Abstract
Background
The emergence of carbapenemase producing bacteria, especially New Delhi metallo-β-lactamase (NDM-1) and its variants, worldwide, has raised amajor public health concern. NDM-1 hydrolyzes a wide range of β-lactam antibiotics, including carbapenems, which are the last resort of antibiotics for the treatment of infections caused by resistant strain of bacteria.
Main body
In this review, we have discussed bla NDM-1variants, its genetic analysis including type of specific mutation, origin of country and spread among several type of bacterial species. Wide members of enterobacteriaceae, most commonly Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae, and gram-negative non-fermenters Pseudomonas spp. and Acinetobacter baumannii were found to carry these markers. Moreover, at least seventeen variants of bla NDM-type gene differing into one or two residues of amino acids at distinct positions have been reported so far among different species of bacteria from different countries. The genetic and structural studies of these variants are important to understand the mechanism of antibiotic hydrolysis as well as to design new molecules with inhibitory activity against antibiotics.
Conclusion
This review provides a comprehensive view of structural differences among NDM-1 variants, which are a driving force behind their spread across the globe.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-017-1012-8) contains supplementary material, which is available to authorized users.
Keywords: Enterobacteriaceae, New Delhi-Metallo-Beta-Lactamases, Carbapenemases, Antibiotic resistance
Background
Although antibiotics were developed to fight infections, the emergence of new resistant markers, especially New Delhi-metallo-beta-lactamases (NDM-1), hampered the capability of all antibiotics of beta lactam group to treat infections caused by microorganisms carrying such resistances. The possible reason for evolving trends of new markers is mutations [1], which may cause delaying in the discovery of new antibiotics for treatments and hence became a great public threat [2]. The overuse of antibiotics is one of the reasons to cause resistance, due to increase selective pressure in a specific population of bacteria allowing the resistant bacteria to bloom and the susceptible bacteria to pass away.
Enzymes are evolving over a period of time by mutations in response to environmental pressure for increased stability and fitness leading to its functional changes. The activity of an enzyme and its future generations success in response to change conditions due to environmental stress and its improved physiological utility for constant perseverance is determined by these evolutionary drivers. Recent reports on antibiotic resistance has made a clear understanding of evolving status of β-lactamase enzymes, which are key player for antibiotic resistance [3].
In Enterobacteriaceae and other Gram-negative bacteria including Pseudomonas and Acinetobacter species, production of carbapenemases has become a noteworthy mechanism for broad-spectrum β-lactam resistance [4]. Carbapenemases may be defined as specific beta-lactamases, which hydrolyze carbapenem group of antibiotics. These are involved in acquired resistance and belong to Ambler molecular classes A, B and D [4].
Intestinal carriage of carbapenemase-producing organisms (CPOs) is an important source of its transmission [5]. However, detection of carbapenemase producing Gram-negative bacteria has become a major concern for the hospital settings to control infections. Use of multiplex PCR analyses and DNA microarray have been reported as rapid detection systems. Most prevalent carbapenemases detected by these systems are KPC and OXA serine carbapenemases [6].
A number of new variants of class A carbapenemases (e.g., KPC and GES enzymes), class B metallo-beta-lactamases (e.g., IMP, VIM and NDM metallo-beta-lactamases), and class D carbapenemases (e.g., OXA-23) are emerging over time scale. Moreover, over-expression of class C beta-lactamases, such as CMY-10 and PDC type beta-lactamases, which are weak carbapenemases, can also lead to carbapenem resistance, especially in combination with other resistance mechanisms [7].
Metallo-beta-Lactamases (MBLs) are class B β-lactamases that hydrolyze almost all clinically-available β-lactam antibiotics and feature the distinctive αβ/βα sandwich fold of the metallo-hydrolase/oxidoreductase superfamily. MBLs possess a shallow active-site groove with one or two divalent zinc ions, bordered by flexible loops [8]. In NDM-1 this flexible hairpin loop moves over the zinc ion for hydrolysis and is later removed after the catalysis [9]. The 3D structure of NDM-1 with active site and Zinc molecules is shown in Fig. 1. MBLs are classified into three subclasses (B1, B2 and B3), according to sequence identity and zinc ion dependence, of which the B1 subclass included most clinically significant enzymes. Not many inhibitors have been successfully designed due to the nature of zinc ligands, catalytic mechanisms and the differences among the active site architecture [8]. The evolution of varied and detrimental range of β-lactamases has lost the effectiveness of β-Lactamase inhibitors (BLIs) which could play an important role in combating β-lactam resistance in Gram-negative bacteria [10]. A triple combination of meropenem/piperacillin/tazobactam β-lactams, has been proved as one of the strategies to kill Methicillin-resistant Staphylococcus aureus (MRSA) in vitro as well as in a mouse model through a novel synergistic mechanism of action [10].
Fig. 1.
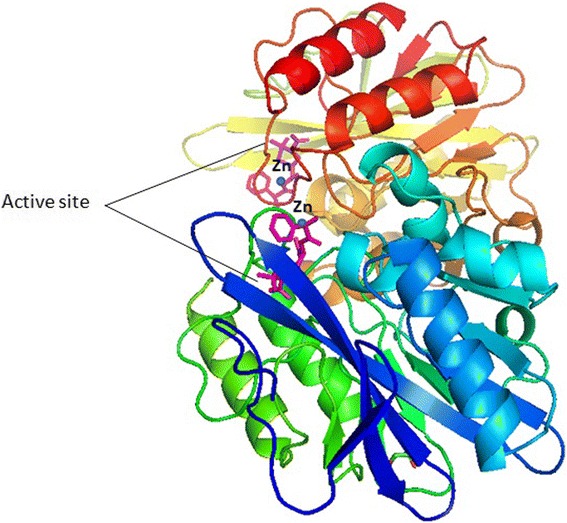
3D structure of NDM-1 protein backbone shown with helices and strands, the two zinc ions at the active sites are shown as blue spheres
A bacterium carrying several antibiotic-resistant genes is called multi-resistant bacteria or informally, a “super bacteria” or “super bug”; infections caused by them are difficult to treat [11]. Most probably, a very rare “genetic fusion” is thought to occur between two previously known antibiotic-resistant genes that evolved to a new mutant called NDM-1.The product of the bla NDM-1 gene is NDM, an enzyme hydrolysing broad range of antibiotics, including the carbapenems, which are considered as last resort of antibiotics. In the last few years, 17 new variants of NDM-1 have been evolved by changing one or two residues at different positions [12–15]. The emergence of bacteria carrying such genes represent a big challenge for physicians to treat infected patients.
Mechanism of resistance
The expression of β-lactamases, efflux pumps and alteration of porins and penicillin binding proteins (PBPs) are the common mechanism for carbapenem resistance in member of Enterobacteriaceae. Combinations of these mechanisms can cause high levels of resistance to carbapenems in certain bacterial species, such as K. pneumoniae, Pseudomonas aeruginosa and A. baumannii. In P. aeruginosa carbapenem resistance is contributed also by the loss of OprD porin leading to decrease in outer membrane permeability, increase in cytoplasmic membrane active efflux pump system, up regulation and alteration in carbapenem hydrolyzing enzymes and penicillin binding proteins [16]. Acquisition of metallo-beta lactamases (MBL), which hydrolyze the carbapenems and all beta lactams except the monobactams, is one of the emerging mechanism of carbapenem resistance [17]. Multi-drug-resistant Pseudomonas aeruginosa (MDRPA) infection risk factors include immunocompromised states, prolonged hospitalization and antimicrobial therapy [18]. NDM-1 producing P. aeruginosa isolates were detected for the first time in Serbia [19]. In all four P. aeruginosa isolates detected, bla NDM-1 genes were present on 50 kb plasmid (Gene Bank accession numbers JX680682, JX680683, JX680684 and JX680685) [20].The co-expression of bla NDM-1 and MexAB-OprM efflux pump occurred into a P. aeruginosa strain upon single dose of meropenem therapy, thus suggesting that both mechanisms contribute to carbapenem resistance, although the efflux system played the major role [21]. One more example of combinatorial effects in A. baumannii harbouring bla NDM is the expression of multiple efflux systems and altered membrane permeability [22].
A distinction exists between resistance to carbapenems in Gram-positive cocci and Gram-negative rods. In Gram-positive cocci, carbapenem resistance is typically the result of substitutions in amino acid sequences of PBPs or acquisition/production of a new carbapenem-resistant PBP. Expression of beta-lactamases and efflux pumps, as well as porin loss and alterations in PBP, are all associated with carbapenem resistance in Gram-negative rods [23]. For example, a clinical strain HPC299 Acinetobacter bereziniae, harbouring bla NDM-1, uses multidrug efflux pumps as its adaptation strategy for survival under different environmental conditions [24]. Carbapenem resistance mechanisms not related to carbapenemase production include increase in efflux pump activity [25] and modifications of outer membrane porin profiles, which regulate access of carbapenems to the cell wall [26].
Multi-drug resistance by ndm-1 producing bacteria
Background of NDM-1 producers
There are hundreds of commensal strains of E. coli bacteria, which are not associated with any infectious diseases. However, emergence of a new mutant strain known as NDM-1 producing E. coli has thrown light on the fact that the development of antibiotic resistance among microorganisms can transform commensals into pathogens. Many NDM-1 variants evolved in Enterobacteriaceae, Vibrionaceae and other non-fermenters by single and double amino acid residue substitutions at different positions [27], for e.g., NDM-1 (major variant), NDM-2, NDM-3, NDM-4and NDM-5 (minor variants), reported worldwide [12, 28, 29]. New Delhi metallo-beta lactamase (NDM) produced by bacterial isolates from the Indian subcontinent are the latest carbapenemases, which hydrolyze all beta lactam antibiotics (except aztreonam), including the broad spectrum antibiotic “carbapenems”, thereby causing havoc in hospitals and community [30]. The gene encoding NDM-1 is often carried by plasmids and hence easily moves to other microorganisms via horizontal gene transfer, thereby increasing the probability of emergence of drug resistant strains of pathogenic microorganisms [31].
Major healthcare risk of NDM producers
NDM-1 strains are particularly hazardous because: (i) most plasmids detected in these bacteria are transferable and capable of wide rearrangement, suggesting a widespread horizontal transmission and flexibility among bacterial populations; (ii) there is lack of a routine standardized phenotypic test for metallo-beta-lactamase (MBL) detection; (iii) there is consequent probable high prevalence of unrecognized asymptomatic carriers; (iv) there is a lack of available effective antibiotics for the treatment of multi-drug resistant NDM-1 expressing bacteria [31].
NDM-1 producing E. coli infects the host by commonly invading sites like, urinary tract, blood, lungs, and wounds, leading to urinary tract infections, septicaemia, pulmonary infections, diarrhoea, peritonitis, device-associated infections and soft tissue infections [12]. These antibiotic resistant bacteria express type IV secretion system as their virulence factor, which allows them to introduce bacterial proteins and enzymes inside the host cell, thereby controlling the host cell metabolism [32]. Mode of transmission of NDM-1 producing strain could either be through cross-contamination during food preparation or via body fluids and may occur in the community or in the hospital setting [33].
Worldwide distribution of NDM variants across the globe
Asian continent serves as the major reservoir of NDM producers, with around 58.15% abundance of NDM-1 variant distributed mostly in China and India. Additional file 1: Table S1 shows detailed prevalence of NDM-1 and its variants in different countries worldwide. Europe shows around 16.8% of the total producers, with the maximum spread of NDM-1 variant in Bulgaria, Romania, Poland, France, Italy, Turkey, Germany, Greece, Serbia, London, Ukraine, Croatia, Azerbaijan and Ireland. NDM-4 is also reported to be distributed in European subcontinent in Italy, while NDM-5 and NDM-7 are prevalent in Denmark and France (Additional file 1: Table S1). American continent shows around 10.8% abundance of the total NDM-1 producers as reported globally, of which subcontinent Brazil serves as the major reservoir while Colorado, Mexico city, California, Georgia, Illinois, Paraguay, Pennsylvania, Florida, Argentina, Jamaica, Uruguay and Ecuador are considered as minor pool (Additional file 1: Table S1). Africa carries around 10.8% pool of the total NDM-1 producers scattered globally. African subcontinent, Algeria showed major distribution, whereas Greater Johannesburg Area, KwaZulu-Natal, Libya, Madagascar, Egypt and Tunisia demonstrated low prevalence of these NDM-1 producers. NDM-5 is also reported to be distributed in Algeria (Additional file 1: Table S1). Australia serves as the 1.6% reservoir of the total NDM-1 producers distributed in Brisbane, Perth and New Zealand. Highest distribution of these NDM variants is detected in K. pneumoniae and E. coli species (Additional file 1: Table S1).
NDM-1 producers were found resistant to imipenem, meropenem, ertapenem, gentamicin, amikacin, tobramycin, and ciprofloxacin, whereas, isolates were found susceptible to colistin (MICs ≤4 mg/L) and to tigecycline (MICs ≤1 mg/L) [34]. Non-clonal Indian isolates from Chennai had bla NDM-1 exclusively on plasmids of size ranging from 50 to 350 kb, whereas another clone of K. pneumoniae isolated in Haryana was found to have plasmid of predominately either 118 kb or 50 kb, suggesting wide environmental spread of bla NDM-1 [34]. Plasmid profiling showed that a plasmid of size 50 kb carries bla NDM-1in Enterobacteriaceae, which were found resistant to almost all antimicrobials except tigecycline and colistin [34].
In Europe, dissemination of NDM-1 has been observed in A. baumannii isolates assigned to international clonal lineage I and to the emerging genotypes ST25 and ST85 [35, 36]. The bla NDM-1 gene was inserted within a Tn125-like transposon which was either chromosomally-located [35] or plasmid-located [35, 36] (Fig. 2).
Fig. 2.
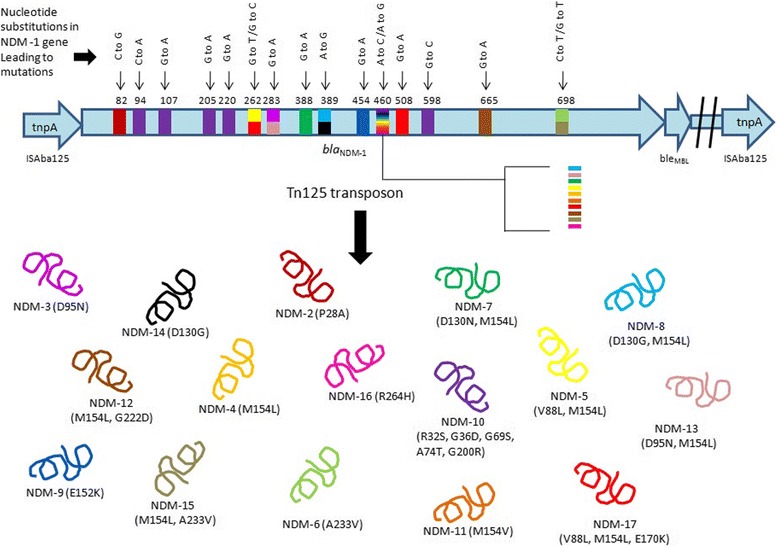
A schematic representation of bla NDM-1 gene carrying Tn125 transposon, showing the mutations at various nucleotide positions leading to the occurrence of NDM variants. Each unique colour of NDM variant in lower panel showing mutant residues at different position and the same is reflected in gene with the same colour at different position and nucleotide (s)
NDM-producing resistant E. coli strains were also found in animal sources [37]. Acinetobacter lwoffii carrying bla NDM-1 gene on plasmid were isolated from chicken rectal swab [38]. The bla NDM gene detection in dairy cattle is a matter of concern because it may lead to spread through food chain. Sequence analysis revealed a gene showing 100% homology with E. coli (JQ348841.1) bla NDM-5 gene and 99% homology with E. coli (JQ348841.1) bla NDM-4 in Pseudomonas aeruginosa (HF546976.1), K. pneumoniae (KC178689.1), Raoultella ornithinolytica (JX680686.1), A. baumannii (KC404829.1, KC347597.1). Apart from this, NDM-1 producing Enterobacter cloacae (EC15) and K. pneumoniae (KP12) strains were isolated from two patients with diabetic foot ulcers in 2010 from northern part of India [39].
The origin of NDM-1 started in the year 2008 when the first case of a NDM-1 episode was reported in a Swedish patient previously hospitalized in New Delhi, suffering from a multidrug-resistant K. pneumoniae, urinary tract infection [40]. Based on the number of victims affected with NDM-1 strains in various parts of the globe, it has been estimated that the Indian subcontinent is the main reservoir of NDM-1 producers [39], next down the line is United Kingdom. On the other hand, Belgium, China, Japan, France, Austria, Germany, Norway, Hong Kong, Sweden, Netherland, Australia and Canada also serve as the secondary reservoirs of bla NDM genes [39] as shown in Additional file 1: Table S1. An average of 1000–1600 patients are admitted daily to the hospitals worldwide as a result of infections due to drug resistant bacteria [41]. It is difficult to predict the rate of spread of the gene encoding NDM- 1, although exchange of the bla NDM-1 gene among unrelated bacterial isolates have been identified already in Enterobacteriaceae and A. baumannii [34]. An increase in population exchange at global level and enhanced medical tourism could play a significant role in spreading uncontrolled NDM-1 related resistance worldwide.
Genetic and Biochemical analysis of NDM variants
The bla NDM-1 gene which encodes for the New Delhi metallo-β-lactamase 1 (NDM-1) is commonly found among members of Enterobacteriaceae and Pseudomonas species [34, 42]. The above bacteria are highly resistant to all antibiotics including carbapenems and aminoglycosides because of co-existence of rmtF methylase gene in most of the isolates [43], but susceptible to tigecycline and colistin [34]. However, bla NDM-9 producing colistin resistant E. coli strain was recently discovered in a chicken meat sample in Guangzhou, China [44]. 16S rRNA methyl transferases responsible for high resistance to antibiotics were reported in bla NDM-1 positive Pseudomonas aeruginosa isolates in co-association with rmtC and rmtF genes on the chromosome [45]. IncR plasmid carrying bla NDM-1 was also reported in Citrobacter koseri acting as a reservoir for multidrug resistance [46]. bla NDM-1 was associated with different plasmid scaffolds (IncFII, IncL/M, IncN, IncR, IncHIB-M/FIB-M), IncF type being the prevalent one. Genetic structures surrounding bla NDM-1 showed its association with at least a remnant of ISAba125 at its 5′-end [47]. Tn125 composite transposon in A. baumannii has been demonstrated to be responsible for bla NDM-1 gene dissemination within Acinetobacter species and Enterobacteriaceae [48]. ble MBL gene, which confers resistance to anti-tumor glycopeptide molecule bleomycin, is found downstream of bla NDM-1 gene [49] (Fig. 2). There has been an exponential increase in resistance among Gram-negative bacteria compared with Gram-positive bacteria [50, 51], while not many new active antibiotics are developed against Gram-negative bacteria [52–54]. Increase in its resistance is mainly due to the presence of mobile elements into conjugative plasmids, which can readily spread through bacterial populations.
The isolates obtained from UK had a more diverse range of plasmid sizes ranging from 80 kb to greater than 500 kb [34]. For example, a RB151 strain was reported to harbour 108 kb plasmid carrying NDM-1 gene on 4.8 Mbp chromosome [55]. E. coli Y5 isolate was found to have bla NDM-1 on chromosome as well [56].The bla NDM-1 was also carried by more than one plasmid in some isolates. Most of the plasmids carrying bla NDM-1 shows transmissibility and plasticity enabling them to diversify and spread among bacterial populations with an alarming potential; many of them were of incompatibility A/C types [34], which is not commonly associated group among multidrug-resistant phenotypes. The emergence of new variants of NDM-1 are taking place in India due to widespread use of antibiotics leading to huge selection pressure. Only few antibiotics against Gram-negative bacteria are available and none of them is active against producers of NDM-1 [57]. Large conjugative plasmids are seen to harbour blaNDM-1 gene along with determinants of antibiotic resistance [58].
In the United States, K. pneumoniae is the most common CRE (carbapenem-resistant Enterobacteriaceae) species, resistant to nearly all available antibiotics encountered, typically as a hospital-acquired infection with high mortality and morbidity rate [59, 60]. ATCC BAA-2146 (Kpn2146) a strain of K. pneumoniae was the first reported U.S isolate encoding NDM-1 along with additional antibiotic resistance determinants on plasmid of size 140.8 kb.
At least one zinc atom is present on the active site of all metallo-carbapenemases, which facilitates bicyclic β-lactam ring hydrolysis [61]. Metallo-carbapenemases have the ability to hydrolyze commercially available carbapenem resistant β-lactamase inhibitors but show sensitivity to metal ion chelators. Along with carbapenems they can also hydrolyze penicillins and cephalosporins, while the ability to hydrolyze aztreonam is lacking. Hydrolysis occurs when zinc ions on active site interacts with β-lactams giving distinctive inhibition trait by EDTA.
It has been reported that the bla NDM is carried by various types of plasmids such as IncA/C, IncF, IncNIncL/M or untypable/IncR, and is rarely found to be chromosomally integrated [62]. Plasmid characterization demonstrated that different mechanism leads to acquisition of NDM gene even if it is located on very closely related plasmids [63]. The sequencing of few plasmids reveals bla NDM association with insertion sequences and transposons, which regulate its horizontal gene transfer and aadB, dfrA12, bla OXA-30 and aacA4 additional resistance markers [64]. Tn3000 transposon has been reported to be responsible for bla NDM-1 dissemination among enterobactericeae [65]. A non-active site residue Trp 93 is found to play role in maintaining the structural integrity of NDM-1, although not being directly involved in recognition and catalysis [66]. Recently, a new plasmid type IncX3 is reported to be responsible for making the spread of NDM gene more effective [67], for example in China IncX3 and InA/C plasmids were reported to be responsible for spread of bla NDM genes [68].
Recent studies demonstrated the coexistence of NDM-1 gene along with other resistant genes, such as IMP-1 in Acinetobacter species [69], co-expression of NDM-1 and OXA-232 in an E. coli isolate was reported from Germany [70], co-production of NDM-5 and MCR-1 in ST648 and ST156 E.coli isolates from fowl in China [71]. Coexistence of bla NDM-1 and bla OXA-48 carrying plasmids was reported in an isolate of K. pneumoniae from China [72]. Similarly, coexistence of bla NDM-1 and bla OXA-23 was reported in A. baumannii from Nepal [73]. K. pneumoniae of type 11 was reported in Greek to be coproducing two metallo beta-lactamses markers, NDM-1 and VIM-1 together [74]. A report on E. coli isolate obtained from a patient in Thailand and another report from China, showed the presence of colistin resistant mcr-1 gene along with beta lactamases genes such as NDM-1 [75–77]. In E. coli ST471 isolated from Turkey, NDM-1 was found present along with CTX-M-9, TEM, SHV and rmtC [78]. Another coexistence of bla SHV-12 along with bla NDM-13 on a self-transferable plasmid of about 54 kb size isolated from E. coli in China was reported [79]. Coproduction of NDM-5 along with mcr-1 in China, NDM-7 along with OXA-48 in Spain and NDM-9 along with MCR-1 in Taiwan was reported recently [80–82]. Also, the presence of both NDM-9 and MCR-1 was reported in Cronobacter sakazakii and E. coli [83].
To date, a number of variants of New Delhi metallo-β-lactamase-1(NDM-1) have been reported. Of these variants, NDM-2 had a substitution of Cysteine to Glycine at position 82, and amino acid being substituted by alanine at position 28 in place of proline, in A. baumannii [84] (Table 1). However, 16S RNA methylase and extended-spectrum- β-lactamases were not detected. Moreover, strains carrying bla NDM-2 lacked detectable plasmids and the bla NDM-2 was not seen to be transferred by conjugation [84].
Table 1.
Genetic variations among the NDM-1 and its variants and its first source of spread
| NDM-1 variants | Amino acid(s) substitution | Source organism(s) |
|---|---|---|
| NDM-2 | Proline 28 to Alanine | Acinetobacter baumannii |
| NDM-3 | Aspartate 95 to Asparagine | Escherichia coli |
| NDM-4 | Methionine 154 to Leucine | Escherichia coli |
| NDM-5 | Valine 88 to Leucine Methionine 154 to Leucine |
Escherichia coli |
| NDM-6 | Alanine 233 to Valine | Escherichia coli |
| NDM-7 | Aspartate 130 to Asparagine Methionine 154 to Leucine |
Escherichia coli |
| NDM-8 | Aspartate 130 to Glycine Methionine 154 to Leucine |
Escherichia coli |
| NDM-9 | Glutamic Acid 152 to Lysine | Klebsiella pneumoniae |
| NDM-10 | Arginine 32 to Serine, Glycine 36 toAspartic acid, Glycine 69 to serine, Alanine 74 to threonine, Glycine 200 to Arginine |
Klebsiella pneumoniae |
| NDM-11 | NA | Escherichia coli |
| NDM-12 | Glycine 222 to Aspartic acid and Methionine 154 to Leucine | Escherichia coli |
| NDM-13 | Aspartic acid 95 to Asparagine and Methionine 154 to Leucine | Escherichia coli |
| NDM-14 | Aspartic acid 130 to Glycine | Acinetobacter lwoffii |
| NDM-15 | Alanine 233 to valine Methionine 154 to Leucine |
Escherichia coli |
| NDM-16 | Arginine 264 to Histidine | Klebsiella pneumoniae |
| NDM-17 | Valine 88 to Leucine, Methionine 154 to Leucine and Glutamic acid 170 to Lysine | Escherichia coli |
Another variant NDM-3 with an amino acid substitution of Aspartate to Asparagine at position 95 was observed in E. coli [85] (Table 1). NDM-3 showed similar enzyme activities against β-lactams like those of NDM-1, although slightly lower Kcat/Km ratios for all the β-lactams tested except for doripenem was seen, which is caused by the lower Kcat values of NDM-3 being 19.0 to 47.5% as compared to NDM-1 [86]. In fact, the decreased Kcat values and the decrease in hydrolysis rate of all tested β-lactams except for doripenem is due to subsitution of Asn from Asp at position 95. Residue 95 is found to be in α1, located on the surface of the protein [86]. The crystal structure study of NDM-1 revealed that the NDM-1 active site is located at the bottom of a shallow groove being enclosed by two important loops named L3 and L10. However, α1 95th residue was not located in these loops and indirectly may affect the interaction of the substrate with the active site [86]. Among 9 NDM variants, substitutions of amino acids were identified at 7 different positions (28, 88, 95, 130, 152, 154, and 233), but which position(s) plays a critical role in the enzymatic activities, remained unclear. For bla NDM-3 the genetic context tnpA-bla NDM-3-ble MBL-trpF-dsbC-tnpA-sulI-qacEdeltaI-aadA2-dfrA1, was present on approximately 250-kb plasmid. The bla NDM-3 and bla NDM-1 gene expression in E. coli DH5α conferred reduced susceptibility and resistance to all cephalosporins, moxalactam, and carbapenems. E. coli expressing NDM-3 showed 2-fold higher MIC of cefpirome than the one expressing NDM-1 in contrast to those of 2-fold lower MIC of cefepime, cefoselis, cefotaxime, cefoxitin, imipenem, meropenem, and penicillin G for NDM-3 than NDM-1. Recombinant NDM-3 and NDM-1 hydrolyzed all tested β-lactams except for aztreonam [86].
NDM-4 variant showed substitution of amino acid from 154th Methionine to Leucine in E. coli [87] (Table 1). NDM-4-producing E. coli isolate from a North Indian hospital sewage was recently reported by Khan and Parvez [15]. Gene expression of bla NDM-1 and bla NDM-4 in E. coli TOP10 conferred lower susceptibility or resistance to all β-lactams except aztreonam. However, the MICs of imipenem and ertapenemwere found to be higher for E. coli expressing NDM-4 than the one expressing NDM-1, suggesting the role of Leu154 residue in the high carbapenemase activity [87]. NDM-4 β-lactamase hydrolyzed all tested β-lactams except for aztreonam, just similar to other MBLs. Kinetic data showed higher level of hydrolysis of imipenem by NDM-4 than by NDM-1. Similarly, catalytic activity of NDM-4 for meropenem was slightly higher than that of NDM-1. NDM-4 showed higher catalytic efficiencies for cefalotin, ceftazidime, and cefotaxime, as cefepime was less hydrolyzed by NDM-4. Higher Kcat values for NDM-4 than NDM-1 for cefalotin and cefotaxime was also observed. K m values of 72 and 181 μM for NDM-4 and NDM-1 was observed, respectively. NDM-4 showed lower affinity for ceftazidime than NDM-1 [87]. bla NDM-4 was found on IncF type plasmid in one of the earlier studies [88]. A remnant of insertion sequence ISAba125 on upstream of the bla NDM-4was found previously by PCR mapping during study of genetic structures surrounding the bla NDM-4 gene [89]. The ble MBL, a bleomycin resistant gene, was identified downstream of the bla NDM-4, similar genetic environment has been observed for most of the analyzed NDM-1 positive enterobacterial isolates [88]. PCR-based replicon typing showed that this bla NDM-4 positive plasmid belongs to the IncFIA incompatibility group. In keeping with this, bla NDM-5 was also found associated with IncFIA [28, 87].
The substitution of Valine by Leucine at position 88 and Methionine by Leucine at position 154 was found in NDM-5, which was first detected in E.coli [28] (Table 1). NDM-5 shows greater hydrolytic activity than NDM-1 toward carbapenems, cefotaxime, cephalotin and ceftazidime [85]. NDM-5 carrying plasmid of size >100 kb reduced susceptibilities of E. coli transformants to carbapenems and cephalosporins [28]. Other detected resistance determinants in NDM-5 producing E. coli included dfrA17 and aadA5 genes, which were found to be located within a class I integron structure, and the 16S rRNA methylase gene, rmtB, which was thought to account for aminoglycoside high-level resistance. The effect of NDM-5 on susceptibility of E. coli to carbapenems and expanded-spectrum cephalosporins appeared to be greater than that of NDM-1. Sequence analysis of 5′-flanking region of bla NDM-5 allele revealed presence of partial ISAba125, likely to be derived from A. baumannii, which generated a hybrid (−35/−10) promoter as described earlier by Poirel et al. in an NDM-1-producing E. coli isolate [90]. NDM-6 showed substitution of Alanine to Valine at 233 position, again first time detected in E. coli [12] (Table 1).
Substitutions of Aspartate to Asparagine at position 130 and Methionine to Leucine at position 154were found in NDM-7, identified in E. coli ST599 [91] (Table 1). TOP10 cells carrying plasmid harbouring bla NDM-7 in E.coli conferred higher resistance to carbapenems than a plasmid carrying bla NDM-1 [91]. A recent report demonstrated the role of Leu154 in enhancing carbapenem MICs in NDM-7 producing E. coli strain [91]. The bla NDM-7 gene was found to be located on a self-transferable IncX3 plasmid of 60 kb.
NDM-8 variant having substitutions at positions 130th (Aspartic acid to Glycine) and 154th (Methionine to Leucine) resulted in enzymatic activities against β-lactams similar to those shown by NDM-1 [92]. NDM-9 differing by a single amino acid substitution (E152K) from NDM-1 was recently identified in K. pneumoniae ST107 strain from China [93].
NDM-10 was first identified in K. pneumoniae isolated from Maharashtra, India and was found to have multiple substitutions at Arginine 32 to Serine, Glycine 36 to Aspartic acid, Glycine 69 to Serine, Alanine 74 to Threonine and Glycine 200 to Arginine [94].
NDM-11 was reported in E. coli KnPEc14 strain (Gene Bank KP265939.1).
NDM-12 has two amino acid substitutions at 154th (Methionine to Leucine) and 222th (Glycine to Aspartic acid). It was first identified on plasmid size 160 kb in E. coli [95] (Table 1). NDM-12 enzymatic activities were similar to those of NDM-1 against β-lactams, although kcat/Km ratios for all β-lactams were tested except doripenem.
NDM-13, a novel New Delhi Metallo-β-lactamase was identified in Nepal from the urine sample of patient showing a carbapenem-resistant E. coli infection [96]. It showed substitutions of Asparagine in place of Aspartic acid at position 95 and Leucine in place of Methionine at position 154 (Table 1) and similar enzymatic activity against β-lactams, but higher Kcat/Km ratios for cefotaxime compared with NDM-1. The bla NDM-13 gene was located into the chromosome within the genetic environment of tnpA-IS30-bla NDM-13-ble MBL-trpF-dsbC-cutA-groES-groL. Recently, complete sequence of pNDM13-DC33 plasmid harbouring bla NDM-13 isolated from E. coli isolate ST5138 in China, was reported, consisting of a backbone of 33 kb size and encoding an antimicrobial resistance region of 21 kb; tra, trb and pil transfer functions; repB plasmid replication gene and stability partitioning. pNDM13-DC33 plasmid harbouring bla NDM-13 gene showed high similarity with pNDM-HN380 IncX3 plasmid harbouring bla NDM-1 gene [79].
NDM-14 was first identified in clinical isolate of Acinetobacter lwoffii with substitution of Aspartic acidat 130th position to Glycine [97] (Table 1). NDM-14 showed higher enzymatic activities than NDM-1 towards carbapenem. NDM-14 have higher affinity for meropenem and imipenem than NDM-1, as indicated by the kinetic data [97].
NDM-15 was reported in an E. coli strain (Gene Bank KP735848.1). It showed substitution of Alanine to valine at 233th position and Methionine to Leucine at 154th position.
NDM-16 variant showed substitution at 264th position of Arginine to Histidine [98].
NDM-17 was reported in E.coli strain from a chicken in China. It showed amino acid subsitution of valine 88 to leucine, methionine 154 to leucine and glutamic acid 170 to lysine [99]. Schematic representation of mutations on various nucleotides leading to formation of new NDM variant is shown in Fig. 2. Phylogenetic analysis among the protein sequence of NDM variants are represented as cladogram in Fig. 3.
Fig. 3.
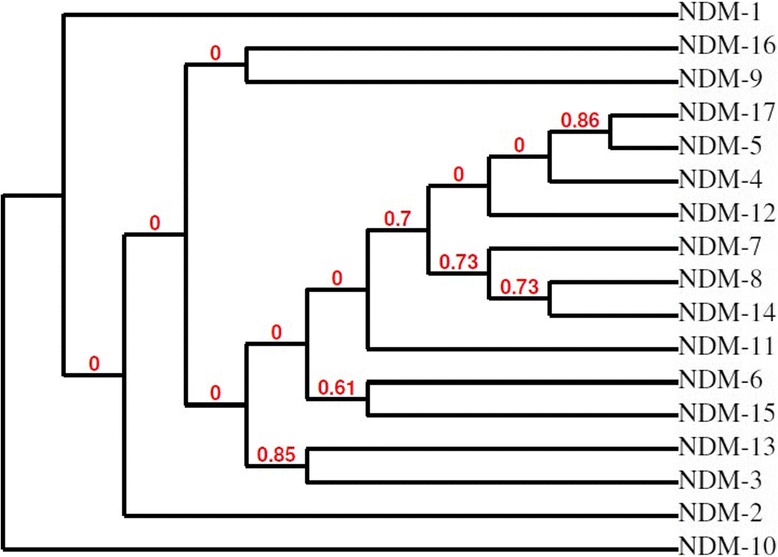
The phylogenetic relationship between protein sequences of NDM variants is shown. The tree construct has been generated using Phylogeny.fr, which used the maximum likelihood method to generate phylogenetic tree [107, 108]
Carbapenem hydrolysing activity was gradually reduced from NDM-7 to NDM-5, NDM-6 and NDM-1. All isolates positive for variants of bla NDM-1 showed resistance to aminoglycosides with MIC greater than 256 mg/L and MIC range of 2–512 mg/L for different lactams, lactams/lactamase inhibitor combinations [100]. Moreover, these variants showed susceptibility to tigecycline and colistin except for KNKp6a isolate, which showed MIC of 1.5 mg/L to tigecycline [100].
NDM variants were found associated with all other groups of antibiotic resistance enzymes encoding genes i.e. ESBL, carbapenemase, AmpC and rRNAmethylase. In bla NDM and its variants, due to genetic co-existence of other antibiotic resistant markers, there is limited options left to treat infections [101].
Recently, an NDM-1 producing Cedecea lapagei isolated from a neonate admitted to the paediatric ICU of a north India hospital was reported from our lab [102]. Also, a recent study demonstrated that 11 out of 55 patients with carbapenem-resistant Enterobacteriaceae nosocomial infections in China showed NDM variants as carbapenemase genes [103]. Recently, metabolite aspergillomarasmine A (AMA) which is found in fungi and its natural LLL isomer were identified to be effective inactivators of NDM-1 enzyme both in vivo and in vitro [104]. Also, the combination of levofloxacin and tigecyclinewas recently reported to successfully treat nosocomial pneumonia caused by NDM-1 producing Raoultella planticola [105].
Conclusion
The continual evolution of resistant markers due to the selection pressure and their spread among the bacteria through horizontal gene transfer is one of the alarming threats to the health worker in the hospital settings in order to control infections. NDM-1 and its variants producing bacteria was one of the challenges, which has became even more urgent since the detection of mcr-1 gene in Chinaand the spread of resistance against colistin has ended all hopes to control infections [77]. Now this is a time to think prudently the ways to check infections from hospital settings and to coordinate globally for surveillance of such resistant markers producing bacteria. Proper infection control guidelines need to be implemented worldwide. Surveillance should also be carried out to identify undetected asymptomatic carriers of carbapenem-resistant bacteria. To discover new drug molecules which could fight with multi-resistant bacteria, Infectious Disease Society of America has launched a “bad bugs need drugs” campaign to promote development of new antibiotics by 2020 [106].
Acknowledgments
Availability of data and materials
All data included in this study are publicly available because they have been published already.
Authors’ contribution
AUK conceived idea of compiling review and completed first draft. LM updated the information on genetics and structure of NDM-1 and helped writing, and preparing figs. RZ reviewed and critically revised it as per the current scenario. All authors read and approved the final manuscript.
Competing interests
Author’s declare that there is no competing interest.
Consent for publication
All authors agree to submit in BMC Microbiology.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- AMA
Aspergillomarasmine A
- BLIs
Beta lactamse inhibitors
- CPOs
Carbapenem producing organisms
- CRE
Carbapenem resistant enterobacteriaceae
- ESBL
Extended spectrum beta lactamase
- MBLs
Metallo beta lactamses
- MDRPA
Muti-drug-resistant pseudomonas aeruginosa
- MRSA
Methicillin resistant Staphylococcus aureus
- NDM
New delhimetallo beta lactamse
- PBPs
Penicillin binding proteins
Additional file
Worldwide distribution of NDM producing bacteria, as per articles available on PubMed database in the time period of Dec 2013 to Feb 2017. (PDF 136 kb)
Contributor Information
Asad U. Khan, Phone: 00919837021912, Email: asad.k@rediffmail.com
Lubna Maryam, Email: lucymary20@gmail.com.
Raffaele Zarrilli, Phone: 00390817463026, Email: rafzarri@unina.it.
References
- 1.Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clin Microbiol Infect. 2007;13:5–18. doi: 10.1111/j.1469-0691.2006.01492.x. [DOI] [PubMed] [Google Scholar]
- 2.Gould IM, Bal AM. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence. 2013;4:185–191. doi: 10.4161/viru.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkler ML, Bonomo RA. SHV-129: A gateway to global suppressors in the SHV beta-Lactamase Family? Mol Biol Evol. 2016;33:429–441. doi: 10.1093/molbev/msv235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viau R, Frank KM, Jacobs MR, Wilson B, Kaye K, Donskey CJ, Perez F, Endimiani A, Bonomo RA. Intestinal carriage of carbapenemase-producing organisms: current status of surveillance methods. Clin Microbiol Rev. 2016;29:1–27. doi: 10.1128/CMR.00108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K, Pannell M, Lock JL, Queenan AM, Jorgensen JH, Lee RM, Lewis JS, Jarrett D. Detection systems for carbapenemase gene identification should include the SME serine carbapenemase. Int J Antimicrob Agents. 2013;41:1–4. doi: 10.1016/j.ijantimicag.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Chia JH, Siu LK, Su LH, Lin HS, Kuo AJ, Lee MH, Wu TL. Emergence of carbapenem-resistant Escherichia coli in Taiwan: resistance due to combined CMY-2 production and porin deficiency. J Chemother. 2009;21:621–626. doi: 10.1179/joc.2009.21.6.621. [DOI] [PubMed] [Google Scholar]
- 8.Mojica MF, Bonomo RA. Fast W.B1-Metallo-beta-Lactamases: Where Do We Stand? Curr Drug Targets. 2016;17:1029–1050. doi: 10.2174/1389450116666151001105622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aitha M, Moller AJ, Sahu ID, Horitani M, Tierney DL, Crowder MW. Investigating the position of the hairpin loop in New Delhi metallo-beta-lactamase, NDM-1, during catalysis and inhibitor binding. J Inorg Biochem. 2016;156:35–39. doi: 10.1016/j.jinorgbio.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bush K. A resurgence of beta-lactamase inhibitor combinations effective against multidrug-resistant Gram-negative pathogens. Int J Antimicrob Agents. 2015;46:483–493. doi: 10.1016/j.ijantimicag.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Padhi S. New Delhi metallo-beta-lactamase: a weapon for the newly emerging drug-resistant bacteria. Indian J Med Sci. 2011;65:317–320. doi: 10.4103/0019-5359.107767. [DOI] [PubMed] [Google Scholar]
- 12.Kaase M, Nordmann P, Wichelhaus TA, Gatermann SG, Bonnin RA, Poirel L. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J Antimicrob Chemother. 2011;66:1260–1262. doi: 10.1093/jac/dkr135. [DOI] [PubMed] [Google Scholar]
- 13.Khan AU, Nordman P. Spread of carbapenemase NDM-1 producers: the situation in India and what may be proposed. Scand J Infect Dis. 2012;44:531–535. doi: 10.3109/00365548.2012.669046. [DOI] [PubMed] [Google Scholar]
- 14.Williamson DA, Sidjabat HE, Freeman JT, Roberts SA, Silvey A, Woodhouse R, Mowat E, Dyet K, Paterson DL, Blackmore T, Burns A, Heffernan H. Identification and molecular characterisation of New Delhi metallo-beta-lactamase-1 (NDM-1)- and NDM-6-producing Enterobacteriaceae from New Zealand hospitals. Int J Antimicrob Agents. 2012;39:529–533. doi: 10.1016/j.ijantimicag.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 15.Khan AU, Parvez S. Detection of bla(NDM-4) in Escherichia coli from hospital sewage. J Med Microbiol. 2014;63:1404–1406. doi: 10.1099/jmm.0.076026-0. [DOI] [PubMed] [Google Scholar]
- 16.Köhler T, Michea-Hamzehpour M, Epp SF, Pechere JC. Carbapenem activities against Pseudomonas aeruginosa: respective contributions of OprD and efflux systems. Antimicrob Agents Chemother. 1999;43:424–427. doi: 10.1128/aac.43.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/AAC.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 19.Jovcic B, Lepsanovic Z, Suljagic V, Rackov G, Begovic J, Topisirovic L, Kojic M. Emergence of NDM-1 metallo-beta-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob Agents Chemother. 2011;55:3929–3931. doi: 10.1128/AAC.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khajuria A, Praharaj AK, Kumar M, Grover N. Emergence of NDM-1 in the clinical isolates of Pseudomonas aeruginosa in India. J Clin Diagn Res. 2013;7:1328–1331. doi: 10.7860/JCDR/2013/5509.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choudhury D, Paul D, Ghosh AS, Das Talukdar A, Dutta Choudhury M, Maurya AP, Dhar Chanda D, Chakravarty A, Bhattacharjee A. Effect of single-dose carbapenem exposure on transcriptional expression of blaNDM-1 and mexA in Pseudomonas aeruginosa. J Glob Antimicrob Resist. 2016;7:72–77. doi: 10.1016/j.jgar.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Ruppé É, Woerther PL, Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann Intensive Care. 2015;5:61. doi: 10.1186/s13613-015-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert P, McBain AJ. Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clin Microbiol Rev. 2003;16:189–208. doi: 10.1128/CMR.16.2.189-208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brovedan M, Marchiaro PM, Morán-Barrio J, Revale S, Cameranesi M, Brambilla L, Viale AM, Limansky AS. Draft genome sequence of Acinetobacter bereziniae HPC229, a carbapenem-resistant clinical strain from Argentina harboring blaNDM-1. Genome Announc. 2016;4:pii:e00117-16. [DOI] [PMC free article] [PubMed]
- 25.Pfeifer Y, Cullik A, Witte W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol. 2010;300:371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Martinez L, Pascual A, Hernández-Allés S, Alvarez-Díaz D, Suárez AI, Tran J, Benedí VJ, Jacoby GA. Roles of beta-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob Agents Chemother. 1999;43:1669–1673. doi: 10.1128/aac.43.7.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright GD. Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv Drug Deliv Rev. 2005;57:1451–1470. doi: 10.1016/j.addr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Hornsey M, Phee L, Wareham DW. A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 2011;55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H, Aitha M, Hetrick AM, Richmond TK, Tierney DL, Crowder MW. Mechanistic and spectroscopic studies of metallo-beta-lactamase NDM-1. Biochemistry. 2012;51:3839–3847. doi: 10.1021/bi300056y. [DOI] [PubMed] [Google Scholar]
- 30.Miriagou V, Cornaglia G, Edelstein M, Galani I, Giske CG, Gniadkowski M, et al. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin Microbiol Infect. 2010;16:112–122. doi: 10.1111/j.1469-0691.2009.03116.x. [DOI] [PubMed] [Google Scholar]
- 31.Rolain JM, Parola P, Cornaglia G. New Delhi metallo-beta-lactamase (NDM-1): towards a new pandemia? Clin Microbiol Infect. 2010;16:1699–1701. doi: 10.1111/j.1469-0691.2010.03385.x. [DOI] [PubMed] [Google Scholar]
- 32.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, et al. Novel plasmid and its variant harboring both a bla(NDM-1) gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother. 2012;56:1698–1702. doi: 10.1128/AAC.06199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogaerts P, Verroken A, Jans B, Denis O, Glupczynski Y. Global spread of New Delhi metallo-beta-lactamase 1. Lancet Infect Dis. 2010;10:831–832. doi: 10.1016/S1473-3099(10)70278-4. [DOI] [PubMed] [Google Scholar]
- 34.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, Nordmann P. Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin Microbiol Infect. 2012;18:E362–E365. doi: 10.1111/j.1469-0691.2012.03928.x. [DOI] [PubMed] [Google Scholar]
- 36.Sahl JW, Del Franco M, Pournaras S, Colman RE, Karah N, Dijkshoorn L, Zarrilli R. Phylogenetic andgenomic diversity in isolates from the globally distributed Acinetobacter baumannii ST25 lineage. Sci Rep. 2015;5:15188. doi: 10.1038/srep15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szmolka A, Nagy B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front Microbiol. 2013;4:258. doi: 10.3389/fmicb.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Wu C, Zhang Q, Qi J, Liu H, Wang Y, et al. Identification of New Delhi metallo-beta-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS One. 2012;7:e37152. doi: 10.1371/journal.pone.0037152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan AU, Nordmann P. NDM-1-producing Enterobacter cloacae and Klebsiella pneumoniae from diabetic foot ulcers in India. J Med Microbiol. 2012;61:454–456. doi: 10.1099/jmm.0.039008-0. [DOI] [PubMed] [Google Scholar]
- 40.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janvier F, Jeannot K, Tessé S, Robert-Nicoud M, Delacour H, Rapp C, Mérens A. Molecular characterization of blaNDM-1 in a sequence type 235 Pseudomonas aeruginosa isolate from France. Antimicrob Agents Chemother. 2013;57:3408–3411. doi: 10.1128/AAC.02334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gamal D, Fernández-Martínez M, Salem D, El-Defrawy I, Montes LÁ, Ocampo-Sosa AA, Martínez-Martínez L. Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int J Infect Dis. 2016;43:17–20. doi: 10.1016/j.ijid.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Yao X, Doi Y, Zeng L, Lv L, Liu JH. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis. 2016;16:288–289. doi: 10.1016/S1473-3099(16)00057-8. [DOI] [PubMed] [Google Scholar]
- 45.Rahman M, Prasad KN, Pathak A, Pati BK, Singh A, Ovejero CM, et al. RmtC and RmtF 16S rRNA Methyltransferase in NDM-1-Producing Pseudomonas aeruginosa. Emerg Infect Dis. 2015;21:2059–2062. doi: 10.3201/eid2111.150271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kocsis E, Gužvinec M, Butić I, Krešić S, Crnek SŠ, Tambić A, et al. blaNDM-1 carriage on IncR plasmid in Enterobacteriaceae strains. Microb Drug Resist. 2016;22:123–128. doi: 10.1089/mdr.2015.0083. [DOI] [PubMed] [Google Scholar]
- 47.Datta S, Mitra S, Chattopadhyay P, Som T, Mukherjee S, Basu S. Spread and exchange of bla NDM-1 in hospitalized neonates: role of mobilizable genetic elements. Eur J Clin Microbiol Infect Dis. 2017;36:255–265. doi: 10.1007/s10096-016-2794-6. [DOI] [PubMed] [Google Scholar]
- 48.Bontron S, Nordmann P, Poirel L. Transposition of Tn125 encoding the NDM-1 carbapenemase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2016;60:7245–7251. doi: 10.1128/AAC.01755-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dortet L, Girlich D, Virlouvet AL, Poirel L, Nordmann P, Iorga BI, Naas T. Characterization of BRPMBL, the bleomycin resistance protein associated with the Carbapenemase NDM. Antimicrob Agents Chemother. 2017;61:pii:e02413-16. Print 2017. [DOI] [PMC free article] [PubMed]
- 50.Cornaglia G, Rossolini GM. Forthcoming therapeutic perspectives for infections due to multidrug-resistant Gram-positive pathogens. Clin Microbiol Infect. 2009;15:218–223. doi: 10.1111/j.1469-0691.2009.02740.x. [DOI] [PubMed] [Google Scholar]
- 51.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baiden F, Owusu-Agyei S, Webster J, Chandramohan D. The need for new antibiotics. Lancet. 2010;375:637–638. doi: 10.1016/S0140-6736(10)60265-6. [DOI] [PubMed] [Google Scholar]
- 53.Heddini A, Cars O, Qiang S, Tomson G. Antibiotic resistance in China-a major future challenge. Lancet. 2009;373:30. doi: 10.1016/S0140-6736(08)61956-X. [DOI] [PubMed] [Google Scholar]
- 54.Thabit AK, Crandon JL, Nicolau DP. Antimicrobial resistance: impact on clinical and economic outcomes and the need for new antimicrobials. Expert Opin Pharmacother. 2015;16:159–177. doi: 10.1517/14656566.2015.993381. [DOI] [PubMed] [Google Scholar]
- 55.Marquez-Ortiz RA, Haggerty L, Sim EM, Duarte C, Castro-Cardozo BE, Beltran M, et al. First complete Providencia rettgeri genome sequence, the NDM-1-producing clinical strain RB151. Genome Announc. 2017;5:pii:e01472-16. [DOI] [PMC free article] [PubMed]
- 56.Shen P, Yi M, Fu Y, Ruan Z, Du X, Yu Y, Xie X. Detection of an Escherichia coli sequence type 167 strain with two tandem copies of blaNDM-1 in the chromosome. J Clin Microbiol. 2016;55:199–205. doi: 10.1128/JCM.01581-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livermore DM. Has the era of untreatable infections arrived? J Antimicrob Chemother. 2009;64(Suppl 1):i29–i36. doi: 10.1093/jac/dkp255. [DOI] [PubMed] [Google Scholar]
- 58.Chen Z, Qlu S, Wang Y, Wang Y, Liu S, Wang Z, et al. Coexistence of blaNDM-1 with the prevalent blaOXA23 and blaIMP in pan-drug resistant Acinetobacter baumannii isolates in China. Clin Infect Dis. 2011;52:692–693. doi: 10.1093/cid/ciq231. [DOI] [PubMed] [Google Scholar]
- 59.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daikos GL, Markogiannakis A, Souli M, Tzouvelekis LS. Bloodstream infections caused by carbapenemase-producing Klebsiella pneumoniae: a clinical perspective. Expert Rev Anti-Infect Ther. 2012;10:1393–1404. doi: 10.1586/eri.12.138. [DOI] [PubMed] [Google Scholar]
- 61.Frere JM, Galleni M, Bush K, Dideberg O. Is it necessary to change the classification of {beta}-lactamases? J Antimicrob Chemother. 2005;55:1051–1053. doi: 10.1093/jac/dki155. [DOI] [PubMed] [Google Scholar]
- 62.Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother. 2011;55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wailan AM, Sidjabat HE, Yam WK, Alikhan NF, Petty NK, Sartor AL, et al. Mechanisms involved in acquisition of blaNDM genes by IncA/C2 and IncFIIY plasmids. Antimicrob Agents Chemother. 2016;60:4082–4088. doi: 10.1128/AAC.00368-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mishra S, Upadhyay S, Sen MR, Maurya AP, Choudhury D, Bhattacharjee A. Genetic acquisition of NDM gene offers sustainability among clinical isolates of Pseudomonas aeruginosa in clinical settings. PLoS One. 2015;10:e0116611. doi: 10.1371/journal.pone.0116611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campos JC, da Silva MJ, dos Santos PR, Barros EM, Pereira Mde O, Seco BM, et al. Characterization of Tn3000, a transposon responsible for blaNDM-1 dissemination among Enterobacteriaceae in Brazil, Nepal, Morocco, and India. Antimicrob Agents Chemother. 2015;59:7387–7395. doi: 10.1128/AAC.01458-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khan AU, Rehman MT. Role of non-active-site residue Trp-93 in the function and stability of New Delhi Metallo-beta-Lactamase 1. Antimicrob Agents Chemother. 2015;60:356–360. doi: 10.1128/AAC.01194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang F, Xie L, Wang X, Han L, Guo X, Ni Y, Qu H, Sun J. Further spread of bla NDM-5 in Enterobacteriaceae via IncX3 Plasmids in Shanghai, China. Front Microbiol. 2016;7:424. doi: 10.3389/fmicb.2016.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.An J, Guo L, Zhou L, Ma Y, Luo Y, Tao C, Yang J. NDM-producing Enterobacteriaceae in a Chinese Hospital, 2014-2015: Identification of NDM-producing Citrobacter werkmanii and acquisition of blaNDM-1-carrying plasmid in vivo in a clinical Escherichia coli isolate. J Med Microbiol. 2016;65:1253–1259. doi: 10.1099/jmm.0.000357. [DOI] [PubMed] [Google Scholar]
- 69.Tran DN, Tran HH, Matsui M, Suzuki M, Suzuki S, Shibayama K, et al. Emergence of New Delhi metallo-beta-lactamase 1 and other carbapenemase-producing Acinetobacter calcoaceticus-baumannii complex among patients in hospitals in Ha Noi, Viet Nam. Eur J Clin Microbiol Infect Dis. 2017;36:219–225. doi: 10.1007/s10096-016-2784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Both A, Huang J, Kaase M, Hezel J, Wertheimer D, Fenner I, et al. First report of Escherichia coli co-producing NDM-1 and OXA-232. Diagn Microbiol Infect Dis. 2016;86:437–438. doi: 10.1016/j.diagmicrobio.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 71.Yang RS, Feng Y, Lv XY, Duan JH, Chen J, Fang LX, et al. Emergence of NDM-5 and MCR-1-producing Escherichia coli clone ST648 and ST156 from a single Muscovy Duck (Cairina moschata) Antimicrob Agents Chemother. 2016;60:6899–6902. doi: 10.1128/AAC.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie L, Dou Y, Zhou K, Chen Y, Han L, Guo X, Sun J. Coexistence of blaOXA-48 and truncated blaNDM-1 on different plasmids in a Klebsiella pneumoniae isolate in China. Front Microbiol. 2017;8:133. doi: 10.3389/fmicb.2017.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joshi PR, Acharya M, Kakshapati T, Leungtongkam U, Thummeepak R, Sitthisak S. Co-existence of blaOXA-23 and blaNDM-1 genes of Acinetobacter baumannii isolated from Nepal: antimicrobial resistance and clinical significance. Antimicrob Resist Infect Control. 2017;6:21. doi: 10.1186/s13756-017-0180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Papagiannitsis CC, Malli E, Florou Z, Sarrou S, Hrabak J, Mantzarlis K, Zakynthinos E, Petinaki E. Emergence of sequence type 11 Klebsiella pneumoniae coproducing NDM-1 and VIM-1 metallo-β-lactamases in a Greek hospital. Diagn Microbiol Infect Dis. 2016;87:295–297. doi: 10.1016/j.diagmicrobio.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 75.Paveenkittiporn W, Kerdsin A, Chokngam S, Bunthi C, Sangkitporn S, Gregory CJ. Emergence of plasmid-mediated colistin resistance and New Delhi metallo-β-lactamase genes in extensively drug-resistant Escherichia coli isolated from a patient in Thailand. Diagn Microbiol Infect Dis. 2017;87:157–159. doi: 10.1016/j.diagmicrobio.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 76.Zhong LL, Zhang YF, Doi Y, Huang X, Zhang XF, Zeng KJ, et al. Coproduction of MCR-1 and NDM-1 by colistin-resistant Escherichia coli isolated from a healthy individual. Antimicrob Agents Chemother. 2017;61:pii:e01962-16. [DOI] [PMC free article] [PubMed]
- 77.Zheng B, Dong H, Xu H, Lv J, Zhang J, Jiang X, Du Y, Xiao Y, Li L. Coexistence of MCR-1 and NDM-1 in clinical isolates. Clin Infect Dis. 2016;63:1393–1395. doi: 10.1093/cid/ciw553. [DOI] [PubMed] [Google Scholar]
- 78.Kapmaz M, Erdem F, Abulaila A, Yeniaras E, Oncul O, Aktas Z. First detection of NDM-1 with CTX-M-9, TEM, SHV and rmtC in Escherichia coli ST471 carrying IncI2, A/C and Y plasmids from clinical isolates in Turkey. J Glob Antimicrob Resist. 2016;7:152–153. doi: 10.1016/j.jgar.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 79.Lv J, Qi X, Zhang D, Zheng Z, Chen Y, Guo Y, et al. First Report of Complete Sequence of a blaNDM-13-harboring plasmid from an Escherichia coli ST5138 clinical isolate. Front Cell Infect Microbiol. 2016;6:130. doi: 10.3389/fcimb.2016.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quan J, Li X, Chen Y, Jiang Y, Zhou Z, Zhang H, Sun L, Ruan Z, Feng Y, Akova M, Yu Y. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17:400–410. doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed] [Google Scholar]
- 81.Lázaro-Perona F, Sarria-Visa A, Ruiz-Carrascoso G, Mingorance J, García-Rodríguez J, Gómez-Gil R. Klebsiella pneumoniae co-producing NDM-7 and OXA-48 carbapenemases isolated from a patient with prolonged hospitalisation. Int J Antimicrob Agents. 2016;49:112–113. doi: 10.1016/j.ijantimicag.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 82.Lai CC, Chuang YC, Chen CC, Tang HJ. Coexistence of MCR-1 and NDM-9 in a clinical carbapenem-resistant Escherichia coli isolate. Int J Antimicrob Agents. 2017;49:517–518. doi: 10.1016/j.ijantimicag.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 83.Liu BT, Song FJ, Zou M, Hao ZH, Shan H. Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and in Escherichia coli from the same animal. Antimicrob Agents Chemother. 2017;61:pii: e02347-16. [DOI] [PMC free article] [PubMed]
- 84.Du H, Chen L, Chavda KD, Pandey R, Zhang H, Xie X, Tang YW, Kreiswirth BN. Genomic characterization of Enterobacter cloacae isolates from China that coproduce KPC-3 and NDM-1 carbapenemases. Antimicrob Agents Chemother. 2016;60:2519–2523. doi: 10.1128/AAC.03053-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rogers BA, Sidjabat HE, Silvey A, Anderson TL, Perera S, Li J, Paterson DL. Treatment options for New Delhi metallo-beta-lactamase-harboring enterobacteriaceae. Microb Drug Resist. 2013;19:100–103. doi: 10.1089/mdr.2012.0063. [DOI] [PubMed] [Google Scholar]
- 86.Tada T, Miyoshi-Akiyama T, Shimada K, Kirikae T. Biochemical analysis of metallo-beta-lactamase NDM-3 from a multidrug-resistant Escherichia coli strain isolated in Japan. Antimicrob Agents Chemother. 2014;58:3538–3540. doi: 10.1128/AAC.02793-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nordmann P, Boulanger AE, Poirel L. NDM-4 metallo-beta-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob Agents Chemother. 2012;56:2184–2186. doi: 10.1128/AAC.05961-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 89.Poirel L, Bonnin RA, Nordmann P. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother. 2011;55:4224–4229. doi: 10.1128/AAC.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dortet L, Nordmann P, Poirel L. Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56:1693–1697. doi: 10.1128/AAC.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gottig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-beta-lactamase with increased carbapenemase activity. J Antimicrob Chemother. 2013;68:1737–1740. doi: 10.1093/jac/dkt088. [DOI] [PubMed] [Google Scholar]
- 92.Tada T, Miyoshi-Akiyama T, Dahal RK, Sah MK, Ohara H, Kirikae T, Pokhrel BM. NDM-8 metallo-beta-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob Agents Chemother. 2013;57:2394–2396. doi: 10.1128/AAC.02553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang X, Li H, Zhao C, Chen H, Liu J, Wang Z, et al. Novel NDM-9 metallo-beta-lactamase identified from a ST107 Klebsiella pneumoniae strain isolated in China. Int J Antimicrob Agents. 2014;44:90–91. doi: 10.1016/j.ijantimicag.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 94.Khajuria A, Praharaj AK, Kumar M, Grover N. Presence of a novel variant NDM-10, of the New Delhi metallo-beta-lactamase in a Klebsiella pneumoniae isolate. Indian J Med Microbiol. 2016;34:121–123. doi: 10.4103/0255-0857.174101. [DOI] [PubMed] [Google Scholar]
- 95.Tada T, Shrestha B, Miyoshi-Akiyama T, Shimada K, Ohara H, Kirikae T, Pokhrel BM. NDM-12, a novel New Delhi metallo-beta-lactamase variant from a carbapenem-resistant Escherichia coli clinical isolate in Nepal. Antimicrob Agents Chemother. 2014;58:6302–6305. doi: 10.1128/AAC.03355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shrestha B, Tada T, Miyoshi-Akiyama T, Shimada K, Ohara H, Kirikae T, Pokhrel BM. Identification of a novel NDM variant, NDM-13, from a multidrug-resistant Escherichia coli clinical isolate in Nepal. Antimicrob Agents Chemother. 2015;59:5847–5850. doi: 10.1128/AAC.00332-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zou D, Huang Y, Zhao X, Liu W, Dong D, Li H, et al. A novel New Delhi metallo-beta-lactamase variant, NDM-14, isolated in a Chinese Hospital possesses increased enzymatic activity against carbapenems. Antimicrob Agents Chemother. 2015;59:2450–2453. doi: 10.1128/AAC.05168-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kazmierczak KM, Rabine S, Hackel M, McLaughlin RE, Biedenbach DJ, Bouchillon SK, Sahm DF, Bradford PA. Multiyear, Multinational Survey of the Incidence and Global Distribution of Metallo-beta-Lactamase-Producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60:1067–1078. doi: 10.1128/AAC.02379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Z, Wang Y, Walsh TR, Liu D, Shen Z, Zhang R, et al. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a ST48 Escherichia coli strain. Antimicrob Agents Chemother. 2017. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 100.Rahman M, Shukla SK, Prasad KN, Ovejero CM, Pati BK, Tripathi A, et al. Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug resistant Enterobacteriaceae from India. Int J Antimicrob Agents. 2014;44:30–37. doi: 10.1016/j.ijantimicag.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 101.Sidjabat HE, Townell N, Nimmo GR, George NM, Robson J, Vohra R, et al. Dominance of IMP-4-producing Enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob Agents Chemother. 2015;59:4059–4066. doi: 10.1128/AAC.04378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ahmad N, Ali SM, Khan AU. First reported New Delhi metallo-β-lactamase-1- producing Cedecea lapagei. Int J Antimicrob Agents. 2017;49:118–119. doi: 10.1016/j.ijantimicag.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 103.Wang Q, Zhang Y, Yao X, Xian H, Liu Y, Li H, et al. Risk factors and clinical outcomes for carbapenem-resistant Enterobacteriaceae nosocomial infections. Eur J Clin Microbiol Infect Dis. 2016;35:1679–1689. doi: 10.1007/s10096-016-2710-0. [DOI] [PubMed] [Google Scholar]
- 104.Albu SA, Koteva K, King AM, Al-Karmi S, Wright GD, Capretta A. Total synthesis of aspergillomarasmine A and related compounds: A sulfamidate approach enables exploration of structure-activity relationships. Angew Chem Int Ed Engl. 2016;55:13259–13262. doi: 10.1002/anie.201606657. [DOI] [PubMed] [Google Scholar]
- 105.Pan Z, Liu R, Zhang P, Zhou H, Fu Y, Zhou J. Combination of tigecycline and levofloxacin for successful treatment of nosocomial pneumonia caused by New Delhi Metallo-β-Lactamase-1-producing Raoultella planticola. Microb Drug Resist. 2016;23:127–131. doi: 10.1089/mdr.2015.0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 107.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data included in this study are publicly available because they have been published already.


