a DEN plus TCPOBOP protocol. *, p < 0.05; **, p < 0.01; n=14 or 9. (C) Representative images of liver tumors in the WT and CAMK2γ−/− mice after DEN plus TCPOBOP injection. (D). Statistical analyses of liver size, tumor number, and tumor size in the mice. *, p < 0.05; **, p < 0.01; n=11 or 12.
Histological analyses revealed that the hepatic tissues from CAMK2γ−/− mice displayed enhanced liver necrosis, apoptosis, inflammatory cell infiltration (Fig. S2A). An increase in compensatory hepatocyte proliferation was observed in the CAMK2γ−/− liver (Fig. S2B). Consistently, quantitative real-time PCR analyses showed an enhanced expression of inflammatory cytokines such as TNFα and IL-6 in the CAMK2γ−/− livers (Fig. S3). Loss of p53 and activation of STAT3 are hallmarks of liver carcinogenesis and represents advanced HCC progression. Indeed, hepatic STAT3 phosphorylation at Tyr705 was increased in both tumor and non-tumor adjacent tissues, and the p53 expression was diminished in CAMK2γ−/− livers (Fig. S4A). As expected, the expression levels of p53 target genes p21 and Bax were decreased (Fig. S4B). In contrast, the expression levels of STAT3 target genes Socs3, Bcl-2, and Mcl-1 were increased by CAMK2γ deletion (Fig S4C).
CAMK2γ deletion enhanced carcinogen-induced hepatocyte deaths
DEN-induced acute liver injury is a commonly used model to study liver cancer initiation because massive liver cell deaths trigger inflammation, the main driver of liver cell proliferation and transformation.31, 35, 44–46 Since deletion of CAMK2γ enhances cell deaths and inflammation in liver tumor and surrounding tissues, we speculate whether deletion of CAMK2γ also promotes liver cancer initiation through elevating the levels of cell deaths and inflammation upon acute liver injury. Therefore, CAMK2γ−/− mice and wild-type littermates were administrated with 100 mg/kg DEN by i.p. injection. As expected, CAMK2γ−/− mice had enhanced body weight loss (Fig. 3A) and significantly higher serum alanine aminotransferase (ALT) levels (Fig 3B), indicating a higher level of liver injury and substantial hepatocyte deaths in CAMK2γ−/− mice. The histological analyses of hematoxylin & eosin (H&E)-stained also demonstrate significantly enhanced necrosis caused by CAMK2γ deletion (Fig. 3C). DEN led to significant necrosis in the lobules that is distant from vein area in addition to enhanced necrosis around portal vein in the CAMK2γ−/− livers. In contrast, the same dose of DEN did not induce necrosis around portal vein in the wild-type mice (Fig. 3D). Furthermore, compared with wild type livers, TUNEL staining showed that CAMK2γ−/− livers exhibited significantly increased hepatocyte apoptosis induced by DEN, especially around portal veins (Fig. 3E,F). It is also worth mentioning that CAMK2γ−/− mice exhibited more spontaneous apoptosis without DEN treatment (Fig. 3F). Severe cell deaths are known to trigger inflammatory responses, which is a key cause of tumor initiation in the liver.5 In fact, CAMK2γ deletion strongly elevated inflammatory cell infiltration caused by DEN-induced liver injury (Fig. 3G).
Figure 3. Enhanced acute responses and compensatory proliferation in CAMK2γ−/− livers.
(A) Body weight loss and serum ALTs (B) 48 h after DEN injection. *, p < 0.05; ***, p < 0.001; n=5 or 6. (C) Grading of necrosis. (D) A representative H&E staining of the liver in the mice. (E) Quantification of apoptotic cells shown in the TUNEL staining. (F) Representative images of TUNEL staining. (G) Grading of inflammatory cell infiltration.
CAMK2γ deletion potentiated DEN-induced mTORC1 activation and enhanced compensatory hepatocyte proliferation
A key event of liver tumor initiation is the continuous compensatory proliferation of the premalignant hepatocytes. Compensatory proliferation plays a critical role in DEN-induced hepatocarcinogenesis, and dysregulated compensatory proliferation results in enhanced malignant transformation of liver cells.31, 35, 44–46 Indeed, CAMK2γ−/− livers displayed greatly enhanced hepatocyte proliferation as shown by PCNA immunostaining (Fig. 4A). In contrast, wild-type livers only displayed limited PCNA stains around portal vein areas (Fig. 4B).
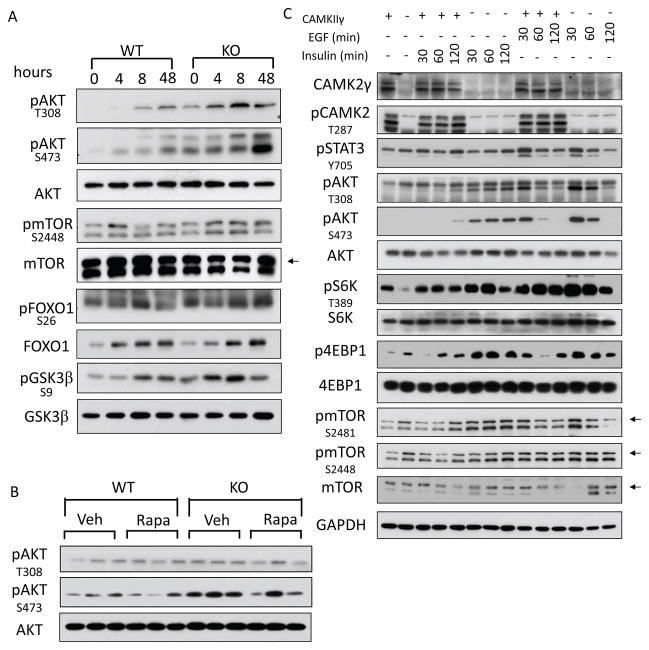
Figure 4. Upregulation of mTORC1 signaling in CAMK2γ−/− livers.
(A) Representative images of PCNA staining. (B) Quantification of hepatocyte compensatory proliferation by PCNA staining. *, p < 0.05; ***, p < 0.001. (C) Key signaling pathways in the liver of DEN-treated mice were analyzed by western blotting. (D) Western blots of mTOR pathway components in the livers of DEN treated mice. Small arrow indicates the mTOR band.
NF-κB, STAT3, and MAPK (p38, ERK, JNK) signaling pathways are known to drive the compensatory proliferation of liver cells during initiation stages. On the other hand, CAMK2γ has been known to affect NF-κB activation and phosphorylation of STAT3 and MAPKs.25, 47, 48 Therefore, we profiled the activation of these pathways in the liver treated with DEN. The results showed that STAT3 and MAPK had marginally increased activation based on the phosphorylation status of STAT3, p38, ERK1/2 and c-Jun. However, no degradation of IκB proteins is observed, indicating that NF-κB may be not involved in the increased compensatory proliferation in CAMK2γ−/− livers (Fig. 4C).
Unexpectedly, mTORC1 signaling was robustly activated in CAMK2γ−/− mice (Fig. 4D). The autophosphorylation of mTOR at S2481 was much higher in the CAMK2γ−/− livers at the 48 h after DEN treatment, indicating that the activities of mTOR were enhanced in the CAMK2γ−/− mice.49 mTORC1 promotes tissue growth by coordinating protein synthesis and inhibiting autophagy. mTORC1 stimulates protein translation by directly phosphorylating S6K and 4E-BP-1.21 Indeed, DEN treatment dramatically increased phosphorylation of S6K at T389 as early as 4 h after DEN injection in the wild-type mice and subsequently diminished at the 48 h (Fig. 4D). In contrast, the initial induction in the CAMK2γ−/− livers was slightly delayed but maintained until the 48 h. This is correlated with the greater enhanced hepatocyte proliferation in the CAMK2γ−/− livers. Though DEN did not dramatically increase the phosphorylation of 4E-BP-1, the overall phosphorylation of 4E-BP-1 at T37/46 and S65 was significantly higher in the CAMK2γ−/− livers. In addition, the phosphorylation of eIF4E at S209 was also slightly increased in the CAMK2γ−/− livers. There was also a slightly reduced autophagosome formation as shown by the LC3 conversion in the CAMK2γ−/− livers. Our results suggest that CAMK2γ may antagonize the exaggerated activation of mTORC1 in the livers after DEN treatment.
Because mTORC1 potentially promotes liver tumor initiation in the CAMK2γ−/− mice through promoting hepatocyte proliferation, we therefore investigated whether rapamycin could abolish the observed effects. Immunoblotting showed that rapamycin effectively decreased the hyperactivation of mTORC1 in the CAMK2γ−/− livers, as shown by the decreased phosphorylation of S6K and 4E-BP-1 (Fig. 5A). Interestingly, rapamycin also significantly increased the autophosphorylation of CAMK2γ as well (Fig. 5A). As expected, rapamycin treatment greatly reduced the hepatocyte proliferation in the CAMK2γ−/− livers (Fig 5B, C). However, rapamycin did not rescue the enhanced inflammatory cell infiltration, centrilobular necrosis, and hepatocyte apoptosis (Fig 5D), indicating that mTORC1 hyperactivation caused by CAMK2γ deletion may contribute to only the enhanced proliferation but not the cell deaths or the inflammation in the liver.
Figure 5. Inhibition of mTORC1 signaling by rapamycin in CAMK2γ−/− mice.
CAMK2γ−/− mice and WT mice received either 5mg/kg rapamycin or 2% vehicle i.p. injection 2 h before the DEN challenge. The mice then received a second and a third rapamycin or vehicle injection 24 h and 46 h after the DEN challenge. At 48 h after the DEN injection, the mice were sacrificed and the liver were harvested for PCNA immunostaining and Western blotting analysis. (A) Immunoblotting analyses of mTORC1 signaling in the liver 48 h after DEN treatment. Small arrow indicates the mTOR band. (B) Representative images of PCNA staining in the liver 48 h after DEN treatment. (C) Quantification of proliferating hepatocytes by PCNA staining. (D) Quantification of hepatic inflammatory cell infiltration, centrilobular necrosis, and hepatocyte apoptosis. **, p <0.01; n=4–7.
CAMK2γ deletion enhanced AKT/mTORC1 signaling
RTK/AKT/mTORC1 pathway plays a broad role in cancer, including liver cancer.7 Therefore, we next examined whether the deletion of CAMK2γ affected AKT activation. DEN induced significant AKT phosphorylation in the wild-type livers (Fig 6A). Surprisingly, deletion of CAMK2γ not only enhanced DEN-induced AKT phosphorylation but also increased the basal AKT phosphorylation in the livers (Fig 6A, Fig. S5A). Consistently, the substrates of AKT, including mTOR (S2448), FoxO1 (S26), and GSK3β (S9), all showed increased phosphorylation in the DEN-treated CAMK2γ−/− livers. On the other hand, ramamycin, which greatly lowers mTORC1 activities in the liver, did not affect AKT phosphorylation significantly. This is consistent with the notion that mTORC1 is at the downstream of AKT (Fig 6B). Hepatocyte proliferation is triggered by hepatomitogens including cytokines, growth factors, or hormones (etc., EGF and insulin), which are known to activate AKT/mTORC1 pathway in the liver. Indeed, we observed that the insulin or EGF-induced activation of AKT and mTORC1 is greatly enhanced by CAMK2γ deletion in primary hepatocytes (Fig 6C).
Figure 6. Suppression of AKT activities by CAMK2γ in liver cells.
(A) AKT was hyperphosphorylated in the DEN-treated CAMK2γ−/− livers. Small arrow indicates the mTOR band. (B) AKT phosphorylation was unchanged in the rapamycin-treated liver 48 h after DEN treatment. (C) EGF (20 ng/ml) or insulin (10 nM) induced exaggerated activation of AKT in CAMK2γ−/− primary hepatocytes at the time points as indicated. Small arrow indicates the mTOR band.
To further assess a direct suppressive effect of CAMK2γ on AKT, we generated a tetracycline-inducible cell system to evaluate the effects of CAMK2γ overexpression on insulin-induced AKT activation. As expected, CAMK2γ overexpression repressed the insulin-induced AKT phosphorylation in both HepG2 (Fig 7A) and Huh7 cells (Fig 7B). A chemical inhibitor of CAMK2γ, KN93, but not its non-functional structural analog KN92, also enhanced insulin-induced AKT phosphorylation (Fig 7C). However, this enhancement was largely abolished by chemical compound that are reported to inhibit PI3K activity, which indicated that CAMK2γ might act at the upstream of PI3K.50 Insulin, EGF, and other hormone/growth factors activate PI3K/AKT pathway by binding to the insulin receptor or EGFR, and then activate the signaling transducer IRS1. We found that DEN could induce strong IRS1 phosphorylation at S612 (Fig 7D). IRS1-S612 is a putative phosphorylation site of PKC and ERK,51, 52 and phosphorylation of this site leads to desensitization of insulin signals to the cells.53 However, in the CAMK2γ−/− livers, this robust induction was completely abolished. In contrast, the PTEN protein levels were unchanged. Consistently, CAMK2γ deletion also abolished the insulin-induced IRS1-S612 phosphorylation in the primary hepatocytes (Fig 7E). It is reported previously that CAMK2γ affects S612 phosphorylation in an ERK-dependent manner.54 However, treating liver cells with ERK inhibitors did not alter the phosphorylation of IRS1 and AKT (Fig. S5B), which suggested that CAMK2γ might affect IRS-S612 activation independent of ERK.
Figure 7. Regulation of AKT/mTOR pathway by CAMK2γ.
(A–B) HepG2 and Huh7 cells with tetracycline-inducible CAMK2γ expression were cultured with Dox for 48 h to induce the expression of CAMK2γ. The cells were then cultured in serum free medium overnight to minimize basal AKT activities. Subsequently, the cells were treated with 10 nM insulin for the indicated time points. (C) Huh7 cells were starved in serum-free medium overnight and pretreated with a combination of DMSO, KN93, and LY294002 for 30 min. Cells were then stimulated with 10 nM insulin and harvested at the indicated time points. The PI3K inhibitor LY294002 suppressed the hyperactivation of AKT induced by CAMK2γ inhibitor KN93. (D) Phosphorylation of IRS1-S612 in the DEN-treated mouse livers. (E) Deletion of CAMK2γ abolished insulin-induced IRS1-S612 phosphorylation in mouse hepatocytes.
Discussion
Kinases are known to play key roles during hepatocarcinogenesis. In this study, we used bioinformatics approaches to analyze the transcriptomics of HCC patients as well as two animal HCC models. We identified several kinases that are potentially involved in hepatocarcinogenesis. Some of these kinases, for instance, MAPK1 and MAPK14, are well studied and known to be crucial for HCC development,55, 56 which confirmed the efficacy and reliability of our computational analyses. Meanwhile, several kinases such as CAMK2γ and CSNK1α1 were less studied and were not clearly linked to hepatocacinogenesis. Specifically, we found that CAMK2γ was activated by HCC associated genotoxic stress. Considering its oncogenic role in hematopoietic malignancies,42 we first hypothesized that CAMK2γ might play an oncogenic role in HCC development. Unexpectedly, deletion of CAMK2γ in mice accelerated HCC development, indicating that CAMK2γ may play a protective role against hepatocarcinogensis.
CAMK2γ was previously implicated as a glucagon effector in the liver and was involved in regulating glucose homeostasis.57 Our previous study showed that, in liver cancer cells, CAMK2γ can promote survival in in vitro cultured liver cancer cells.41 This survival-promoting function of CAMK2γ is consistent with the results that we obtained from animal studies here (Fig 3; Fig. S2A, B). Indeed, CAMK2γ prevents hepatocyte death from tissue damage, thereby suppressing the compensatory cell proliferation and expansion of the transformed hepatocytes.5, 58 Similar animal studies on NF-κB and JNK have also shown that survival signaling for hepatocytes is required to prevent chemical-induced hepatocarcinogenesis.45, 59 In this scenario, the dual roles of CAMK2γ in liver cancer are similar with those of NF-κB, JNK, and SHP2 in hepatocytes.58 That is, they prevent malignant transformation by suppressing hepatocyte death and compensatory liver growth in the tumor initiation and early stages. However, in cancer cells, the same functional role may promote the survival and expansion of the transformed cells and tumor progression at the later stages of cancer progression. This phenomenon seems to be common for HCC and has many implications in human HCC pathogenesis and therapy.58
Mechanistically, we have identified a novel role of CAMK2γ in antagonizing mTORC1 signaling. It is well documented that activation of AKT/mTORC1 plays essential role during both the initiation and later stage development of HCC.7, 60 It is observed that AKT/mTORC1pathway is gradually activated from pre-malignant liver tissue towards HCC tissues in both patient and animal models.61, 62 Abnormal activation of mTORC1 pathway itself is also sufficient to drive the spontaneous development of HCC in transgenic mouse models.63, 64 Sirolimus and everolimus, two derivatives of rapamycin are believed to suppress liver fibrosis which is an important risk factor of HCC.65 Administration of Metformin, which can suppress mTORC1 activity through both AMPK dependent and AMPK independent mechanism, show an overall 62% reduction in the risk of liver cancer for patient with type 2 diabetes.66 It is interesting that members of the RSK family of serine/threonine kinases (RPS6Kα1, RPS6Kα2, RPS6Kα3, RPS6Kα4), well-known regulators of mTORC1,67 are also identified as key kinases that are involved in hepatocarcinogenesis by our bioinformatics analyses, which further implied the importance of mTORC1 pathway in HCC. However, mTORC1 itself was not identified in our analyses probably due to the fact that mTORC1 is mainly a post-transcriptional regulator. mTORC1 is positively regulated by hormones (e.g. insulin) and growth factors (e.g. IGF1, EGF),68 but suppressed by stress signaling (genotoxic, oxidative, ER stress, etc.). However, the molecular mechanisms by which mTORC1 is suppressed by stress signaling remain unclear. Because CAMK2s are well-established stress-responsive kinases,24, 25 our studies thus establish a molecular link between the stress signaling and AKT/mTORC1 regulation. The receptor tyrosine kinases, which are activated by insulin and growth factors, function through AKT and ERK to phosphorylate TSC267, 69 and ERK/RSK1 to phosphorylate Raptor to activate mTORC1.70 In the CAMK2γ−/− mice, the MAPKs (p38/ERK/JNK) were not significantly changed. In contrast, AKT activation became exaggerated after CAMK2γ deletion. Furthermore, our in vitro experiments clearly demonstrate that CAMK2γ simultaneously inhibits the activation of AKT and mTORC1 by insulin or EGF. A previous report shows that CAMK2γ suppresses AKT by phosphorylating p38 in liver, thereby activating a direct AKT inhibitor TRB3.71 However, we have not observed any difference of p38 phosphorylation between wild-type and CAMK2γ−/− livers after DEN treatment. Instead, we found that CAMK2γ suppressed AKT, possibly by enhancing IRS1-S612 phosphorylation, which negatively regulated the RTK/IRS/PI3K module (Fig. 7C–E).54 Because IRS1-S612 is a PKC phosphorylation site,72 it would be interesting to know whether CAMK2γ affects PKC activities. Alternatively, IRS1-S612 could be a direct CAMK2γ phosphorylation site because CAMK2γ and PKC share many substrate sites such as GluR1-S831 and STAT1-S727.73–75 In summary, we provide clear evidence to show that activation of CAMK2γ suppresses AKT/mTORC1 activation during HCC initiation in both mouse models and human cell lines. Though we also observed a potential negative correlation between CAMK2 γ and AKT/mTORC1 expression in human pre-malignant HCC samples (data not shown), further studies to confirm this mechanism in human HCC are required to extend its clinical relevance.
Last but not least, through one of our bioinformatics approaches, KEA, we also identified that CAMK2γ is exclusively linked to one of the top 7 TFs, CREM (Fig. S7A). It is reported that CREM can be directly phosphorylated by CAMK2γ at a site that is important for its transactivation.76 Previous publications have shown that CREM is highly expressed in human HCC cell lines while inhibition of CREM can suppress HCC cell proliferation (Fig S7B, C).77 This effect of CREM is consistent with the role of CAMK2γ at the later stages of cancer progression.41 However, knocking down CREM in human HCC cell lines has no significant effect on insulin-induced Akt activation (Fig. S7D). More importantly, the Akt inhibition induced by CAMK2γ overexpression cannot be rescued by knocking down CREM (Fig. S7E). In this case, the effect of CAMK2γ of Akt/mTOR activation is independent from CREM; the CAMK2γ/CREM signaling may play independent roles in hepatocarcinogenesis. In addition, we also cannot exclude the possibility that CAMK2γ may also affect HCC initiation and development through other downstream targets. Further study is required in the future to investigate how CAMK2γ/CREM axle and other CAMK2γ-related signal pathways are involved in early and late stage liver cancer development.
In summary, this study identifies a novel mechanism by which CAMK2γ antagonizes AKT/mTORC1 signaling in liver during hepatocarcinogenesis. Our findings not only provided a better mechanistic insight into our understanding of compensatory hepatocyte proliferation during hepatocarcinogenesis, but also have important implications for developing new HCC therapies.
Materials and methods
Microarray and computational Kinase Enrichment Analysis
Our previously published microarray data 31 from FXR−/− mice versus wild-type mice was used for this study. Two additional microarray datasets were downloaded from Gene Expression Omnibus (GEO): GEO accession numbers GSE3224434 and GSE12941.36 For the GSE32244 dataset, the gene expression in liver from wild-type mice treated with the carcinogen DEN was compared to liver from control mice. For the GSE12941 dataset, gene expression from HCC patient liver tissue was compared to surrounding non-tumorous liver tissue. Microarray data was analyzed with GEO2R. Differential gene expression was assessed by limma.37 P-values were adjusted using Benjamini & Hochberg False Discovery Rate method. Genes that were significantly up- or down-regulated (adjusted p-value < 0.01) were selected. Additional details of KEA and ChEA analysis are provided in the supporting documents.
Animals
Camk2γ knockout mice were kindly provided by Dr. Johannes Backs at University of Heidelberg, and were backcrossed to C56BL6/J background. The mice were maintained in a pathogen-free animal facility under standard 12:12-h light/dark cycle, and were fed with standard rodent chow and water ad libitum. To induce hepatocellular carcinogenesis, 25 mg/kg DEN (Sigma, Santa Louis, MO) was i.p. injected into 2-weeks-old male mice. The mice were euthanized after 10 months. The DEN and TCPOBOP-induced HCC rodent models were generated according to a previous report.43 Briefly, 100 mg/kg DEN was i.p. injected into 4-weeks-old male mice, and after 2 weeks, 3 mg/kg TCPOBOP (Sigma) was i.p. injected into the mice every two weeks for 8 times. Six months after the DEN treatment, mice were euthanized. All procedures followed the NIH guidelines for the care and use of laboratory animals.
Liver histology, TUNEL, and PCNA staining
Liver specimens were prepared and analyzed by pathologists at City of Hope Pathology Core Lab. Livers were fixed in 4% PBS-buffered formalin, dehydrated and embedded in paraffin, sectioned and processed for H&E and immunostaining. Necrosis and leukocyte infiltration (inflammation) were graded as described previously and below.78 TUNEL and PCNA staining were used to quantify liver cell apoptosis and proliferation using kits from Roche (San Diego, CA) and Invitrogen (San Diego, CA), respectively.
The necrosis grading standard:
Score 0: No focus of necrosis;
Score 0.5: One focus of necrosis
Score 1: Greater than 1 focus of necrosis in at least two regions, but not diffusely scattered throughout the liver lobe
Score 2: Diffuse small foci of CN across an entire liver lobe
Score 3: Diffuse and large area of geographic necrosis
The inflammation grading standard:
Score 0: No inflammation
Score 1: At least one focus or neutrophil-rich inflammation in at least two low-power [×4] fields
Score 2: Greater than two foci of neutrophil-rich inflammation in at least two low-power fields
Score 3: Diffuse and large area of neutrophil-rich inflammation
Cell line and in vitro cell assay
Human HCC cell line Huh7 and HepG2 were purchased from ATCC. Huh7 and HepG2 cell lines with CAMK2γ overexpression was as described elsewhere. For in vitro insulin stimulation assay, HCC cell lines were starved overnight to decrease basal Akt/mTOR activity. The cells were pre-incubated with differently types of kinase inhibitors, including KN-93 (Calbiochem), LY294002 (LC labs), PD98058 (LC labs), and Rapamycin (LC labs) for 30 minutes. The cells then were treated with human recombinant insulin (10 nM) for indicated time. Treated cells were subject to lysis on ice and saved for further analysis.
Antibodies and Western blot
Anti-pCAMK2 and anti-CAMK2γ antibodies were ordered from Santa Cruz Biotechnology (Santa Cruz, CA). All the other antibodies were purchased from Cell Signaling Technology (Danvers, MA).
ALT Analysis
Serum was obtained by centrifuging mouse blood at 3,500 rpm at 4°C for 10 min. Serum ALT were measured at the City of Hope Helford Research Hospital.
Statistical analysis
Data were presented as mean±SEM. Two-tailed Student’s t test was used to determine the significance of differences between data groups.
Supplementary Material
Acknowledgments
We thank Dr. Johannes Backs for providing the Camk2γ KO mice. We also thank Dr. Richard Ermel and the Animal Resource center for the technique support in animal experiments. Similarly, we thank Dr. Yun Yen and Yafan Wang and the Translational Research Core for providing reagents.
This work is supported in part by NCI 1R01-CA139158 and the National Natural Science Foundation of China (81270601 and 81328016).
Footnotes
Conflict of interest
The authors in this manuscript have no conflicts of interest to declare.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Thomas MB, Zhu AX. Hepatocellular carcinoma: the need for progress. J Clin Oncol. 2005;23:2892–2899. doi: 10.1200/JCO.2005.03.196. [DOI] [PubMed] [Google Scholar]
- 2.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–399. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 4.Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 5.He G, Karin M. NF-kappaB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21:159–168. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 7.Bhat M, Sonenberg N, Gores GJ. The mTOR pathway in hepatic malignancies. Hepatology. 2013;58:810–818. doi: 10.1002/hep.26323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pernicova I, Korbonits M. Metformin--mode of action and clinical implications for diabetes and cancer. Nat Rev Endocrinol. 2014;10:143–156. doi: 10.1038/nrendo.2013.256. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Chance MR. Integrating phosphoproteomics in systems biology. Computational and structural biotechnology journal. 2014;10:90–97. doi: 10.1016/j.csbj.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim MS, Zhong J, Pandey A. Common errors in mass spectrometry-based analysis of post-translational modifications. Proteomics. 2016;16:700–714. doi: 10.1002/pmic.201500355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cretich M, Damin F, Pirri G, Chiari M. Protein and peptide arrays: recent trends and new directions. Biomolecular engineering. 2006;23:77–88. doi: 10.1016/j.bioeng.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Reimer U, Reineke U, Schneider-Mergener J. Peptide arrays: from macro to micro. Current opinion in biotechnology. 2002;13:315–320. doi: 10.1016/s0958-1669(02)00339-7. [DOI] [PubMed] [Google Scholar]
- 13.Arsenault R, Griebel P, Napper S. Peptide arrays for kinome analysis: new opportunities and remaining challenges. Proteomics. 2011;11:4595–4609. doi: 10.1002/pmic.201100296. [DOI] [PubMed] [Google Scholar]
- 14.Chen WM, Danziger SA, Chiang JH, Aitchison JD. PhosphoChain: a novel algorithm to predict kinase and phosphatase networks from high-throughput expression data. Bioinformatics. 2013;29:2435–2444. doi: 10.1093/bioinformatics/btt387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen EY, Xu H, Gordonov S, Lim MP, Perkins MH, Ma’ayan A. Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics. 2012;28:105–111. doi: 10.1093/bioinformatics/btr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azeloglu EU, Hardy SV, Eungdamrong NJ, Chen Y, Jayaraman G, Chuang PY, et al. Interconnected network motifs control podocyte morphology and kidney function. Science signaling. 2014;7:ra12. doi: 10.1126/scisignal.2004621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasudevan HN, Soriano P. SRF regulates craniofacial development through selective recruitment of MRTF cofactors by PDGF signaling. Developmental cell. 2014;31:332–344. doi: 10.1016/j.devcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Minicis S, Seki E, Uchinami H, Kluwe J, Zhang Y, Brenner DA, et al. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology. 2007;132:1937–1946. doi: 10.1053/j.gastro.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 20.Zhang SJ, Steijaert MN, Lau D, Schutz G, Delucinge-Vivier C, Descombes P, et al. Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron. 2007;53:549–562. doi: 10.1016/j.neuron.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 23.Hell JW. CaMKII: claiming center stage in postsynaptic function and organization. Neuron. 2014;81:249–265. doi: 10.1016/j.neuron.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swaminathan PD, Purohit A, Soni S, Voigt N, Singh MV, Glukhov AV, et al. Oxidized CaMKII causes cardiac sinus node dysfunction in mice. J Clin Invest. 2011;121:3277–3288. doi: 10.1172/JCI57833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Timmins JM, Ozcan L, Seimon TA, Li G, Malagelada C, Backs J, et al. Calcium/calmodulin-dependent protein kinase II links ER stress with Fas and mitochondrial apoptosis pathways. The Journal of clinical investigation. 2009;119:2925–2941. doi: 10.1172/JCI38857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Florian MC, Nattamai KJ, Dorr K, Marka G, Uberle B, Vas V, et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature. 2013;503:392–396. doi: 10.1038/nature12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Midorikawa R, Takei Y, Hirokawa N. KIF4 motor regulates activity-dependent neuronal survival by suppressing PARP-1 enzymatic activity. Cell. 2006;125:371–383. doi: 10.1016/j.cell.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 29.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lachmann A, Ma’ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25:684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu N, Meng Z, Lou G, Zhou W, Wang X, Zhang Y, et al. Hepatocarcinogenesis in FXR−/− mice mimics human HCC progression that operates through HNF1alpha regulation of FXR expression. Mol Endocrinol. 2012;26:775–785. doi: 10.1210/me.2011-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 33.Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 36.Satow R, Shitashige M, Kanai Y, Takeshita F, Ojima H, Jigami T, et al. Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin Cancer Res. 2010;16:2518–2528. doi: 10.1158/1078-0432.CCR-09-2214. [DOI] [PubMed] [Google Scholar]
- 37.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 38.Rabinowitz KM, Wang Y, Chen EY, Hovhannisyan Z, Chiang D, Berin MC, et al. Transforming growth factor beta signaling controls activities of human intestinal CD8(+)T suppressor cells. Gastroenterology. 2013;144:601–612. e601. doi: 10.1053/j.gastro.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldman A, Roy J, Bodenmiller B, Wanka S, Landry CR, Aebersold R, et al. The calcineurin signaling network evolves via conserved kinase-phosphatase modules that transcend substrate identity. Molecular cell. 2014;55:422–435. doi: 10.1016/j.molcel.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rexer BN, Ham AJ, Rinehart C, Hill S, de Granja-Ingram NM, Gonzalez-Angulo AM, et al. Phosphoproteomic mass spectrometry profiling links Src family kinases to escape from HER2 tyrosine kinase inhibition. Oncogene. 2011;30:4163–4174. doi: 10.1038/onc.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng Z, Li T, Ma X, Wang X, Van Ness C, Gan Y, et al. Berbamine inhibits the growth of liver cancer cells and cancer-initiating cells by targeting Ca(2)(+)/calmodulin-dependent protein kinase II. Mol Cancer Ther. 2013;12:2067–2077. doi: 10.1158/1535-7163.MCT-13-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu Y, Chen T, Meng Z, Gan Y, Xu X, Lou G, et al. CaMKII gamma, a critical regulator of CML stem/progenitor cells, is a target of the natural product berbamine. Blood. 2012;120:4829–4839. doi: 10.1182/blood-2012-06-434894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, et al. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol Endocrinol. 2005;19:1646–1653. doi: 10.1210/me.2004-0520. [DOI] [PubMed] [Google Scholar]
- 44.Meng Z, Wang X, Gan Y, Zhang Y, Zhou H, Ness CV, et al. Deletion of IFNgamma enhances hepatocarcinogenesis in FXR knockout mice. J Hepatol. 2012;57:1004–1012. doi: 10.1016/j.jhep.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 46.Sakurai T, Maeda S, Chang L, Karin M. Loss of hepatic NF-kappa B activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10544–10551. doi: 10.1073/pnas.0603499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishiguro K, Green T, Rapley J, Wachtel H, Giallourakis C, Landry A, et al. Ca2+/calmodulin-dependent protein kinase II is a modulator of CARMA1-mediated NF-kappaB activation. Mol Cell Biol. 2006;26:5497–5508. doi: 10.1128/MCB.02469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Si J, Collins SJ. Activated Ca2+/calmodulin-dependent protein kinase IIgamma is a critical regulator of myeloid leukemia cell proliferation. Cancer research. 2008;68:3733–3742. doi: 10.1158/0008-5472.CAN-07-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soliman GA, Acosta-Jaquez HA, Dunlop EA, Ekim B, Maj NE, Tee AR, et al. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J Biol Chem. 2010;285:7866–7879. doi: 10.1074/jbc.M109.096222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdul-Ghani R, Serra V, Gyorffy B, Jurchott K, Solf A, Dietel M, et al. The PI3K inhibitor LY294002 blocks drug export from resistant colon carcinoma cells overexpressing MRP1. Oncogene. 2006;25:1743–1752. doi: 10.1038/sj.onc.1209201. [DOI] [PubMed] [Google Scholar]
- 51.De Fea K, Roth RA. Protein kinase C modulation of insulin receptor substrate-1 tyrosine phosphorylation requires serine 612. Biochemistry. 1997;36:12939–12947. doi: 10.1021/bi971157f. [DOI] [PubMed] [Google Scholar]
- 52.Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mothe I, Van Obberghen E. Phosphorylation of insulin receptor substrate-1 on multiple serine residues, 612, 632, 662, and 731, modulates insulin action. J Biol Chem. 1996;271:11222–11227. doi: 10.1074/jbc.271.19.11222. [DOI] [PubMed] [Google Scholar]
- 54.Illario M, Monaco S, Cavallo AL, Esposito I, Formisano P, D’Andrea L, et al. Calcium-calmodulin-dependent kinase II (CaMKII) mediates insulin-stimulated proliferation and glucose uptake. Cell Signal. 2009;21:786–792. doi: 10.1016/j.cellsig.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 55.Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- 56.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 57.Ozcan L, Wong CC, Li G, Xu T, Pajvani U, Park SK, et al. Calcium signaling through CaMKII regulates hepatic glucose production in fasting and obesity. Cell Metab. 2012;15:739–751. doi: 10.1016/j.cmet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng GS. Conflicting roles of molecules in hepatocarcinogenesis: paradigm or paradox. Cancer Cell. 2012;21:150–154. doi: 10.1016/j.ccr.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakurai T, Maeda S, Chang L, Karin M. Loss of hepatic NF-kappa B activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc Natl Acad Sci U S A. 2006;103:10544–10551. doi: 10.1073/pnas.0603499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matter MS, Decaens T, Andersen JB, Thorgeirsson SS. Targeting the mTOR pathway in hepatocellular carcinoma: current state and future trends. J Hepatol. 2014;60:855–865. doi: 10.1016/j.jhep.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calvisi DF, Wang C, Ho C, Ladu S, Lee SA, Mattu S, et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology. 2011;140:1071–1083. doi: 10.1053/j.gastro.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horie Y, Suzuki A, Kataoka E, Sasaki T, Hamada K, Sasaki J, et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004;113:1774–1783. doi: 10.1172/JCI20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Menon S, Yecies JL, Zhang HH, Howell JJ, Nicholatos J, Harputlugil E, et al. Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in mice. Science signaling. 2012;5:ra24. doi: 10.1126/scisignal.2002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kenerson HL, Yeh MM, Kazami M, Jiang X, Riehle KJ, McIntyre RL, et al. Akt and mTORC1 have different roles during liver tumorigenesis in mice. Gastroenterology. 2013;144:1055–1065. doi: 10.1053/j.gastro.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patsenker E, Schneider V, Ledermann M, Saegesser H, Dorn C, Hellerbrand C, et al. Potent antifibrotic activity of mTOR inhibitors sirolimus and everolimus but not of cyclosporine A and tacrolimus in experimental liver fibrosis. J Hepatol. 2011;55:388–398. doi: 10.1016/j.jhep.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 66.Zhang ZJ, Zheng ZJ, Shi R, Su Q, Jiang Q, Kip KE. Metformin for liver cancer prevention in patients with type 2 diabetes: a systematic review and meta-analysis. The Journal of clinical endocrinology and metabolism. 2012;97:2347–2353. doi: 10.1210/jc.2012-1267. [DOI] [PubMed] [Google Scholar]
- 67.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 68.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nature reviews Molecular cell biology. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 70.Carriere A, Cargnello M, Julien LA, Gao H, Bonneil E, Thibault P, et al. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol. 2008;18:1269–1277. doi: 10.1016/j.cub.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 71.Ozcan L, Cristina de Souza J, Harari AA, Backs J, Olson EN, Tabas I. Activation of calcium/calmodulin-dependent protein kinase II in obesity mediates suppression of hepatic insulin signaling. Cell Metab. 2013;18:803–815. doi: 10.1016/j.cmet.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ravichandran LV, Esposito DL, Chen J, Quon MJ. Protein kinase C-zeta phosphorylates insulin receptor substrate-1 and impairs its ability to activate phosphatidylinositol 3-kinase in response to insulin. J Biol Chem. 2001;276:3543–3549. doi: 10.1074/jbc.M007231200. [DOI] [PubMed] [Google Scholar]
- 73.Boehm J, Kang MG, Johnson RC, Esteban J, Huganir RL, Malinow R. Synaptic incorporation of AMPA receptors during LTP is controlled by a PKC phosphorylation site on GluR1. Neuron. 2006;51:213–225. doi: 10.1016/j.neuron.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 74.DeVries TA, Kalkofen RL, Matassa AA, Reyland ME. Protein kinase Cdelta regulates apoptosis via activation of STAT1. J Biol Chem. 2004;279:45603–45612. doi: 10.1074/jbc.M407448200. [DOI] [PubMed] [Google Scholar]
- 75.Nair JS, DaFonseca CJ, Tjernberg A, Sun W, Darnell JE, Jr, Chait BT, et al. Requirement of Ca2+ and CaMKII for Stat1 Ser-727 phosphorylation in response to IFN-gamma. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:5971–5976. doi: 10.1073/pnas.052159099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Groot RP, den Hertog J, Vandenheede JR, Goris J, Sassone-Corsi P. Multiple and cooperative phosphorylation events regulate the CREM activator function. The EMBO journal. 1993;12:3903–3911. doi: 10.1002/j.1460-2075.1993.tb06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ella E, Heim D, Stoyanov E, Harari-Steinfeld R, Steinfeld I, Pappo O, et al. Specific genomic and transcriptomic aberrations in tumors induced by partial hepatectomy of a chronically inflamed murine liver. Oncotarget. 2014;5:10318–10331. doi: 10.18632/oncotarget.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papa S, Zazzeroni F, Fu YX, Bubici C, Alvarez K, Dean K, et al. Gadd45beta promotes hepatocyte survival during liver regeneration in mice by modulating JNK signaling. J Clin Invest. 2008;118:1911–1923. doi: 10.1172/JCI33913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.