Abstract
Efficient hematopoietic stem cell (HSC) homing is important for hematopoietic cell transplantation (HCT), especially when HSC numbers are limited, as with the use of cord blood (CB). In a screen of small molecule compounds, we identified glucocorticoid hormone signaling as an activator of CXCR4 expression in human CB HSCs and hematopoietic progenitor cells (HPCs). Short-term glucocorticoid (GC) pretreatment of human CB HSC/HPCs promoted SDF-1/CXCR4 mediated chemotaxis, homing, and long-term engraftment when transplanted into primary and secondary NSG mice. Mechanistically, activated glucocorticoid receptor binds directly to a glucocorticoid response element (GRE) in the CXCR4 promoter and recruits the SRC1/p300 complex to promote H4K5 and H4K16 histone acetylation, facilitating transcription of CXCR4. These results suggest a new and readily available means to enhance the clinical efficacy of CB HCT.
Hematopoietic stem cells (HSCs), which give rise to all blood cell types, are essential cells for successful hematopoietic cell transplantation (HCT)1. Cord blood (CB) is an increasingly used source of HSC for HCT2, which is used to treat patients with malignant and non-malignant hematological disorders. One limitation to greater usage of CB for HCT is that HSCs are present in only limited numbers in a single unit of CB3. While ex vivo expansion of HSCs is currently being assessed for enhancement of CB HCT3–5, another means to enhance single CB unit HCT is to enhance the homing, and thus the engraftment efficiency of HSCs.
Intravenously infused HSCs traffic to bone marrow (BM) and implant in microenvironmental niches, where they are nurtured for self-renewal and differentiation6. The SDF-1/CXCR4 chemotactic axis is a major pathway directing the migration and homing of HSC from peripheral blood to BM niches7,8. By modulating the interplay between SDF-1 and CXCR4, HSC homing efficiency can be improved. For example, DPP4 inhibition blocks proteolytic inactivation of SDF-1 and enhances engraftment of HSCs9, treatment with PGE2 or valporic acid facilitates HSC chemotaxis towards SDF-1 gradients by upregulating CXCR4 surface expression10–12, and mild heat exposure promotes incorporation of CXCR4 into lipid rafts enhancing HSC chemotaxis and engraftment13. However, there is still a need for other methods to enhance homing and engraftment of HSCs.
Small synthetic molecules have been evaluated for their effects on HSC function3–5. To identify compounds that might be useful for increasing HSC homing efficiency, we performed a small scale compound screen for molecules that can upregulate surface expression of CXCR4 on human CB CD34+ cells. From a nuclear hormone ligand library including 74 chemical compounds (Supplementary Table 1), we found that treatment of CB CD34+ cells for 16 hours with dexamethasone (Dex), a synthetic glucocorticoid, greatly promoted cell surface expression of CXCR4 (Fig. 1a). Expression of CXCR4 on CB CD34+ cells was also increased after treating cells with other glucocorticoids (which were not present in the library) including Flonase (Fluticasone propionate), cortisol (Hydrocortisone), and Medrol (Methylprednisolone) (Fig. 1b). We focused on Flonase, which of these compounds forms the most stable activated complex with glucocorticoid receptor (GR)14. Flonase treatment enhanced CXCR4 expression at concentrations as low as 10 nM (Supplementary Fig. 1a). Flow cytometry and confocal imaging analysis demonstrated a dramatic increase in surface CXCR4 expression on CB CD34+ cells treated with Flonase, compared to the vehicle control (Fig. 1c,d and Supplementary Fig. 1b). Flonase also enhanced CXCR4 surface expression on HSCs (CD34+CD38−CD45RA−CD49f+CD90+), multipotential progenitors (MPPs, CD34+CD38−CD45RA−CD49f−CD90−), and CD34+CD38− cells (Fig. 1e and Supplementary Fig. 1c,d).
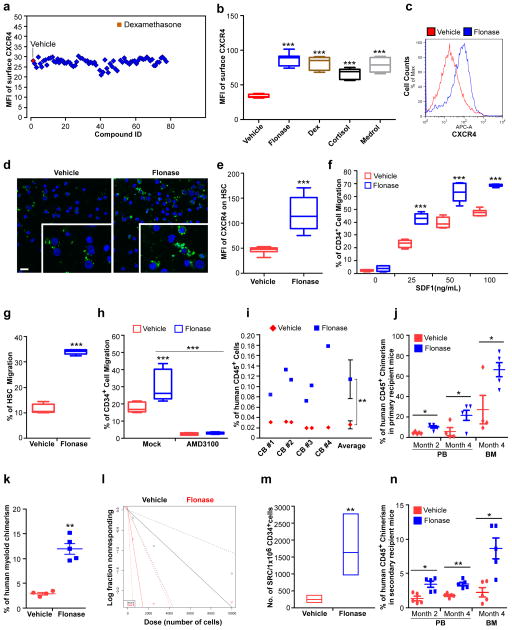
Figure 1. Glucocorticoids increase surface expression of CXCR4 and promote SDF-1/CXCR4 axis mediated chemotaxis, homing and long term engraftment of human hematopoietic stem and progenitor cells.
(a) Mean fluorescence intensity (MFI) of surface CXCR4 of human cord blood (CB) CD34+ cells after treatment of the cells for 16 hours with compounds from a nuclear receptor ligand library. The concentration of all compounds used in this study was 1 μM unless otherwise stated.
(b) Quantification of mean fluorescence intensity (MFI) of surface CXCR4 of human CB CD34+ cells treated with vehicle, Flonase, dexamethasone (Dex), cortisol or methylprednisolone (Medrol). Data pooled from three independent experiments are shown (n=9 cultures per group, one-way ANOVA).
(c) Histogram of surface CXCR4 expression of human CB CD34+ cells treated with vehicle or Flonase. Representative histograms from three independent experiments are shown.
(d) Confocal imaging analysis of surface CXCR4 expression of human CB CD34+ cells treated with vehicle or Flonase. FITC (green) indicates CXCR4 expression; DAPI (blue) labels the cell nucleus. Representative images from two independent experiments are shown (the inset shows the amplified part of the image). Scale bar: 20 μm.
(e) Quantification of mean fluorescence intensity (MFI) of surface CXCR4 of human CB HSCs (CD34+CD38−CD45RA−CD90+CD49f+) treated with vehicle or Flonase (n=9 cultures per group). Data pooled from three independent experiments are shown.
(f) Migration of human CB CD34+ cells towards human recombinant SDF-1α, as quantified by flow cytometry. The cells were cultured in the presence of vehicle or Flonase for 16 hours and then allowed to migrate towards the indicated concentrations of SDF-1α for 4 hours. Data pooled from two independent experiments are shown (n=6 cultures per group, two-way ANOVA).
(g) Migration of human phenotypic HSCs in CB CD34+ cells towards human recombinant SDF-1α (50 ng/mL), as quantified by flow cytometry. The migration percentage of HSCs was calculated as the HSC (CD34+CD38−CD45RA−CD90+CD49f+) frequency among migrated CD34+ cells. Data pooled from two independent experiments are shown (n=6 cultures per group).
(h) Migration of vehicle or Flonase treated human CB CD34+ cells in the presence of the CXCR4 antagonist, AMD3100 (5 μg/mL), as quantified by flow cytometry. Data pooled from three independent experiments are shown (n=9 cultures per group, one-way ANOVA).
(i) The percentage of human CD45+ cells in the bone marrow of NSG mice 24 h after transplantion with 500,000 CB CD34+ cells that had been treated with vehicle or Flonase. CD34+ cells from four cord blood samples (CB#1-4) were tested. (n=6 mice per group).
(j) The percentage of human CD45+ cells in peripheral blood (PB) and BM of NSG mice at the indicated time points after transplantation with 10,000 CB CD34+ cells that had been treated with vehicle or Flonase. (n=4 mice in vehicle group and n=5 mice in Flonase group).
(k) The percentage of human CD33+ myeloid cells in BM of NSG mice 4 months after transplantation with 10,000 CB CD34+ cells that had been treated with vehicle or Flonase (n=4 mice in vehicle group and n=5 mice in Flonase group).
(l,m) The frequency of human SRCs in CB CD34+ cells treated with vehicle or Flonase, as determined by transplantations of graded doses of treated cells into NSG mice and determination of human CD45+ cell chimerism 3 months after transplantation (n=4–5 mice per group, see Supplementary Table 2). (l) Poisson statistical analysis of data from Supplementary Table 2. Shapes (circle or triangle in the plot) represent the percentage of negative mice for each dose of cells. The inverted triangles indicate that all tested mice were positive in this group. Solid lines indicate the best-fit linear model for each data set. Group A (black line) indicates vehicle group, Group B (red line) indicates Flonase group. Dotted lines represent 95% confidence intervals. (m) HSC frequencies (line in the box) and 95% confidence intervals (box) presented as the number of SRCs in 1×106 CD34+ cells.
(n) Human CD45+ cell chimerism in the PB and BM of secondary recipient NSG mice, which had been transplanted with 5×106 bone marrow cells from primary recipient NSG mice (n=5 mice per group). Data are shown as dot plots (mean±s.e.m.) in j, k, n, or as box-and-whisker plots (the lines indicate median values, the whiskers indicate minimum and maximum values, the boxes indicate interquartile range) in b and e–h. *p<0.05. **p<0.01. ***p<0.001.
Next, we assessed if GCs can enhance chemotaxis of HSCs and HPCs towards SDF-1. Flonase pre-treatment greatly enhanced chemotaxis of CB CD34+ cells to graded doses of SDF-1 (Fig. 1f), as well as chemotaxis to SDF-1 of CD34+CD38− cells, HSCs and MPPs (Fig. 1g and Supplementary Fig. 1e,f). Chemotaxis of CB CD34+ cells to SDF-1 was also enhanced by treating the cells with Dex, cortisol or Medrol (Supplementary Fig. 1g). The glucocorticoid receptor (GR) was expressed most highly in human CB HSCs as compared to other CB cell types tested (Supplementary Fig. 1h,i); moreover, expression of GR on CB HSC, CD34+ or CD34+CD38− cells was higher than on CB CD34− cells (Supplementary Fig. 1h,i), suggesting that GC signaling may be of significance for the physiological functioning of HSCs and HPCs.
To evaluate whether facilitated chemotaxis of CB HSCs and HPCs towards SDF-1 by GC pretreatment depends on CXCR4, we used AMD3100, a CXCR4 antagonist15. Treatment with AMD3100 fully blocked the effects of Flonase on chemotaxis of CB CD34+ cells to SDF-1 (Fig. 1h), demonstrating that GC acts through the SDF-1/CXCR4 axis. Since activation of GR increased surface expression of CXCR4 on CB HSCs and HPCs and enhanced SDF-1/CXCR4 mediated chemotaxis, we next evaluated whether inactivation of GR would have opposite effects. RU486, a widely used GR antagonist, did not change the basal level of CXCR4 surface expression of CB CD34+ cells, and had no effect on the chemotaxis of CB CD34+ cells towards SDF-1 (Supplementary Fig. 1j,k). However, RU486 neutralized the increased surface expression of CXCR4 and the enhanced chemotaxis induced by GC (Supplementary Fig. 1j,k). This suggests that GR signaling, while dispensable for basal levels of CXCR4 expression, was important for increased surface expression of CXCR4 on CB HSCs and HPCs, thus facilitating their response to SDF-1 induced chemotaxis.
We next tested if GC treatment enhances in vivo homing. We treated human CB CD34+ cells with vehicle or Flonase for 16 hours and evaluated homing in sublethally irradiated NSG mice. Flonase, cortisol and Dex treatment significantly enhanced the homing efficiency of CB CD34+ cells by 4.4-fold, 2.5-fold and 3.0-fold respectively (Fig. 1i and Supplementary Fig. 2a,b). To assess the long-term reconstituting ability of GC-pretreated human CB CD34+ cells, we performed limiting dilution analysis to calculate the frequency of SCID repopulating cells (SRC), a measure of functional human HSCs1. Flonase-treated CB cells demonstrated significantly increased engraftment in primary NSG recipients compared with that of the vehicle control group, with increased human myeloid, B cell and T cell chimerism (Fig. 1j,k and Supplementary Fig. 2c–h). A Poisson distribution analysis revealed an SRC frequency of 1/4162 for vehicle-treated CB CD34+ cells and an enhanced SRC frequency of 1/611 for Flonase-treated CB CD34+ cells, indicating the presence of 240 SRCs and 1637 SRCs in vehicle- and Flonase-treated cells, respectively (Fig. 1l,m and Supplementary Table 2); A second Poisson distribution analysis from another independent experiment revealed an SRC frequency of 1/7344 for vehicle-treated CB CD34+ cells and an enhanced SRC frequency of 1/1576 for Flonase-treated CB CD34+ cells, indicating the presence of 136 SRCs and 635 SRCs in vehicle- and Flonase-treated cells, respectively (Supplementary Fig. 2i,j and Supplementary Table 3). Enhanced engraftment of Flonase-treated as compared to vehicle-treated cells was also apparent in transplanted secondary recipients (Fig. 1n), demonstrating effects of Flonase treatment on a long-term repopulating and self-renewing HSC population. Thus, short-term glucocorticoid treatment of human CB HSCs and HPCs enhances their homing and long-term engraftment.
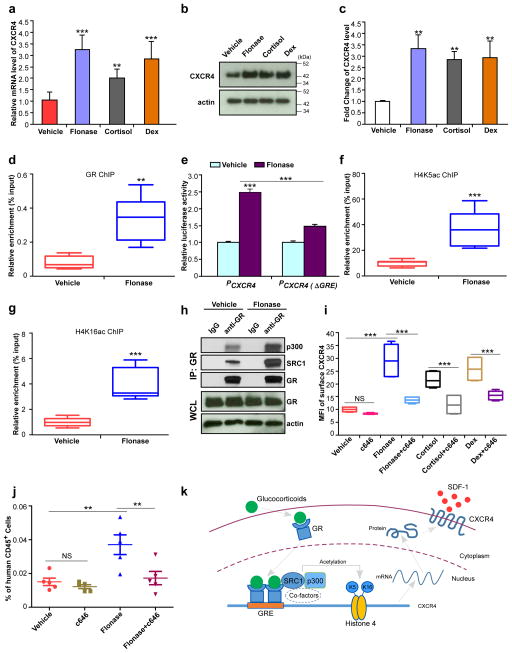
Mechanistically, we hypothesized that GC might increase CXCR4 expression in human CB CD34+ cells via effects on CXCR4 transcription. Upon activation by agonists, GR translocates into the nucleus and forms a homodimer that regulates transcription by binding to glucocorticoid response elements (GREs) in the promoter region of target genes16. We found that GC (Flonase, cortisol, Dex) pretreatment significantly increased CXCR4 mRNA levels in human CB CD34+ cells, as assessed by quantitative real-time PCR analysis (Fig. 2a). CXCR4 protein levels were also upregulated in Flonase-, cortisol- or Dex-treated human CB CD34+ cells compared to control cells (Fig. 2b,c). We searched for the consensus GR binding sequence AGAACAnnnTGTNCN17 and found a putative glucocorticoid response element (GRE) AGAACATTCTGTGCA in the human CXCR4 promoter region (from −1,662 bp to −1,648 bp upstream of the transcription start site). Chromatin immunoprecipitation analysis indicated enrichment for the GR was in this region of the CXCR4 promoter upon GC treatment (Fig. 2d and Supplementary Fig. 3a). Using a dual luciferase reporter assay system to examine whether GC administration directly enhances promoter activity of CXCR4, we found that the luciferase signal was significantly enhanced by Flonase treatment, and that this effect was abrogated by disruption of the GRE in the CXCR4 promoter (Fig. 2e).
Figure 2. Glucocorticoids enhance H4K5 and H4K16 acetylation associated with the CXCR4 promoter, facilitate expression of CXCR4 and promote homing of human CB CD34+ cells.
(a) CXCR4 mRNA levels in Flonase, cortisol or Dex treated human CB CD34+ cells, relative to vehicle-treated cells, as assessed by quantitative realtime-PCR. Data pooled from three independent experiments are shown (n=9 replicates per group, one-way ANOVA).
(b,c) Total CXCR4 protein levels in Flonase, cortisol or Dex treated human CB CD34+ cells, relative to vehicle-treated cells, as assessed by western blotting. A representative blot is shown in b, and quantification of CXCR4 protein levels from three independent western blot assays is shown in c (n=3 experiments, one-way ANOVA). Actin was used as a loading control. Uncropped images of blots are shown in Supplementary Figure 4a.
(d) GR levels at the CXCR4 promoter in vehicle or Flonase treated human CB CD34+ cells, as assessed by a chromatin immunoprecipitation (ChIP) assay. Data pooled from two independent experiments are shown (n=6 replicates per group).
(e) Promoter activities of the full length CXCR4 promoter (PCXCR4) and a glucocorticoid response element (GRE) defective form of the CXCR4 promoter (PCXCR4 (ΔGRE)) treated by vehicle or Flonase, as determined by a dual-luciferase reporter assay system. The relative luciferase activity of vehicle treated full length CXCR4 promoter group was set to 1 (n=3 replicates per group, one-way ANOVA). Representative data from three independent experiments are shown.
(f,g) Acetylated H4K5 (H4K5ac, f) and H4K16 (H4K16ac, g) levels at the CXCR4 promoter in vehicle or Flonase treated human CB CD34+ cells, as assessed by a ChIP assay. Data pooled from two independent experiments are shown (n=8 replicates per group).
(h) Levels of SRC1 and p300 that are co-immunoprecipitated with receptor GR in vehicle or Flonase treated human CB CD34+ cells. Extracts of treated cells were immunoprecipitated with anti-GR antibody and the resulting precipitates were analyzed by western blot. GR and actin in the whole cell lysate (WCL) serve loading controls. Representative data from three independent experiments are shown. Uncropped images of blots are shown in Supplementary Figure 4b.
(i) Mean fluorescence intensity of surface CXCR4 in vehicle, c646 (30 μM), Flonase (10 nM), Flonase+c646, cortisol (1μM), cortisol+c646, Dex (100 nM), or Dex+c646 treated human CB CD34+ cells, as assessed by flow cytometry. Data pooled from two independent experiments are shown (n=6 cultures per group, one-way ANOVA).
(j) The percentage of human CD45+ cells in the bone marrow of NSG mice 24 h after transplantion with 500,000 CB CD34+ cells that had been treated with vehicle, c646 (30 μM), Flonase (10 nM) or Flonase+c646. (n=5 mice per group, one-way ANOVA). Data are shown as dot plots (mean±s.e.m.) in j, or as box-and-whisker plots (the lines indicate median values, the whiskers indicate minimum and maximum values, the boxes indicate interquartile range) in d, f, g and i. NS, not significant. **p<0.01. ***p<0.001.
(k) Model for the role of glucocorticoids in regulating CXCR4 expression in human HSC/HPCs. Glucocorticoid binding to GR in the cytoplasm results in GR activation, translocation to the nucleus and dimerization. The GR homodimer recognizes and binds to a GRE in CXCR4 promoter. Activated GR recruits SRC1, p300 and other co-factors with histone acetyltransferase activity to enhance acetylation of histone 4 on lysine 5 and lysine 16. Acetylation of histone 4 facilitates chromatin remodeling and promotes expression of CXCR4, and thus enhances HSC/HPC homing and engraftment.
Next, we assessed how GR might activate transcription of CXCR4 upon binding to the GRE in the promoter region. Nuclear hormone receptors (NRs) can directly or indirectly associate with coactivators, including SRC1 (steroid receptor coactivator-1), CBP (CREB binding protein) and p300 (E1A binding protein p300), which have histone acetyltransferase activity and acetylate histones associated with target gene promoters18,19. Accordingly, we investigated histone acetylation levels in the CXCR4 promoter region and found that, compared to the vehicle control, Flonase treatment dramatically enhanced the acetylation levels of histone H4 on lysines 5 and 16 (H4K5 and H4K16) (Fig. 2f,g), whereas the acetylation levels of histone H3 (H3K9 and H3K14) were unchanged (Supplementary Fig. 3b). Flonase treatment did not affect acetylation levels of total H4 (Supplementary Fig. 3c), suggesting that GR signaling specifically targets CXCR4. The interaction between GR and SRC1/p300 complex was increased by Flonase treatment, as assessed by co-immunoprecipitation analysis (Fig. 2h). Notably, treatment of CB CD34+ cells with c646, a histone acetyltransferase p300 inhibitor, or knockdown of SRC1 or p300, suppressed the effects of Flonase, cortisol and Dex on CXCR4 expression (Fig. 2i and Supplementary Fig. 3d,e) and suppressed the effects of Flonase pre-treatment on SDF-1/CXCR4 mediated chemotaxis (Supplementary Fig. 3f). Knockdown of CBP did not suppress Flonase-induced cell surface expression of CXCR4 (Supplementary Fig. 3d,e), suggesting that p300 but not CBP is involved in the selective histone acetylation induced by GC. Treatment with the p300 inhibitor also suppressed the enhanced homing of Flonase-treated CB CD34+ cells (Fig. 2j). These data demonstrate that the increased homing efficiency resulting from activation of GR signaling depends on enhanced histone acetylation at CXCR4.
Notably, we could not identify a consensus GRE in the mouse Cxcr4 promoter, and glucocorticoid treatment did not affect Cxcr4 mRNA levels or Cxcr4 surface expression of mouse hematopoietic stem and progenitor cells and did not affect their chemotaxis to SDF-1 (Supplementary Fig. 3g–i).
Our findings show that upon GC treatment, activated GR binds to the GRE at the CXCR4 promoter region, transcriptionally up-regulating CXCR4 expression and thereby enhancing the homing efficiency and engraftment capability of human CB CD34+ cells (Fig. 2k).
Histone acetylation is one of the major epigenetic chromatin remodeling pathways that regulate gene transcription. Acetylation, which targets the amino terminal regions of core histones and results in chromatin decondensation that prevents chromatin from forming higher-order structures, allows access to transcription factors or regulators20. Valporic acid, a histone deacetylase (HDAC) inhibitor, upregulates expression of CXCR4 and enhances SDF-1/CXCR4 axis-mediated chemotaxis11,12. However, direct modulation of HDAC activity may lead to changes in the expression profiles of many target genes, which could potentially result in detrimental effects.
Our studies demonstrate that glucocorticoid-bound GR recruits the SRC1/p300 acetyltransferase complex to the CXCR4 promoter in CB CD34+ cells. Consistent with the notion that transcriptional regulation by GR is promoter- and cell type-dependent, GC administration did not increase global acetylation levels of H4, but specifically enhanced acetylation of H4K5 and H4K16 associated with the CXCR4 promoter. Many types of synthetic GCs, including Flonase and dexamethasone, are widely used FDA-approved drugs for anti-inflammation and immunosuppressive therapy16. Our study indicates that GC pre-treatment of cells has the potential to significantly enhance the engraftment of human CB HSC and HPC, designating GCs as promising drug candidates to facilitate clinical HCT.
METHODS
Mice
NSG (NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ) mice (6–8 weeks old) were obtained from the In Vivo Therapeutics Core of the Indiana University School of Medicine (IUSM). All animal experiments followed protocols approved by The Institutional Animal Care and Use Committee of IUSM.
Isolation of CB CD34+ cells and cell culture
Mononuclear cells from normal human cord blood (CB) (CordUse, Orlando, FL, USA) were isolated by density gradient centrifugation with Ficoll-Paque Plus (GE Healthcare, Piscataway, NJ, USA). CD34+ cells were collected using an immunomagnetic selection kit (Miltenyi Biotec, Auburn, CA, USA). Briefly, mononuclear cells were resuspended in MACS buffer (0.5% BSA, 2mM EDTA in PBS, pH7.2). FcR blocking reagent (Miltenyi Biotec, #130-046-702) and CD34 microbeads (Miltenyi Biotec, #130-046-702) were added sequentially, and then the cells were incubated for 30 min in the refrigerator. The cells were washed with MACS buffer and centrifuged at 300 g for 10 min. The supernatant was aspirated and the cells were resuspended in 1 mL MACS buffer. Magnetic separation was performed with an appropriate MACS column (Miltenyi Biotec, #130-042-401). This procedure yields CD34+ cells with a purity of 90–98%. CB CD34+ cells were cultured in RPMI-1640 medium with 10% fetal bovine serum, 100 ng/mL stem cell factor (SCF) (R&D Systems, #7466-SC-010/CF), thrombopoietin (TPO) (R&D Systems, #288-TP-200/CF), and Fms-like tyrosine kinase 3 ligand (Ftl3L) (BioLegend, # 710802). For screening, 50,000 CD34+ cells were cultured for 16 hours in the above medium in the presence of compounds (1 μM) from the Nuclear Receptor Ligand Library (Enzo Life Sciences, Farmingdale, NY, USA). For administration of glucocorticoids, Flonase (S1992, 1 μM), cortisol (S1696, 1 μM), dexamethasone (S1322, 1 μM) or methylprednisolone (S1733, 1 μM) (Selleck Chemicals, Houston, TX, USA) was added to the medium and the cells were incubated for 16 hours unless stated otherwise. Dimethyl sulfoxide (DMSO) (Sigma, # D2650) was used as vehicle control.
Mouse lineage negative bone marrow cell isolation and culture
Mouse bone marrow lineage cell depletion was performed using the mouse lineage cell depletion kit following the manufacturer’s protocol (Miltenyi Biotec, Auburn, CA, USA). Lineage negative mouse hematopoietic stem and progenitor cells were cultured in RPMI-1640 medium with 10% fetal bovine serum, 100 ng/mL mouse stem cell factor (mSCF) (R&D Systems, #455-MC-010), mouse thrombopoietin (mTPO) (R&D Systems, # 488-TO-005/CF), and mouse Fms-like tyrosine kinase 3 ligand (mFtl3L) (BioLegend, #550706).
Immunostaining and flow cytometry
Cells were collected by centrifuging at 300 g for 10 min, washed twice with cold PBS, resuspended in 200 μL PBS, and stained with fluorescence conjugated antibodies at 4 °C for 30 min. The cells were washed with cold PBS and the samples were fixed with 1% formaldehyde. The samples were analyzed on an LSRII flow cytometer (BD Biosciences). For intracellular staining, after cell surface staining, the cells were fixed and permeabilized using the Cell Permeabilization Kit (Thermo Fisher, Florence, KY, USA). The cells were washed with cold PBS and then stained with primary antibodies at room temperature for 20 min. APC or FITC-conjugated secondary antibodies were added and the cells were incubated for another 20 min. The cells were washed and fixed for flow cytometry analysis. The following antibodies from BD Bioscience (San Jose, CA, USA) were used for cell surface staining: CXCR4-APC (12G5), CD34-FITC (581), CD38-PE (HIT2), CD45RA-PE-CF594 (HI100), CD90-PEcy7 (5E10) and CD49f-PerCPcy5.5 (GoH3), CD3-FITC (UCHT1), CD19-PE (HIB19), CD33-PEcy7 (WM53) and CD45-APC (HI30). The following antibodies were used for intracellular staining: anti-Glucocorticoid Receptor [BuGR2] (Abcam, Cambridge, MA, USA), anti-H4K5ac (9672) and anti-H4K16ac (13534) (Cell Signaling Technology, Beverly, MA, USA), anti-H3K9ac (#06-942) and anti-H3K14ac (#07-353) (Millipore, Kankakee, IL, USA). For mouse staining, the following antibodies were used: APC anti-mouse Cxcr4 (BD Bioscience, #558644), Brilliant Violet 421 anti-mouse Lineage cocktail (Biolegend, #133311), PE/Cy7 anti-mouse Ly-6A/E (Sca-1) (Biolegend, #122513), APC anti-mouse CD117 (c-Kit) (Biolegend, #105811), FITC anti-mouse CD48 (Biolegend, #103404), and PerCP/Cy5.5 anti-mouse CD150 (Biolegend, #115922).
Confocal imaging
Human CB CD34+ cells treated with vehicle or glucocorticoids were seeded on a Nunc glass bottom dish (Thermo Fisher Scientific, #150680) coated with poly-L-lysine. Immunostaining was performed using anti-CXCR4 [UMB2] (Abcam, Cambridge, MA, USA) and FITC labeled secondary antibodies. Cells were then washed, fixed and permeabilized. Samples were covered with mounting medium containing DAPI (Vector Laboratories). Fluorescence was examined using an Olympus FV-1000 confocal microscope.
Chemotaxis assay
Costar 24-well Transwell plates with 6.5 mm diameter inserts with 5.0 μm pores (Corning Inc, Corning, NY, USA) were used for the chemotaxis assay. 650 μL pre-warmed serum free IMDM medium containing SDF1 (50 ng/mL, unless otherwise indicated) was added to the lower chamber. Cells (1×105 cells/100 μL) were resuspended in IMDM with 0.5% bovine serum albumin (Sigma-Aldrich, Miamisburg, OH, USA). The cell suspension (100 μL) was placed in the upper chamber of the transwell. Transwell plates were placed in a 37 °C incubator with 5% CO2 and 95% humidity for 4 hours. The percentage migration was determined using flow cytometry with number of migrated cells in the lower chamber divided by number of cells placed in the upper chamber. A no-SDF-1 group was used to subtract background migration and served as a negative control group. For HSC migration assay, phenotypic HSC frequency was determined by surface staining and analyzed by flow cytometry. CD34+CD38−CD45RA−CD90+CD49f+ cells were defined as HSCs. The migration percentage of human CB HSCs was calculated as the HSC (CD34+CD38−CD45RA−CD90+CD49f+) frequency among migrated CD34+ cells. For AMD3100 treatment, cells were pretreated with AMD3100 (Sigma, # 239820) for half hour before chemotaxis assay.
CD34+ CB cell homing assay
NSG mice were sublethally irradiated (with a single dose of 350 cGy, 137Cs) before transplantation. Glucocorticoid or vehicle treated CD34+ CB cells (500,000) were intravenously injected into each recipient NSG mouse. After 24 hours, mice were sacrificed and bone marrow mononuclear cells from a femur of each mouse were collected. Immunostaining with anti-human CD45 was performed. Samples were fixed in 1% formaldehyde buffer and analyzed by flow cytometry to determine the percentage of human CD45+ cells. Bone marrow cells from non-transplanted NSG mice were used as the negative control.
Limiting dilution analysis
The frequency of human SCID repopulating cells (SRCs) was analyzed by LDA as previously reported3. Increasing doses of vehicle or glucocorticoid treated CD34+ cells (500, 2500, or 10,000 cells) were intravenously injected into NSG recipient mice that had been sublethally irradiated. Three or four months after transplantation, the mice were sacrificed and the percentage of human CD45+ cell, B cell, T cell and myeloid cell chimerism was determined by immunostaining and flow cytometry. For the long-term engraftment assay, 5×106 bone marrow cells from the primary recipients of the 10,000 cell group were infused into secondary recipient mice. The HSC frequency was calculated using L-Calc software (Stem Cell Technologies Inc, Vancouver, BC, Canada) and plotted using ELDA software (bioinf.wehi.edu.au/software/elda/).
RNA extraction and Real-time PCR
RNA was extracted using the RNA Mini Kit following the manufacturer’s protocol (QIAGEN, #74106). Total RNA was reverse transcribed using the Invitrogen Superscript III kit. Quantitative real-time PCR reactions were performed using the SYBR Green PCR Master Mix (Thermo Fisher, Florence, KY, USA) and an Applied Biosystems 7500 Real-Time PCR System. GAPDH was used as an internal control. The data are shown as relative mRNA level normalized to the level in vehicle control, which was set to 1. CXCR4 primers were: sense primer: 5′-TCTATGTTGGCGTCTGGATCC-3′, anti-sense primer: 5′-CTTGGAGTGTGACAGCTTGG-3′. GAPDH primers were: sense primer: 5′-TTCGTCATGGGTGTGAACCA-3′, anti-sense primer: 5′-TGGCAGTGATGGCATGGACT-3′. For mouse Cxcr4 amplification, the following primers were used. mCxcr4 sense primer: 5′-GAGCATGACGGACAAGTACC -3′, anti-sense primer: 5′-TGGACAATAGCGAGGTACCG -3′. Actin sense primer: 5′-CTTCTACAATGAGCTGCGTG-3′, anti-sense primer: 5′-ATCACAATGCCTGTGGTACG-3′.
Western blotting
Cells were lysed in RIPA buffer (25mM Tris-HCl pH 7.6, 150mM NaCl, 1% sodium deoxycholate, 0.1% SDS) with Halt TM Protease Inhibitor Cocktail (Thermo Fisher, Florence, KY, USA) on ice for 30 mis. Samples were centrifuged at 4 °C, 13,000 g for 15 min. Collected supernatants and protein concentrations of the supernatants were quantified using the Protein Quantitation Kit (Abcam, Cambridge, MA, USA). Equal levels of total protein from different samples were subjected to SDS-PAGE analysis and signals were detected using corresponding primary and secondary antibodies. The following antibodies (1:1000 dilution) were used: anti-CXCR4 (Abcam, #ab181020) anti-SRC-1 (Cell Signaling Technology, 128E7, #2191); anti-Glucocorticoid Receptor (Cell Signaling Technology, D8H2, #3660); anti-p300 (Cell Signaling Technology, C-20, sc-585); anti-actin (Sigma, A3853).
Chromatin immunoprecipitation
The chromatin immunoprecipitation (ChIP) assay was performed using EZ-ChIP kit according to the manufacturer’s protocol (EMD Millipore, #17-371). CD34+ CB cells were cultured with vehicle or glucocorticoids for 16 hours. 1% formaldehyde was then added to the culture medium and the cells were incubated at 37 °C for 10 min. Cells were washed with cold PBS twice and then lysed in SDS lysis buffer with protease inhibitor for 10 min on ice. The cells were sonicated and then centrifuged at 4 °C, 13, 000 g for 15 min. The supernatant was diluted 1:10 with ChIP dilution buffer in the kit (EMD Millipore, #17-371). 1% of the diluted sample from each group was used as the input control. Protein A agarose beads were added to the diluted sample and the sample was incubated at 4 °C with rotation for 30 min to eliminate the non-specific binding. The supernatant was collected and the beads were discarded. Control IgG or antibody was added to the samples which were incubated at 4 °C with rotation overnight. 60 μL Protein A agarose beads were added to each sample and the cells incubated at 4 °C with rotation for 1 hour. The samples were washed following the manufacturer’s instructions (EMD Millipore, #17-371). DNA-protein complex was eluted from the beads twice using elution buffer (1% SDS, 0.1M NaHCO3) in the kit (EMD Millipore, #17-371). Samples and the corresponding input were reverse cross-linked following the manufacturer’s protocol (EMD Millipore, #17-371). DNA was purified and subjected to real-time quantitative PCR analysis. Data are shown as the percentage of input. The primers used for CXCR4 qPCR were as follows: sense primer: 5′-TTCCAGTGGCTGCATGTGTC-3′, anti-sense primer: 5′-CAGACAATGTAACTCGCTCC-3′. The following antibodies were used for ChIP: anti-Glucocorticoid Receptor [BuGR2] (Abcam, Cambridge, MA, USA), anti-H4K5ac (9672), and anti-H4K16ac (13534) (Cell Signaling Technology, Beverly, MA, USA), anti-H3K9ac (#06-942) and anti-H3K14ac (#07-353) (Millipore, Kankakee, IL, USA).
Co-immunoprecipitation
Cells were lysed in immunoprecipitation (IP) buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and protease inhibitor cocktail) on ice for 30 min. Lysates were centrifuged at 4 °C, 13,000 g for 15 min and the supernatant was subjected to immunoprecipitation using primary antibodies or IgG control and protein A agarose beads (Cell Signaling Technology, Beverly, MA, USA). Precipitated immunocomplexes were washed five times with washing buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) and subjected to immunoblotting analysis. The following antibodies were used: anti-SRC-1 (Cell Signaling Technology, 128E7, #2191); anti-Glucocorticoid Receptor (Cell Signaling Technology, D8H2, #3660); anti-p300 (Cell Signaling Technology, C-20, sc-585); anti-actin (Sigma, A3853).
Dual luciferase reporter assay
The promoter of human CXCR4 (NM_001008540.1) was cloned into pGL4.10[luc2] (Promega, #E6651) between the Xho1 and HindIII restriction sites, generating pGL4.10-PCXCR4[luc2]. Glucocorticoid response element (GRE) deletion, generating pGL4.10-PCXCR4(ΔGRE)[luc2] was performed by deletion PCR amplification using sense primer 5′-CTGTTTGCAAACGTACAAGTGCAGAGAAGGCGTGC-3′ and anti-sense primer 5′-CCTTCTCTGCACTTGTACGTTTGCAAACAGCGTGC-3′. pGL4.10-PCXCR4[luc2] or pGL4.10-PCXCR4(ΔGRE)[luc2] was co-transfected with pGL4.75[hRluc/CMV] (Promega, #E6931) into human CB CD34+ cells using nucleofector (Lonza). After 24 hours, vehicle or Flonase was added and the cells were incubated for 16 hours. Promoter activity was examined using the Promega Dual-Luciferase Reporter Assay System (#E1910), following the manufacturer’s instructions. Briefly, cells were washed with PBS and lysed in passive lysis buffer for 15 min. 100 μL of LAR II substrate was predispensed into each well of white microtiter plates. 20 μL of cell lysate was carefully transferred into the firefly luciferase assay reagent and mixed by pipetting 3 times. After a 5-min reaction, the plate was placed in the luminometer for reading of the luminescent signal generated by Firefly luciferase. 100 μL of Stop & Glo Reagent was dispensed into each sample and vortexed briefly to mix. A second reading was initiated for the luminescent signal generated by Renilla luciferase. The relative promoter activity was calculated as Readout[luc2]/Readout[hRluc].
Statistical analysis
Data are shown as mean values ± standard deviation (SD), unless stated to show standard error of the mean (SEM). Statistical analysis was performed using Microsoft Excel and GraphPad Prism 5.0. Two-tailed Student’s t-tests were performed for statistical analysis between two groups. One-way or two-way analysis of variance (ANOVA) was used to compare the difference in means between more than two groups where indicated. P-value less than 0.05 was considered as statistically significant.
Supplementary Material
Editorial summary.
Glucocorticoid treatment of human cord blood hematopoietic stem cells increases expression of the receptor CXCR4 by chromatin remodeling, thereby enhancing hematopoietic stem cell homing and engraftment.
Acknowledgments
This work was supported by US Public Health Service Grants from the NIH to HEB: (R01 HL112669, R01 HL056416, U54 DK106846). We thank other members in the Broxmeyer laboratory for helpful discussion and assistance, and Anthony L. Sinn from the In Vivo Therapeutics Core of the Indiana University School of Medicine for transplantation assistance.
Footnotes
AUTHOR CONTRIBUTIONS
BG and HEB conceived the research, designed and performed experiments, interpreted data and wrote the manuscript. XXH and SC performed i.v. injection and interpreted data.
COMPETING FINANCIAL INTERESTS
Dr. Broxmeyer is a member of the Medical Scientific Advisory Board of CordUse, a cord blood banking company based in Orlando, Florida.
References
- 1.Doulatov S, Notta F, Laurenti E, Dick JE. Cell Stem Cell. 2012;3:120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Ballen KK, Gluckman E, Broxmeyer HE. Blood. 2013;122:491–498. doi: 10.1182/blood-2013-02-453175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fares I, et al. Science. 2014;345:1509–1512. doi: 10.1126/science.1256337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boitano AE, et al. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner JE, et al. Cell Stem Cell. 2016;18:144–155. doi: 10.1016/j.stem.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison SJ, Scadden DT. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peled A, et al. Science. 1999;283:845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- 8.Capitano ML, Broxmeyer HE. Encylopedia of Cell Biology. 2016;3:624–631. [Google Scholar]
- 9.Christopherson KW, Hangoc G, Mantel CR, Broxmeyer HE. Science. 2004;305:1000–1003. doi: 10.1126/science.1097071. [DOI] [PubMed] [Google Scholar]
- 10.Hoggatt J, Singh P, Sampath J, Pelus LM. Blood. 2009;113:5444–5455. doi: 10.1182/blood-2009-01-201335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaurasia P, Gajzer DC, Schaniel C, D’Souza S, Hoffman R. J Clin Invest. 2014;124:2378–2395. doi: 10.1172/JCI70313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gul H, Marquez-Curtis LA, Jahroudi N, Lo J, Turner AR, Janowska-Wieczorek A. Stem Cells Dev. 2009;18:831–838. doi: 10.1089/scd.2008.0235. [DOI] [PubMed] [Google Scholar]
- 13.Capitano ML, Hangoc G, Cooper S, Broxmeyer HE. Stem Cells. 2015;33:1975–1984. doi: 10.1002/stem.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Högger P, Rohdewald P. Steriods. 1994;59:597–602. doi: 10.1016/0039-128x(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 15.Broxmeyer HE, et al. J Exp Med. 2005;201:1307–1318. doi: 10.1084/jem.20041385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadmiel M, Cidlowski JA. Trends Pharmacol Sci. 2013;34:518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson LC, et al. Nat Struct Mol Biol. 2013;20:876–883. doi: 10.1038/nsmb.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna NJ, O’Malley BW. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 19.Lonard DM, O’Malley BW. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Eberharter A, Becker PB. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.