Abstract
Calmodulin (CaM) uniquely promotes signaling of oncogenic K-Ras; but not N-Ras or H-Ras. How CaM interacts with K-Ras and how this stimulates cell proliferation are among the most challenging questions in KRAS-driven cancers. Earlier data pointed to formation of a ternary complex consisting of K-Ras, PI3Kα and CaM. Recent data point to phosphorylated CaM binding to the SH2 domains of the p85 subunit of PI3Kα and activating it. Modeling suggests that the high affinity interaction between the phosphorylated CaM tyrosine motif and PI3Kα, can promote full PI3Kα activation by oncogenic K-Ras. Our up-to-date review discusses CaM’s role in PI3K signaling at the membrane in KRAS-driven cancers. This is significant since it may help development of K-Ras-specific pharmacology.
Keywords: K-Ras, calcium, proliferation, phosphorylated tyrosine motif, plasma membrane, drug discovery
The Puzzle of Calmodulin’s Critical Action in KRAS-Driven Cancers
How does calmodulin (CaM) act to promote oncogenic KRAS-driven cell proliferation? This question is highly significant, since CaM interacts only with K-Ras; but not with the other Ras isoforms, H-Ras or N-Ras [1–7]. K-Ras is among the most frequently mutated oncogenes in human tumors [8–12], particularly in pancreatic (almost 100%), lung (35%), and colorectal (45%) cancers [13, 14]. K-Ras has two alternative splicing variants: K-Ras4A and the most abundant K-Ras4B. Elucidating CaM’s mode of action will enable developing K-Ras4B–specific medications [5, 12, 15–18]. Despite some clues [2, 6, 19, 20], this question has proven immensely challenging. NMR and modeling [2, 12] made it clear that CaM, a protein with a negatively charged interdomain linker can bind favorably to the hypervariable region (HVR) of K-Ras4B, and possibly also to that of K-Ras4A. Both HVRs are highly positively charged–unlike those of H-Ras or N-Ras [21]. Experimental data indicated that K-Ras4B’s farnesyl group, at the C-terminal of the HVR, can dock into a hydrophobic pocket in CaM [6, 19]. Additional interaction elements were also proposed [22]. However, obtaining the experimental structure of the complex of CaM and K-Ras4B has been elusive, likely due to the multiple ways through which CaM can interact with the catalytic domain of active K-Ras4B. This hampers structural determination.
There also exists the mechanistic enigma: how does CaM contribute to K-Ras4B proliferative signaling? This question is of paramount significance since proliferation is the first hallmark of cancer [23, 24]. Several hypotheses were proposed toward this aim. One posited that K-Ras binding to CaM reduces the activity of Ca2+-dependent protein kinase II (CaMKII) and expression of frizzled-8 precursor (Fzd8) protein. Depletion of Fzd8 in H-RasG12V-transformed cells stimulates tumor initiation [15]; the thesis of another [5, 12] was that CaM is an integral component of a K-Ras4B/phosphatidylinositide-3-kinase α (PI3Kα) ternary complex, where CaM molecules bind the Src homology 2 (SH2) domains of the p85 subunit of PI3Kα. Both hypotheses raise questions. As to the first hypothesis, the expected relative concentration of CaM in the tumor cell appears to be much higher than that of K-Ras. Thus, the question is to what extent K-Ras/CaM binding would deplete cellular CaM, such that it would affect its activation of CaMKII. As to the second hypothesis, the ternary complex model indicated a direct interaction between CaM molecules and the SH2 domains of the p85 subunit of PI3Kα; CaM was Ca2+-bound; but not phosphorylated in line with earlier publications [25]. However, as we discuss below, newer compelling observations, as well as a body of data chronicling phosphorylated CaM molecules and receptor tyrosine kinases binding to SH2 domains, suggest that a phosphorylated tyrosine motif constitutes the major CaM element recognized by p85 SH2 domains.
CaM’s action is critical for KRAS-driven signaling in cell proliferation. Due to CaM’s specific binding to the K-Ras isoform, the community has invested an immense effort to identify the structure and understand how the binding affects oncogenic K-Ras activity. Here we point out that CaM may be phosphorylated in this action. This expectation, which is inspired by a recent report [26], coupled with earlier ones on CaM’s phosphorylation and signaling [27, 28], is supported by considerable additional literature, and as we discuss below, is in line with numerous CaM/SH2 mechanisms. Our thesis is that CaM acts primarily through full activation of the PI3K/Akt pathway by binding of its phosphorylated tyrosine (Tyr99) to the two SH2 domains of PI3Kα (Figure 1, Key Figure). Even though a non-phosphorylated CaM can also interact and activate PI3Kα [25], CaM’s high affinity interaction appears to be via a phosphorylated tyrosine motif. Although not always the case, SH2 domains tend to interact with phosphorylated substrates. Full activation of PI3Kα is essential for tumor proliferation because of its critical role in regulation of cell growth [24].
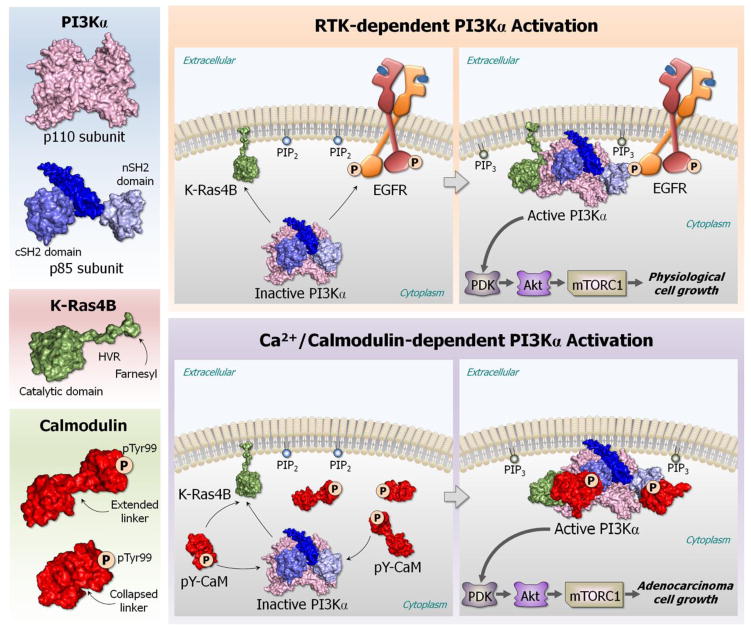
Figure 1. Key Figure.
The role of calmodulin (CaM) with phosphorylated tyrosine 99 (pTyr99) in KRAS-driven adenocarcinoma. In physiological cell growth, receptor tyrosine kinase (RTK) signal activates phosphatidylinositol 3-kinase (PI3K) through binding of the RTK’s pYXXM motif to the n/cSH2 domains of the p85 subunit of PI3Kα. In KRAS-driven adenocarcinomas, CaM with pTyr99 acts by fully activating PI3Kα through binding of the pTyr99 to the two SH2 domains of PI3Kα. The CaM’s pTyr99 binding site acts as the RTK’s pYXXM motif, substituting the missing RTK signal, thus stimulating the PI3Kα/Akt/mTOR pathway.
CaM Interacts Directly with PI3Kα
In a seminal paper published almost two decades ago, Sacks and coauthors [25] pointed to CaM’s interacting directly with PI3Kα through its p85 subunit. By incubating CaM-Sepharose with glutathione S-transferase (GST) fusion proteins containing various regions of p85, the authors identified the CaM-binding region of p85, observing that the carboxyl-terminal SH2 domain (cSH2) bound to the CaM-Sepharose. Longer exposure revealed the binding of the N- terminal SH2 domain (nSH2) as well, suggesting that nSH2 binds with lower affinity than cSH2. At the time, the mode of CaM interaction was unknown; however, insulin has been known to activate PI3K by promoting the interaction of the SH2 domains with specific phosphotyrosine-containing motifs of insulin receptor substrate 1 (IRS-1) adapter proteins [29]. The phosphorylated motif was the pYXXM peptide (pY608, amino acids 605–615 of IRS-1) [30]. Sacks and his colleagues [25] noticed that only the phosphorylated peptide (Y608) displaced the p85 from p85/CaM-Sepharose. This pointed to the location in SH2 and a possible mode of CaM’s binding to the SH2 at (or near) where the phosphorylated YXXM motifs bind. The authors thus suggested that CaM competes with the pYXXM motif to bind to p85 SH2 domains and thereby it either directly activates or modulates PI3K’s activity. Combined with the low amount of the catalytic p110 subunit which was bound to CaM, these pointed to a significant enhancement of PI3K activity by CaM’s binding to p85. Of note, the activation of PI3K by CaM was comparable to that obtained by pYXXM-bound SH2 domains, indicating that CaM does indeed bind to the p85 SH2 domains and stimulates PI3K activation, altogether leading the authors to conclude that increased intracellular [Ca2+] acts through CaM to modulate PI3K activation.
Additional support for CaM’s binding to PI3K comes from tuberculosis toxin blocking phagosome maturation which inhibited a Ca2+/CaM/PI3K human vacuolar protein sorting 34 (hVPS34) cascade [31]; CaM/Ca2+ is required for production of phosphatidylinositol 3-phosphate (PI3P) on liposomes in vitro and on phagosomes in vivo. CaM-PI3Kα binding is also supported by observations of altered CaM degradation and signaling in non-neuronal cells from Alzheimer’s disease (AD) patients [32]. The increased levels of CaM synergize with serum to overactivate PI3K/Akt in AD patient cells by direct binding of CaM to p85, leading the authors to suggest that failure of CaM degradation, and thus of Ca2+/CaM-dependent signaling, may be important in the development of AD.
CaM/Ca2+ binding to PI3Kα stimulates PI3Kα/Akt/mTOR signaling, and thereby regulates cell proliferation and growth [28, 33]. Via CaM, increase of intracellular free Ca2+ can activate PI3K/Akt, localizing Akt in the plasma membrane [34–36]. CaM also modulates epidermal growth factor receptor (EGFR)’s tyrosine kinase activity [37] which activates Ras and PI3Kα and has essential roles in programmed cell death and autophagy [28].
CaM Phosphorylation at Tyr99 Regulates PI3Kα Activation
Finally, and most tellingly, recently Chaudhuri et al. [26] observed that inhibiting the phosphorylation of CaM at Tyr99 decreased the interaction of the p85 subunit with CaM and PI3K activation. This interfered with PI3K production of phosphatidylinositol (3,4,5)-trisphosphate (PIP3), which promotes the insertion of endocellular transient receptor potential cation channel subfamily C member 6 (TRPC6, which forms a receptor-activated calcium channel in the cell membrane) into the cell membrane, and permitted the translocation of endothelial cells by lysophosphatidylcholine (lysoPC). LysoPC stimulates protein tyrosine kinase activity. Tyrosine phosphorylation is followed by elevation of intracellular Ca2+ concentration. Inhibition of the tyrosine kinase with genistein decreased the rise in the intracellular Ca2+ [38]. Pretreatment of bovine aortic endothelial cells (BAECs) with an inhibitor for Src-family kinases (PP2) blocked lysoPC-induced CaM phosphorylation at Tyr99, supporting lysoPC-stimulated CaM phosphorylation at Tyr99 by a Src family kinase [26]. The authors concluded that lysoPC induces Tyr99 phosphorylation by a Src family kinase and, importantly, that subsequently the phosphorylated CaM/Ca2+ activates PI3K to produce PIP3. CaM’s phosphorylation at Tyr99 forms a motif to which the nSH2 and cSH2 domains of the p85 subunit bind, activating PI3K, as postulated by Joyal et al. [25]. The conformational change induced by binding of a phosphorylated substrate can activate PI3K [39] by releasing the p110 catalytic subunit from the inhibitory effects of the nSH2, and allosterically promote p110’s catalytic activity through the cSH2 binding [5, 12]. This is further in line with Joyal et al.’s outcompeted CaM/SH2 interaction by the pYXXM peptide.
CaM Phosphorylation
CaM is phosphorylated by multiple protein serine/threonine kinases, and by receptor and non-receptor protein tyrosine kinases [40]. However, among the over fifty members in the superfamily of receptors with tyrosine kinase activity, the only ones shown to phosphorylate CaM are the insulin receptor (InsR) [41–43] and the EGFR [44–46]. Among the non-receptor protein-tyrosine kinases known to phosphorylate CaM are some members of the Src family kinases [27], Janus kinase 2 (JAK2), and p38Syk, and protein tyrosine kinase-III (c-Fyn and c-Fgr). All phosphorylate CaM at Tyr99 [40, 47] and to a lesser extent at Tyr138 [40]. Early on [48, 49] it was already observed that a highly basic peptide corresponding to the HVR of K-Ras stimulates the phosphorylation of CaM by the insulin receptor, which led to the suggestion that the HVR of K-Ras, but not of N-Ras, may stimulate the phosphorylation of CaM. This was later proposed to relate to CaM’s interaction with K-Ras, but not with N-Ras or H-Ras as observed by Villalonga et al. [1], although CaM was not phosphorylated in those experiments. Nonetheless, this raised the possibility of the coupling of the two events–CaM’s phosphorylation and binding to K-Ras at the membrane–however, it has been unclear how [40]. The phosphorylation of CaM by the InsR is sensitive to Ca2+; lower Ca2+ concentration stimulates phosphorylation whereas higher (above 1–10 μM) gradually inhibits it [48, 50–54]. Experiments using CaM-(Y99D/Y138D) and CaM-(Y99E/Y138E), where the negative charges of aspartic and glutamic acids mimic the phosphates [27], suggest that diphospho-CaM with pTyr99/pTyr138 can interact with Src, as can non-phosphorylated CaM, but not monophospho-CaM with pTyr138. CaM with pTyr138 also does not co-immunoprecipitate with Src [55], confirming pTyr99-CaM as a key player. In line with these observations, Chaudhuri et al. [26] found that CaM’s activity and the peptide binding affinity are altered by CaM’s phosphorylation state and bound Ca2+, and that Ca2+ can regulate the phosphorylation of CaM.
Finally, the inhibition of the interaction of calmodulin with Src-SH2 domain was also proposed to be an attractive strategy to inhibit cell proliferation of pancreatic cancer. Screening of compound libraries identified some initial leads that targeted the calmodulin-binding region on the SH2 domain and inhibited proliferation in in vitro assays [56].
How Ca2+/CaM Can Help K-Ras to Fully Activate PI3Kα
PI3Kα is regulated through its membrane binding ability and the population of effective phosphate transfer transition complexes [57–59]. These events are structurally coupled, as reflected in the Km, the PIP2 concentration at which PI3Kα is at half of its maximal catalytic rate (ka). The kinetic efficiency (ka/Km) of the Michaelis-Menten model is a direct measurement of enzyme activity. PI3Kα is recruited to the signaling complexes at the membrane by activated phosphorylated receptor tyrosine kinases (RTKs), which bind p85 SH2 domains. RTK-activated Ras binds p110; together with RTK’s (such as EGFR) phosphorylated C-terminal motif, Ras acts to fully activate PI3Kα (Figure 1). Although oncogenic K-Ras is active independently of the RTK signal, in the absence of RTK’s phosphorylation motifs, PI3Kα is not fully activated. There are two distinct mechanisms, through which the activated RTK pYXXM motif (or an associated protein, e.g. IRS-1 [30]) can activate PI3Kα [60–63]. In the first [64], the activated RTK pYXXM motif binds to the nSH2 domain [65] (Figure 2A). This releases the autoinhibition of p110α by nSH2, since the binding of nSH2 to p110α and to the pYXXM peptide cannot take place simultaneously [66]. The ‘hot spot’ mutations E542K and E454K–with the same charge repulsion instead of favorable salt-bridges–release the nSH2 autoinhibition of the p110 [67]. In the second mechanism, binding of the phosphorylated RTK motif to the cSH2 domain (Figure 2B) allosterically activates p110. In addition to the RTK pYXXM motif, PI3Kα activity is further stimulated allosterically by binding of the p110 Ras binding domain (RBD) to Ras-GTP [68, 69]. Ras appears to act similarly to the PI3Kα ‘hot spot’ (H1047R) mutation in p110 [63], with both inducing conformational changes in the p110 C-lobe at the membrane interface [67]. Both may enhance membrane binding which would promote accessibility to phosphatidylinositol (4,5)-bisphosphate (PIP2) on the membrane surface, making the oncogenic H1047R PI3Kα mutant independent of Ras-GTP [63]. The release of nSH2-p85α domain from p110α, which permits the effective formation of phosphate transfer transition complex, may correspond to an increase of ka. Experimental data indicate that both membrane binding capability and effective formation of the phosphate transfer transition complex are required for a fully active PI3Kα.
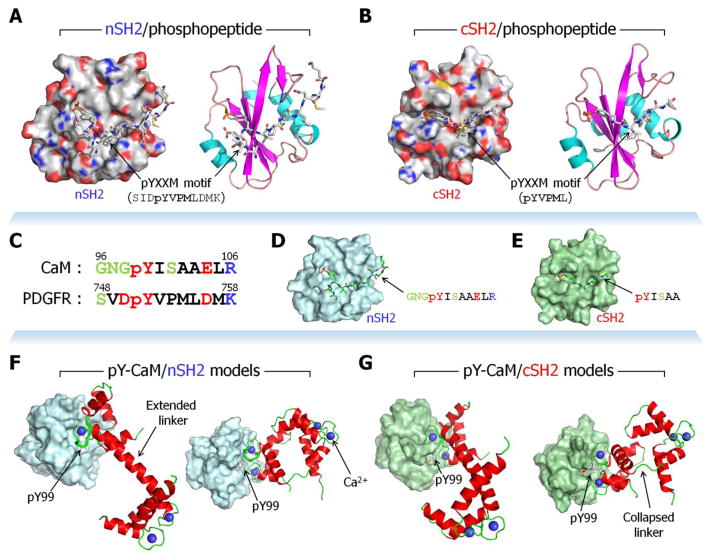
Figure 2.
The binding mode of the receptor tyrosine kinase (RTK)’s pYXXM motif to SH2 domains of the p85 subunit of phosphatidylinositol 3-kinase (PI3K). Crystal structures of (A) nSH2 (PDB code: 2IUI) and (B) cSH2 (PDB code: 1H9O) domains of PI3K interacting with RTK’s pYXXM peptides. The phosphotyrosyl peptides with pYVPM (the pYXXM motif) sequence were derived from the platelet-derived growth factor receptor (PDGFR) residues 751–754. SH2 domains are shown as surface (left) and cartoon (right) representations. The phosphotyrosyl peptides are shown as stick representation. In the surface representation, gray, green, red, and blue denote hydrophobic, polar, acidic, and basic sidechains of the SH2 residues, respectively. Model structures of phosphorylated calmodulin (CaM) at Tyr99 interacting with SH2 domains of the p85 subunit of phosphatidylinositol 3-kinase (PI3K). (C) Sequence comparison of phosphorylated CaM (residues 96–106) with PDGFR (residues 748–758) containing the pYXXM motif. In the sequence, black, green, red, and blue letters denote hydrophobic, polar, acidic, and basic residues, respectively. 73% of the residues are similar. (D) Modeling nSH2 with CaM’s phosphotyrosyl peptide (GNGpYISAAELR) by converting sequences from the RTK’s pYXXM peptide (PDB code: 2IUI). (E) Modeling cSH2 with CaM’s phosphotyrosyl peptide (pYISAA) by converting sequences from the RTK’s pYXXM peptide (PDB code: 1H9O). Two crystal CaMs with stretched linker (PDB code: 1CLL) and collapsed linker (PDB code: 1CDL) were phosphorylated at Tyr99 and aligned with the CaM’s phosphotyrosyl peptide, generating (F) CaM/nSH2 and (G) CaM/cSH2 complexes. Thick tubes in CaM’s cartoons represent the aligned portions, and pTyr99 sidechains are shown as sticks.
CaM can bind the n/cSH2 domains and substitute for the missing RTK signal (Figure 1). Genetically-engineered mouse models indicate that even without an RTK signal, oncogenic K-Ras can induce senescence (oncogene induced senescence, OIS) or proliferation and differentiation [70]; but not full PI3Kα activation. To get insight into the structural activation mechanism [60, 71], we constructed a structural model of the PI3Kα heterodimer by superimposing common features in six available PI3K crystal structures [5, 12]. The model includes the p110α catalytic subunit and the three p85α n/cSH2 domains as well as the GNP-bound H-Ras interacting with the PI3K’s RBD, ATP in the p110 cleft between the N- and C- lobes, and two RTK-derived phosphorylated peptides interacting with nSH2 and cSH2 (see Figure 4 in ref. [12]). The PIP2 lipid substrate head is at the mouth of the active site of the catalytic domain.
Modeling the Phosphorylated CaM Interaction and Ternary Complex
The model that we constructed represented one possible organization of the K-Ras4B/CaM/PI3Kα ternary complex (see Figure 3 in ref. [12]). In that model CaM is not phosphorylated. However, recent data pointing to Tyr99 phosphorylation and its interaction with SH2, and subsequent PI3K activation, suggest an additional, likely higher affinity binding mode. Two crystal structures are available for the SH2 domain complexed with an RTK phosphorylated peptide, with shorter and longer peptides (Figure 2A,B), raising the question of whether phosphorylated CaM at Tyr99 can bind an SH2 domain following these modes of interactions (Figure 2C–G). If so, a conformational change in CaM is likely to take place. Alternatively, the phosphorylated CaM can bind at the same site as the non-phosphorylated site. It has already been observed that phosphorylated and non-phosphorylated CaM share the same binding site (645RRRHIVRKRTLRRLLQ660) at the cytosolic juxtamembrane region of the EGFR [72].
To date the experimental data available for K-Ras/CaM interaction comes from NMR [2, 6] as well as fluorescence, imaging and biochemical data [22]. However, piecing together available structures and exploiting overlapped components, we are able to model a ternary complex composed of K-Ras4B-GTP, phosphorylated CaM at Tyr99, and PI3Kα (Figure 3). CaM with pTyr99 interacts with both K-Ras4B-GTP and cSH2 domain at the same time. A second CaM molecule with pTyr99 can interact with the nSH2 domain, which would relieve nSH2’s autoinhibitory action on the p110 catalytic domain. The ternary model that we constructed suggests that two Ca2+/CaM molecules with pTyr99 can bind both nSH2 and cSH2 to activate the PI3Kα/Akt pathway in KRAS-driven adenocarcinoma.
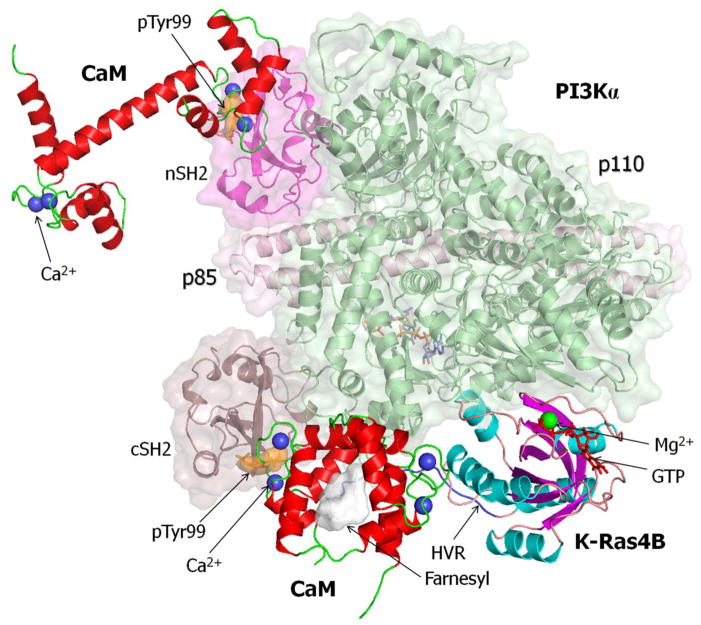
Figure 3.
A predicted ternary complex model constructed by K-Ras4B-GTP, phosphorylated calmodulin (CaM) at Tyr99, and phosphatidylinositol 3-kinase α (PI3Kα). The ternary complex was reconstituted by replacing pTyr99 CaM with unphosphorylated CaM in our previous model constructions [5, 12]. Full-length K-Ras4B-GTP with the post-translational modification (PTM) was modeled from the crystal structure of K-Ras4B catalytic domain (PDB code: 3GFT). CaM with the flexible linker (PDB code: 1CDL) was used to model the interactions of CaM with the hypervariable region (HVR) and farnesyl group of K-Ras4B-GTP and with cSH2 domain. CaM with the stretched linker (PDB code: 1CLL) was used in the interaction of nSH2 domain. The PI3Kα structure was derived from the crystal structures of p110 and p85 subunits (PDB code: 4OVV).
Regulation at the Membrane: K-Ras4B Phosphorylation and CaM Interaction
The interaction of K-Ras with the plasma membrane is mediated by farnesylation and a polybasic region, both at the C-terminal HVR. Phosphorylation of Ser181 within the polybasic region by protein kinase C (PKC) promotes rapid dissociation of K-Ras from the plasma membrane [73]. The polybasic region associates with the phospholipid anionic heads at the inner plasma leaflet and the hydrophobic farnesyl inserts into it, transiently anchoring K-Ras at the membrane. The phosphorylated K-Ras appears to remain at the plasma membrane [74], however, at microdomains different than those of non-phosphorylated K-Ras [75, 76]. Phosphorylation of the HVR modifies the affinity of K-Ras to acidic plasma membranes favored by its positively charged HVR, with the phosphoryl - lipid head groups charge repulsion preventing the K-Ras association [75, 76]. CaM’s direct interactions with the positively charged HVR and the farnesyl (Figure 3) similarly prevent the Ras-membrane association [77]. CaM’s binding also inhibits K-Ras phosphorylation, as its interaction sterically blocks the PKC phosphorylation site. Attachment to the membrane is typically by GTP-bound K-Ras; however, active state-like GDP-bound K-Ras, a certain population of which is present in the GDP-bound ensemble, can also bind [78]. The equilibrium between the attached and free state shifts in the presence of PKC and CaM. Whether PKC binds and phosphorylates K-Ras or CaM binds depends on the environment. Increase in Ca2+ and CaM concentration at the plasma membrane would shift the equilibrium toward a K-Ras/CaM bound state, which prevents K-Ras phosphorylation.
K-Ras4A May Also Interact with phosphorylated CaM
Above we focused on the highly abundant oncogenic K-Ras4B. However, oncogenic K-Ras4A may act similarly to K-Ras4B [10, 21]. This KRAS splice isoform has been observed particularly in K-Ras4B and N-Ras cancers [79]. Its HVR is also positively charged although not to the same extent as K-Ras4B. Like N-Ras, K-Ras4A HVR is farnesylated and palmitoylated; K-Ras4B is only farnesylated. However, palmitoylation is reversible, suggesting that K-Ras4A can exist in two states; K-Ras4B-like and N-Ras-like [21]. Although the initial results demonstrated that CaM specifically binds K-Ras4B [1], it is possible that depalmitoylated K-Ras4A may also bind CaM, albeit with lower affinity, and cooperate with CaM to stimulate PI3Kα activation and signaling. This argues that medications designed to block K-Ras4B proliferative signaling by targeting phosphorylated CaM’s binding and full activation of PI3Kα in the ternary complex of CaM, PI3Kα and K-Ras4B, may also be effective against K-Ras4A driven adenocarcinomas. Adenocarcinomas are predominantly KRAS-driven cancers.
Concluding Remarks
A major aim of the community is to obtain medications to target Ras-driven cancers [80]. The pharmacological landscape to curtail Ras cancers is formidable, and to date with little success. Major questions exist (see Outstanding Questions), and despite some promising leads, none reached the clinic. Targeting K-Ras4B cancers, the most abundant oncogenic Ras isoform, has been a particularly coveted goal. To realize this therapeutic challenge, the scientific community has been focusing on structural and mechanistic features that differentiate K-Ras from other isoforms, N-Ras and H-Ras. Efforts largely centered on those molecules that bind uniquely to K-Ras - but not to the other isoforms - and are key to its signaling, as well as K-Ras association with the membrane [18] and nanocluster formation [81]. Two K-Ras interactions are believed to be specific to this isoform: CaM and PDEδ, the chaperon that helps transport K-Ras4B from the endoplasmic reticulum (ER) to the plasma membrane. However, it is still unclear to what extent designing drugs to inhibit the interaction of K-Ras4B with PDEδ would accomplish this goal. The recent structure [82] coupled with earlier observations [83] and modeling suggest that PDEδ may be able to bind other depalmitoylated isoforms, and geranyl-geranylated K-Ras is also a PDEδ substrate; not only farnesylated K-Ras. CaM’s interaction with K-Ras4B and its role in KRAS-driven cancers have long been considered an option [5]. However, for decades the structure of the CaM with K-Ras and the mechanism of how CaM acts eluded the community. This Review suggests that combining literature reports with modeling reveal that the clue may rest with PI3Kα, a kinase involved in signaling pathways established to be key in cancer [84, 85].
Outstanding Questions Box.
The sequence of the hypervariable region of K-Ras and its lipid post-translational modifications differ from those of other Ras isoforms. Calmodulin binds only to K-Ras. A key question is how does calmodulin bind? After years of attempts, still no crystal or NMR structures, likely due to the multiple states through which calmodulin can interact with the K-Ras catalytic domain.
Calmodulin binding to K-Ras promotes cancers, particularly in the pancreas, lungs and colorectal. Why are these cancers particularly prone to K-Ras is a significant and still unresolved question.
How does calmodulin contribute to K-Ras driven tumor proliferation? This question is of paramount importance since proliferation is the first hallmark of cancer.
The mutational spectrum are not uniform across the different isoforms and cancers, and the clinical consequences differ. In K-Ras, some mutations result in worse patient outcome than others. How to understand these differential behaviors of single point mutations? Does it relate to more exposed effector binding site or effector specificity at the membrane?
How to drug Ras has been a daunting question. Ras has a flat surface, devoid of druggable pockets. Within this framework, an isoform-specific drug would reduce toxicity thus be favored. Drugging K-Ras is a particularly significant aim, due to its relative abundance in adenocarcinomas. A ternary complex of calmodulin/K-Ras4B/PI3Kα could constitute a K-Ras specific target; an important question is where should it be designed to bind? Should it be allosteric drug?
Experimental data pointed to the interaction of CaM with p85 SH2 domains [25]; it also pointed to the formation of a ternary complex involving K-Ras, CaM and PI3K [86]. Substantial data exist that Src family tyrosine kinases phosphorylate CaM at Tyr99 [27, 40, 56] and that SH2 domains commonly bind to tyrosine-phosphorylated substrates. Finally, there exist the recent telling observations: blocking phosphorylation of CaM at Tyr99 reduced the association of CaM with the p85 subunit and activation of PI3Kα [26]. Earlier, we proposed a ternary complex, and based on existing crystal structures and additional biophysical data and built a possible model [5, 12]. In this model, CaM was unphosphorylated. However, the weight of the accumulated data, capped by the direct recent finding, makes us believe that there are at least two possible ways of formation of ternary complexes: low affinity where the CaM interacts with the SH2 in an unphosphorylated state as attested by the Joyal et al. data [25], and a high affinity state where CaM is phosphorylated at Tyr99 (Figure 2F,G). We believe that this is the major state.
CaM is established to bind to only K-Ras; but not to N-Ras or H-Ras [1]. Key questions have thus been how does CaM bind to K-Ras–particularly K-Ras4B which to date appears the major splicing isoform in adenocarcinomas–and what is its exact role in KRAS-driven cell proliferation. The ternary complex model is important in providing plausible clues to both. In the model, the abundant Ca2+/CaM molecules can bind to both the nSH2 and cSH2 p85 domains of PI3Kα, as observed by Sacks and his colleagues [25]. This would respectively release the catalytic kinase domain autoinhibition and allosterically lead to full PI3Kα activation, in agreement with phosphorylated RTK actions [64–67], while binding to the K-Ras farnesylated HVR [68, 69]. Most importantly, it can also explain how oncogenic K-Ras–with CaM’s help–can fully activate PI3K in the absence of a signal from RTKs (Figure 1). Forward looking, due to the fundamental importance of CaM in KRAS-driven adenocarcinomas in drug discovery, efforts center on elucidating the K-Ras/CaM structure. The ternary complex concept [5, 12], which is based on available literature reports, crystal structure data, NMR and modeling, opens new avenues in studies of signaling and KRAS-specific drug discovery. It also provides clues into how CaM stimulates cell proliferation in cancer [87]. Nonetheless, the ambitious and arduous road toward obtaining beneficial drugs that work in the clinic to abolish CaM’s action still lies ahead. We note that recently, ophiobolin A was established to work by targeting K-Ras signaling in a stemness context by blocking CaM [88], which can be viewed as a step toward this aim.
Trends Box.
K-Ras, the most abundant oncogenic isoforms, differs from other Ras isoforms. This leads to altered interactions with the cell membrane, mode of translocation through the cytoplasm and signaling. Revealing K-Ras specific interactions with its partners is important to understand its biology and design therapeutics.
Downstream signaling stimulated by mitogenic cues through activated K-Ras differs from oncogenic mutant K-Ras. Uncovering the signaling alterations between physiological and oncogenic K-Ras is significant since it can be translated to K-Ras specific therapeutics.
Revealing how the relatively locally-abundant calcium-bound calmodulin acts to promote oncogenic K-Ras driven cancers is significant since it would clarify how mutant K-Ras signals efficiently in cancer.
Structural studies of Ras, its mutants and interactions may reveal new pharmacological opportunities.
Acknowledgments
We gratefully acknowledge the generous support from the American Cancer Society Grant RGS-09-057-01-GMC and the National Cancer Institute Grant R01 CA135341 to V.G. This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was supported [in part] by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Villalonga P, et al. Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol Cell Biol. 2001;21(21):7345–54. doi: 10.1128/MCB.21.21.7345-7354.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham SJ, et al. The hypervariable region of K-Ras4B is responsible for its specific interactions with calmodulin. Biochemistry. 2009;48(32):7575–83. doi: 10.1021/bi900769j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez-Moya B, et al. K-Ras4B phosphorylation at Ser181 is inhibited by calmodulin and modulates K-Ras activity and function. Oncogene. 2010;29(44):5911–22. doi: 10.1038/onc.2010.298. [DOI] [PubMed] [Google Scholar]
- 4.Wu LJ, et al. Both the C-terminal polylysine region and the farnesylation of K-RasB are important for its specific interaction with calmodulin. PLoS One. 2011;6(7):e21929. doi: 10.1371/journal.pone.0021929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nussinov R, et al. K-Ras4B/calmodulin/PI3Kα: A promising new adenocarcinoma-specific drug target? Expert Opin Ther Targets. 2016;20(7):831–42. doi: 10.1517/14728222.2016.1135131. [DOI] [PubMed] [Google Scholar]
- 6.Chavan TS, et al. Application of reductive 13C-methylation of lysines to enhance the sensitivity of conventional NMR methods. Molecules. 2013;18(6):7103–19. doi: 10.3390/molecules18067103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu S, et al. Ras conformational ensembles, allostery, and signaling. Chem Rev. 2016;116(11):6607–65. doi: 10.1021/acs.chemrev.5b00542. [DOI] [PubMed] [Google Scholar]
- 8.Mageean CJ, et al. Absolute quantification of endogenous Ras isoform abundance. PLoS One. 2015;10(11):e0142674. doi: 10.1371/journal.pone.0142674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung SI, et al. Comparison of liver oncogenic potential among human Ras isoforms. Oncotarget. 2016;7(6):7354–66. doi: 10.18632/oncotarget.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakrabarti M, et al. Comparison of the Conformations of KRAS Isoforms, K-Ras4A and K-Ras4B, Points to Similarities and Significant Differences. J Phys Chem B. 2016;120(4):667–79. doi: 10.1021/acs.jpcb.5b11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chavan TS, et al. High-affinity interaction of the K-Ras4B hypervariable region with the Ras active site. Biophys J. 2015;109(12):2602–13. doi: 10.1016/j.bpj.2015.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nussinov R, et al. The key role of calmodulin in KRAS-driven adenocarcinomas. Mol Cancer Res. 2015;13(9):1265–73. doi: 10.1158/1541-7786.MCR-15-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prior IA, et al. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72(10):2457–67. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCormick F. K-Ras protein as a drug target. J Mol Med (Berl) 2016;94(3):253–8. doi: 10.1007/s00109-016-1382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang MT, et al. K-Ras promotes tumorigenicity through suppression of non-canonical Wnt signaling. Cell. 2015;163(5):1237–51. doi: 10.1016/j.cell.2015.10.041. [DOI] [PubMed] [Google Scholar]
- 16.Lu S, et al. Drugging Ras GTPase: A comprehensive mechanistic and signaling structural view. Chem Soc Rev. 2016;45(18):4929–52. doi: 10.1039/c5cs00911a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcus K, Mattos C. Direct attack on Ras: Intramolecular communication and mutation-specific effects. Clin Cancer Res. 2015;21(8):1810–8. doi: 10.1158/1078-0432.CCR-14-2148. [DOI] [PubMed] [Google Scholar]
- 18.Cox AD, et al. Targeting Ras membrane association: Back to the future for anti-Ras drug discovery? Clin Cancer Res. 2015;21(8):1819–27. doi: 10.1158/1078-0432.CCR-14-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee A, et al. The disordered hypervariable region and the folded catalytic domain of oncogenic K-Ras4B partner in phospholipid binding. Curr Opin Struct Biol. 2016;36:10–7. doi: 10.1016/j.sbi.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chavan TS, et al. Plasma membrane regulates Ras signaling networks. Cell Logist. 2015;5(4):e1136374. doi: 10.1080/21592799.2015.1136374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nussinov R, et al. A new view of Ras isoforms in cancers. Cancer Res. 2016;76(1):18–23. doi: 10.1158/0008-5472.CAN-15-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Alcala C, et al. Identification of essential interacting elements in K-Ras/calmodulin binding and its role in K-Ras localization. J Biol Chem. 2008;283(16):10621–31. doi: 10.1074/jbc.M706238200. [DOI] [PubMed] [Google Scholar]
- 23.Nussinov R, et al. Independent and core pathways in oncogenic KRAS signaling. Expert Rev Proteomics. 2016;13(8):711–6. doi: 10.1080/14789450.2016.1209417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nussinov R, et al. Oncogenic KRAS signaling and YAP1/β-catenin: Similar cell cycle control in tumor initiation. Semin Cell Dev Biol. 2016;58:79–85. doi: 10.1016/j.semcdb.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Joyal JL, et al. Calmodulin activates phosphatidylinositol 3-kinase. J Biol Chem. 1997;272(45):28183–6. doi: 10.1074/jbc.272.45.28183. [DOI] [PubMed] [Google Scholar]
- 26.Chaudhuri P, et al. Membrane translocation of TRPC6 channels and endothelial migration are regulated by calmodulin and PI3 kinase activation. Proc Natl Acad Sci U S A. 2016;113(8):2110–5. doi: 10.1073/pnas.1600371113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stateva SR, et al. Ca2+/calmodulin and apo-calmodulin both bind to and enhance the tyrosine kinase activity of c-Src. PLoS One. 2015;10(6):e0128783. doi: 10.1371/journal.pone.0128783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berchtold MW, Villalobo A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim Biophys Acta. 2014;1843(2):398–435. doi: 10.1016/j.bbamcr.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 29.Sun XJ, et al. Expression and function of IRS-1 in insulin signal transmission. J Biol Chem. 1992;267(31):22662–72. [PubMed] [Google Scholar]
- 30.Backer JM, et al. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992;11(9):3469–79. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vergne I, et al. Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J Exp Med. 2003;198(4):653–9. doi: 10.1084/jem.20030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esteras N, et al. Altered calmodulin degradation and signaling in non-neuronal cells from Alzheimer’s disease patients. Curr Alzheimer Res. 2012;9(3):267–77. doi: 10.2174/156720512800107564. [DOI] [PubMed] [Google Scholar]
- 33.Chin D, Means AR. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000;10(8):322–8. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 34.Zhou TT, et al. SP6616 as a new Kv2.1 channel inhibitor efficiently promotes βcell survival involving both PKC/Erk1/2 and CaM/PI3K/Akt signaling pathways. Cell Death Dis. 2016;7:e2216. doi: 10.1038/cddis.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, et al. Subtoxic N-methyl-D-aspartate delayed neuronal death in ischemic brain injury through TrkB receptor- and calmodulin-mediated PI-3K/Akt pathway activation. Hippocampus. 2007;17(7):525–37. doi: 10.1002/hipo.20289. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Garcia MJ, et al. Glial cell line-derived neurotrophic factor increases intracellular calcium concentration. Role of calcium/calmodulin in the activation of the phosphatidylinositol 3-kinase pathway. J Biol Chem. 2004;279(7):6132–42. doi: 10.1074/jbc.M308367200. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez-Gonzalez P, et al. Calmodulin-mediated regulation of the epidermal growth factor receptor. FEBS J. 2010;277(2):327–42. doi: 10.1111/j.1742-4658.2009.07469.x. [DOI] [PubMed] [Google Scholar]
- 38.Legradi A, et al. Lysophosphatidylcholine is a regulator of tyrosine kinase activity and intracellular Ca2+ level in Jurkat T cell line. Immunol Lett. 2004;91(1):17–21. doi: 10.1016/j.imlet.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 39.Shoelson SE, et al. Specific phosphopeptide binding regulates a conformational change in the PI 3-kinase SH2 domain associated with enzyme activation. EMBO J. 1993;12(2):795–802. doi: 10.1002/j.1460-2075.1993.tb05714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benaim G, Villalobo A. Phosphorylation of calmodulin. Functional implications. Eur J Biochem. 2002;269(15):3619–31. doi: 10.1046/j.1432-1033.2002.03038.x. [DOI] [PubMed] [Google Scholar]
- 41.Joyal JL, et al. Identification of insulin-stimulated phosphorylation sites on calmodulin. Biochemistry. 1996;35(20):6267–75. doi: 10.1021/bi9600198. [DOI] [PubMed] [Google Scholar]
- 42.Wong EC, et al. Characteristics of calmodulin phosphorylation by the insulin receptor kinase. Endocrinology. 1988;123(4):1830–6. doi: 10.1210/endo-123-4-1830. [DOI] [PubMed] [Google Scholar]
- 43.Williams JP, et al. Tyrosine-phosphorylated calmodulin has reduced biological activity. Arch Biochem Biophys. 1994;315(1):119–26. doi: 10.1006/abbi.1994.1479. [DOI] [PubMed] [Google Scholar]
- 44.Benguria A, et al. Phosphorylation of calmodulin by the epidermal-growth-factor-receptor tyrosine kinase. Eur J Biochem. 1994;224(3):909–16. doi: 10.1111/j.1432-1033.1994.00909.x. [DOI] [PubMed] [Google Scholar]
- 45.Benaim G, et al. Comparative phosphorylation of calmodulin from trypanosomatids and bovine brain by calmodulin-binding protein kinases. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;120(1):57–65. doi: 10.1016/s0742-8413(98)00006-1. [DOI] [PubMed] [Google Scholar]
- 46.Palomo-Jimenez PI, et al. A method for the purification of phospho(Tyr)calmodulin free of nonphosphorylated calmodulin. Protein Expr Purif. 1999;16(3):388–95. doi: 10.1006/prep.1999.1092. [DOI] [PubMed] [Google Scholar]
- 47.Stateva SR, et al. Characterization of phospho-(tyrosine)-mimetic calmodulin mutants. PLoS One. 2015;10(4):e0120798. doi: 10.1371/journal.pone.0120798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sacks DB, et al. The carboxyl terminal segment of the c-Ki-ras 2 gene product mediates insulin-stimulated phosphorylation of calmodulin and stimulates insulin-independent autophosphorylation of the insulin receptor. Biochem Biophys Res Commun. 1989;161(2):399–405. doi: 10.1016/0006-291x(89)92612-0. [DOI] [PubMed] [Google Scholar]
- 49.Fujita-Yamaguchi Y, et al. In vitro tyrosine phosphorylation studies on Ras proteins and calmodulin suggest that polylysine-like basic peptides or domains may be involved in interactions between insulin receptor kinase and its substrate. Proc Natl Acad Sci U S A. 1989;86(19):7306–10. doi: 10.1073/pnas.86.19.7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graves CB, et al. The insulin receptor and calmodulin. Calmodulin enhances insulin-mediated receptor kinase activity and insulin stimulates phosphorylation of calmodulin. J Biol Chem. 1986;261(22):10429–38. [PubMed] [Google Scholar]
- 51.Sacks DB, McDonald JM. Insulin-stimulated phosphorylation of calmodulin by rat liver insulin receptor preparations. J Biol Chem. 1988;263(5):2377–83. [PubMed] [Google Scholar]
- 52.Sacks DB, McDonald JM. Calmodulin as substrate for insulin-receptor kinase. Phosphorylation by receptors from rat skeletal muscle. Diabetes. 1989;38(1):84–90. doi: 10.2337/diab.38.1.84. [DOI] [PubMed] [Google Scholar]
- 53.Sacks DB, et al. Tyrosine-specific phosphorylation of calmodulin by the insulin receptor kinase purified from human placenta. Biochem J. 1989;263(3):803–12. doi: 10.1042/bj2630803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sacks DB. Alteration of calmodulin-protein interactions by a monoclonal antibody to calmodulin. Biochim Biophys Acta. 1994;1206(1):120–8. doi: 10.1016/0167-4838(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 55.Wu J, et al. β-Adducin siRNA disruption of the spectrin-based cytoskeleton in differentiating keratinocytes prevented by calcium acting through calmodulin/epidermal growth factor receptor/cadherin pathway. Cell Signal. 2015;27(1):15–25. doi: 10.1016/j.cellsig.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Tzou YM, et al. Identification of initial leads directed at the calmodulin-binding region on the Src-SH2 domain that exhibit anti-proliferation activity against pancreatic cancer. Bioorg Med Chem Lett. 2016;26(4):1237–44. doi: 10.1016/j.bmcl.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gabelli SB, et al. Activation of PI3Kα by physiological effectors and by oncogenic mutations: structural and dynamic effects. Biophys Rev. 2014;6(1):89–95. doi: 10.1007/s12551-013-0131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burke JE, et al. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110α (PIK3CA) Proc Natl Acad Sci U S A. 2012;109(38):15259–64. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27(41):5486–96. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vadas O, et al. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci Signal. 2011;4(195):re2. doi: 10.1126/scisignal.2002165. [DOI] [PubMed] [Google Scholar]
- 61.Miled N, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317(5835):239–42. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 62.Huang CH, et al. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science. 2007;318(5857):1744–8. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 63.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110α of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105(7):2652–7. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geering B, et al. Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. Proc Natl Acad Sci U S A. 2007;104(19):7809–14. doi: 10.1073/pnas.0700373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carpenter CL, et al. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J Biol Chem. 1993;268(13):9478–83. [PubMed] [Google Scholar]
- 66.Carson JD, et al. Effects of oncogenic p110α subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem J. 2008;409(2):519–24. doi: 10.1042/BJ20070681. [DOI] [PubMed] [Google Scholar]
- 67.Mandelker D, et al. A frequent kinase domain mutation that changes the interaction between PI3Kα and the membrane. Proc Natl Acad Sci U S A. 2009;106(40):16996–7001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kodaki T, et al. The activation of phosphatidylinositol 3-kinase by Ras. Curr Biol. 1994;4(9):798–806. doi: 10.1016/s0960-9822(00)00177-9. [DOI] [PubMed] [Google Scholar]
- 69.Gupta S, et al. Binding of ras to phosphoinositide 3-kinase p110α is required for RAS-driven tumorigenesis in mice. Cell. 2007;129(5):957–68. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 70.Xu Y, et al. Emerging roles of the p38 MAPK and PI3K/AKT/mTOR pathways in oncogene-induced senescence. Trends Biochem Sci. 2014;39(6):268–76. doi: 10.1016/j.tibs.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vanhaesebroeck B, et al. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 72.Stateva SR, et al. The activating role of phospho-(Tyr)-calmodulin on the epidermal growth factor receptor. Biochem J. 2015;472(2):195–204. doi: 10.1042/BJ20150851. [DOI] [PubMed] [Google Scholar]
- 73.Bivona TG, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21(4):481–93. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 74.Barcelo C, et al. Oncogenic K-ras segregates at spatially distinct plasma membrane signaling platforms according to its phosphorylation status. J Cell Sci. 2013;126(Pt 20):4553–9. doi: 10.1242/jcs.123737. [DOI] [PubMed] [Google Scholar]
- 75.Alvarez-Moya B, et al. CaM interaction and Ser181 phosphorylation as new K-Ras signaling modulators. Small GTPases. 2011;2(2):99–103. doi: 10.4161/sgtp.2.2.15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jang H, et al. Mechanisms of membrane binding of small GTPase K-Ras4B farnesylated hypervariable region. J Biol Chem. 2015;290(15):9465–77. doi: 10.1074/jbc.M114.620724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sperlich B, et al. Regulation of K-Ras4B membrane binding by calmodulin. Biophys J. 2016;111(1):113–22. doi: 10.1016/j.bpj.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jang H, et al. The higher level of complexity of K-Ras4B activation at the membrane. FASEB J. 2016;30(4):1643–55. doi: 10.1096/fj.15-279091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsai FD, et al. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci U S A. 2015;112(3):779–84. doi: 10.1073/pnas.1412811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ryan MB, et al. Targeting RAS-mutant cancers: is ERK the key? Trends Cancer. 2015;1(3):183–198. doi: 10.1016/j.trecan.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou Y, et al. Nonsteroidal anti-inflammatory drugs alter the spatiotemporal organization of Ras proteins on the plasma membrane. J Biol Chem. 2012;287(20):16586–95. doi: 10.1074/jbc.M112.348490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dharmaiah S, et al. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc Natl Acad Sci U S A. 2016;113(44):E6766–E6775. doi: 10.1073/pnas.1615316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chandra A, et al. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol. 2011;14(2):148–58. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 84.Thapa N, et al. The hidden conundrum of phosphoinositide signaling in cancer. Trends Cancer. 2016;2(7):378–390. doi: 10.1016/j.trecan.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guri Y, Hall MN. mTOR signaling confers resistance to targeted cancer drugs. Trends Cancer. 2016;2(11):688–97. doi: 10.1016/j.trecan.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 86.Liao J, et al. Growth factor-dependent AKT activation and cell migration requires the function of c-K(B)-Ras versus other cellular ras isoforms. J Biol Chem. 2006;281(40):29730–8. doi: 10.1074/jbc.M600668200. [DOI] [PubMed] [Google Scholar]
- 87.Nussinov R, et al. Principles of K-Ras effector organization and the role of oncogenic K-Ras in cancer initiation through G1 cell cycle deregulation. Expert Rev Proteomics. 2015;12(6):669–82. doi: 10.1586/14789450.2015.1100079. [DOI] [PubMed] [Google Scholar]
- 88.Najumudeen AK, et al. Cancer stem cell drugs target K-ras signaling in a stemness context. Oncogene. 2016;35(40):5248–62. doi: 10.1038/onc.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]