Abstract
The first discovered member of the mammalian FABP family, liver fatty acid binding protein (FABP1, L-FABP), occurs at high cytosolic concentration in liver, intestine and in the case of humans also in kidney. While the rat FABP1 is well studied, the extent these findings translate to human FABP1 is not clear—especially in view of recent studies showing that endocannabinoids and cannabinoids represent novel rat FABP1 ligands and FABP1 gene ablation impacts the hepatic endocannabinoid system, known to be involved in non-alcoholic fatty liver (NAFLD) development. Although not detectable in brain, FABP1 ablation nevertheless also impacts brain endocannabinoids. Despite overall tertiary structure similarity, human FABP1 differs significantly from rat FABP1 in secondary structure, much larger ligand binding cavity, and affinities/specificities for some ligands. Moreover, while both mouse and human FABP1 mediate ligand induction of peroxisome proliferator activated receptor-α, (PPARα), they differ markedly in pattern of genes induced. This is critically important because a highly prevalent human SNP (26–38% minor allele frequency and 8.3±1.9% homozygous) results in a FABP1 T94A substitution that further accentuates these species differences. The human FABP1 T94A variant is associated with altered body mass index (BMI), clinical dyslipidemias (elevated plasma triglycerides and LDL cholesterol), atherothrombotic cerebral infarction, and non-alcoholic fatty liver disease (NAFLD). Resolving human FABP1 and the T94A variant’s impact on the endocannabinoid and cannabinoid system is an exciting challenge due to the importance of this system on hepatic lipid accumulation as well as behavior, pain, inflammation, and satiety.
Keywords: Liver, fatty acid binding protein (FABP1), triglyceride
INTRODUCTION
Liver fatty acid binding protein (FABP1, L-FABP), the first discovered member of the FABP family (1–4), is highly a prevalent soluble protein in rodent (2–6% of cytosol protein; 200–400 μM) and even more so in humans (7–10% of cytosolic protein (700–1000 μM) liver cytosol (5, 6). Nevertheless, most studies of FABP1 structure, ligand specificity and function have focused on the rat and murine FABP1. Although the human (7, 8) and rat (9, 10) FABP1 share in common an overall tertiary structure composed of a ten-β-sheet β-barrel along with two α-helices and turns between them, nearly 20% of the amino acid sequence of human FABP1 is non-identical to that of the rat FABP1 (11). In fact, nearly half of these amino acid substitutions are non-identical nonconservative replacements (11). As a result, the secondary structure of the human FABP1 is less α-helical (12), has higher thermal stability (12), and differs in conformational flexibility and mode of LCFA binding (7, 13–16). Further, recent X-ray and NMR studies show that the binding cavity of human FABP1 is larger and is the largest of any mammalian FABP which suggests potential differences in ligand affinity, specificity, and/or function (7, 8, 13, 14, 17). Rat and human FABP1 are unique among the FABP family in terms of both the size of its binding cavity as well as much broader ligand specificity. Unlike other FABPs, the binding cavity of FABP1 is much larger—accommodating up to two lipophilic ligands rather than only one (7, 8, 13, 14, 17, 18).
More important, FABP1 has much broader ligand specificity. For example, rat FABP1 binds both straight- and branched-chain LCFA (19–21), long chain fatty acyl CoA (LCFA-CoA), acyl-carnitines, LCFA oxidation products, prostaglandins, lysophospholipids, and many other LCFA-like lipophilic ligands (rev. in (4, 5, 9, 22, 22–27)). Rat FABP1 also accommodates a single larger molecule (e.g. cholesterol, bile acid), thereby functioning as the primary cytosolic chaperone for secretion of bile acids and HDL-derived cholesterol into bile (6, 28–34). While early studies of the human FABP1 confirmed significant qualitative overlap in specificity for many lipophilic ligands with that of the rat FABP1, specificity of the human FABP1 was even broader as indicated by the binding of steroid hormones (testosterone, estradiol), fatty alcohols (eicosanol, retinol), retinoic acid, and vitamins (D3, E, K1) (12, 27, 35–37). Importantly, direct comparison of the ligand binding affinities of the human and rat FABP1 within the same study showed that the human FABP1 has slightly higher affinities for saturated LCFA (palmitic and stearic acids) and monounsaturated LCFA (oleic acid), 2.2-fold higher affinity for oleoyl-CoA, and 3 to 200-fold higher affinities for lysophosphatidic acid, 1-palmitoyl-2-oleoyl phosphatidic acid, and fenofibric acid (12, 37). In contrast, while both human and rat FABP1 bind cholesterol, the human FABP1 has 3.5-fold weaker affinity for cholesterol as compared to rat FABP1 (38, 39). Taken together these findings indicate the limitations of assuming similar ligand specificities and/or specificities for the human FABP1 based on those established for the rat FABP1. This caveat is consistent with the structural differences between the human and rat FABP1 binding cavities noted above. Due to its ability to bind fibrates and a broad variety of other xenobiotics, FABP1 is a target of active therapeutic interest (7, 8, 13–16, 40–42). Yet, the above studies underscore the need to examine not just rodent liver and hepatocyte functional models but also extend them to the respective human FABP1, liver, and hepatocytes.
HUMAN AND MURINE FABP1 ENHANCE LCFA UPTAKE
While there have been no reports of complete loss of FABP1 in humans, the impact of human and murine FABP1 expression level has been examined in a variety of tumor cell lines including cloned human HepG2 hepatoma cells, transfected ‘Chang liver’ cells overexpressing human FABP1, and transfected L-cell fibroblasts overexpressing FABP1. Rat FABP1 overexpression in cultured mouse L-cell fibroblasts stimulates fatty acid uptake and trafficking (43–47). Likewise, the expression level of human FABP1 in human liver-derived HepG2 cells correlated directly with uptake of radiolabeled monounsaturated LCFA (48). Overexpression of human FABP1 in ‘Chang liver’ cells also stimulates uptake of LCFA (49). Conversely, the impact of complete loss of FABP1 has been studied extensively in mouse FABP1 gene ablated models. FABP1 gene ablation inhibits uptake of a variety of fluorescent saturated fatty acids (NBD-stearic acid, C18:0; BODIPY-C16) and/or radiolabeled saturated fatty acids (C18:0), branched-chain saturated (phytanic acid), and monounsaturated fatty acids (C18:1) in vivo (50, 51) and in cultured primary mouse hepatocytes (52–54). Concomitantly, FABP1 ablation decreased liver cytosol LCFA binding capacity >80% in vivo (50) and decreased cytosolic transport/diffusion 2-fold (52). LCFA are membrane-bound, and cytoplasm is 10-fold more viscous than aqueous due to cytoskeleton, organelles, and proteins (55). FABP1 overcomes these barriers by desorbing membrane-bound LCFA into the cytosol and decreasing ‘tortuosity’ of diffusional paths (55). It should be noted that FABP1 gene ablation was not compensated for by upregulation of other liver cytosol LCFA binding proteins (SCP-2, FABP7, FABP3, FABP2, FABP5, CRABP1, CRABP2, FABP4) or membrane LCFA transport proteins (50, 53, 56, 57).
HUMAN AND MURINE FABP1 INDUCE LCFA OXIDATION
FABP1 directly targets LCFA-CoA to oxidative organelles for oxidation. FABP1 ablation inhibits LCFA β-oxidation in vitro (58), in mouse hepatocytes (52, 54) and decreased serum β-hydroxybutyrate (in vivo LCFA β-oxidation) in mice (2, 59). Rat FABP1 binds and alters CPT1 conformation to transfer bound LCFA–CoA into mitochondria for β-oxidation (58, 60). Conversely, rat FABP1 overexpression increased LCFA targeting to mitochondria and peroxisomes for oxidation (52).
Recent studies in vitro, transfected cells, and cultured primary mouse and human hepatocytes have established that both human and murine FABP1 also elicit longer term impact on LCFA oxidation by facilitating ligand activation of nuclear receptors such as PPARα and HNF4α. Ligand (LCFA, n-3 polyunsaturated LCFA, fibrates) binding to human and murine FABP1 redistributes the FABP1 into the nucleus, thereby also co-transporting the bound ligands into the nucleus, a process impaired by FABP1 gene ablation (61–63). Within the nucleus these FABP1s directly bind to and alter PPARα conformation (17, 61, 64–66)--thereby facilitating transfer of FABP1-bound ligand to PPARα for transcriptional activation FABP1 gene ablation or chemical inhibition, like PPARα gene ablation, abolishes ligand (fibrates, n-3 polyunsaturated fatty acids) activation of PPARα transcription of multiple genes involved in LCFA uptake (FATP), intracellular transport (FABP1), and oxidation (CPT1A, CPT2, ACOX1) in cultured primary mouse hepatocytes (61, 62, 67). Concomitantly, FABP1 ablation decreased/abolished the ability of synthetic (fibrate) and natural (branched-chain LCFA) peroxisome proliferators to lower serum and hepatic TAG (57, 68), but also exacerbated toxicity of dietary PPARα agonists (52, 57, 69, 70).
Finally, it is important to note that in view of the differences in human and rodent FABP1 structures and ligand specificities noted above, fibrate and other activators of PPARα do not induce the same target genes in human compared to mouse cultured primary hepatocytes. While there is significant overlap in inducing transcription of target genes in LCFA oxidation, nearly half of the ligand-induced PPARα target genes differ between human and mouse cultured primary hepatocytes (71, 72). Recently, it was also shown that rodent FABP1 binds and potentiates ligand (LCFA-CoA) activation of hepatocyte nuclear factor 4α (HNF4α ), another nuclear receptor involved in hepatic LCFA and glucose metabolism (73).
FABP1’s ROLE IN HEPATIC LIPID ACCUMULATION
Rat FABP1 in vitro and overexpression in cultured L-cell fibroblasts markedly enhanced long chain fatty acid (LCFA) intracellular targeting to endoplasmic reticulum for esterification (43–46, 74–77). Conversely, all FABP gene ablated mouse models generated to date have exhibited increased TAG accumulation in liver in vivo (50, 78–81) and in hepatocytes (52–54, 80). Hepatic TAG accumulation in FABP1 gene ablated mice was not associated with altered intestinal fat absorption and food intake was only slightly or not increased (2, 81, 82). It is important to note that these findings with primary hepatocytes in culture and liver in vivo differed significantly from cultured transformed cell models. For example, overexpression of human FABP1 enhanced LCFA targeting to TAG to elicit TAG accumulation in transfected human ‘Chang liver cells’ (49). Contrary to their name, however, human ‘Chang liver’ cells are not of hepatic origin but instead are derived from human cervical cancer cells (83).
FABP1 impacts hepatic lipid accumulation not only by decreasing hepatic LCFA β-oxidation (see above), but also in part by its ability to influence biliary secretion of HDL-derived cholesterol and alter bile acid profile (34, 84, 85). FABP1 gene ablation decreases hepatic uptake and biliary secretion of high density lipoprotein (HDL)-derived NBD-cholesterol (34). Furthermore, FABP1 ablation significantly decreases hepatic bile acid concentration while increasing biliary bile acid and altering biliary bile acid composition towards increased hydrophobicity and lower indices of cholesterol solubility in biliary bile. Concomitantly, FABP1 ablation increases serum TAG (2, 39, 59, 80, 86, 87)—associated not only with reduced hepatic LCFA oxidation but also reduced VLDL clearance by lipoprotein lipase (LPL) but not increased hepatic VLDL secretion (80).
HUMAN AND MURINE FABP1 INTERACT WITH THE ENDOCANNABINOID PRECURSOR ARACHIDONIC ACID
The endogenous endocannabinoids (EC) such as arachidonoylethanolamide (anandamide, AEA), 2-arachidonoylglycerol (2-AG) are both derived from arachidonic acid (ARA)-containing phospholipids. Although FABP1 is not detectable in brain, recent studies suggest that hepatic FABP1 may impact EC formation not only in liver but also in brain by regulating plasma availability (88, 89).
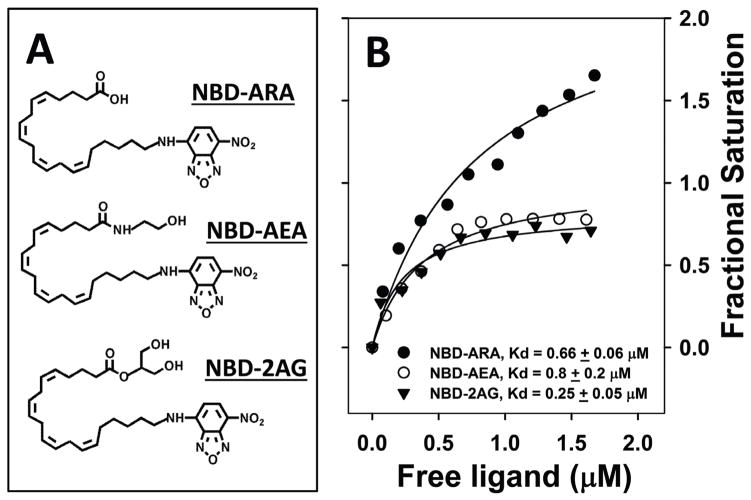
FABP1 has high affinity for arachidonic acid (ARA, C20:4n-6), the precursor of phospholipids from which endocannabinoids AEA and 2-AG are derived. Human as well as rat FABP1 bind ARA with higher affinity than saturated and monounsaturated fatty acids (18, 47, 90). Direct comparison in a single study using an ANS fluorescence displacement assay showed that human and rat FABP1 both bind ARA with high affinity, Kids of 0.113±0.006 and 0.110±0.006 μM, respectively (37). Rat FABP1 affinity for ARA was confirmed by direct binding A5C, a novel metabolizable fluorescent ARA developed in collaboration with Dr. Bill Smith (U. Michigan), that was bound with high affinity, (Kd=77±6nM) (47). NBD-ARA, a novel NBD-ARA probe (Fig 2A) developed in collaboration with Drs. W. Shaw, S. Burgess, and S. Li (Avanti). FABP1 exhibited two NBD-ARA binding sites with average affinity of Kd=0.66±0.06 μM (Fig 2B) (89). Taken together with the high level of human and rat FABP1 in liver cytosol, these findings suggest FABP1 is a major contributor to hepatic cytosolic ARA binding capacity—analogous to its comprising >80% of cytosolic binding of other LCFA (50, 90). Nearly 3/4 of LCFA binding sites are occupied in native FABP1 isolated from rat liver (90). Consistent with FABP1’s higher affinity for ARA than saturated or unsaturated fatty acids, ARA comprises 25% of the total FABP1 bound LCFA despite the fact that other LCFAs are much more prevalent in liver (90). As shown below, FABP1 gene ablation has important consequences not just for liver but also brain levels of ARA-containing endocannabinoids.
Figure 2. Direct binding of NBD-labeled ARA, AEA, and 2AG to rat FABP1.
Panel A: Structures of NBD-arachidonic acid (NBD-ARA), NBD-arachidonoyl-ethanolamide (NBD-AEA) and NBD-arachidonoyl-2-glycerol (NBD-2-AG). Panel B: Binding of the NBD labeled endocannabinoids AEA, 2AG and their precursor ARA to rat L-FABP was measured based on the fluorescence increase of NBD group upon binding to the hydrophobic binding pocket as in (89). Briefly, NBD fluorescence emission spectra were obtained by scanning from 515–600nm with 490 nm excitation. Forward titrations (500nM L-FABP titrated with 0–2.5μM total ligand), and reverse titrations (100nM NBD-labeled ligands titrated with 0–3μM FABP1 were performed. Signals from corresponding NBD labeled ligands without FABP1 were used as background and subtracted from each data point. From the curve fitting of the reverse titration, the fluorescence intensity (at emission wavelength 540nm) of NBD-labeled ligand (per nM) when fully bound to FABP1 was calculated. This parameter was then used to calculate the fractional saturation and free ligand concentration in forward titration. Binding curves were constructed by plotting fractional saturation (Y) vs free ligand concentration (X), from which Kd and Bmax were calculated by curve fitting. Kd was the mean ± SE (n=3).
FABP1 enhances uptake of the endocannabinoid precursor arachidonic acid (ARA) (47). Although nothing is known about the impact of human FABP1 on ARA uptake, the impact of rat FABP1 overexpression has been examined in murine L-cell fibroblasts. Overexpressing rat FABP1 in L-cells increased the uptake of cis-parinaric acid (43, 44, 91, 92). While cis-parinaric acid, the first naturally-occurring fluorescent LCFA discovered (93), has four double bonds as does ARA, neither the 18-carbon chain length nor the methyl-terminal location of the double bonds tetraene reflect that of ARA (47). In contrast, both the chain-length and the double bond localization of A5C much more accurately reflect those of ARA (47). Rat FABP1 overexpression increased ARA uptake as shown by real-time multiphoton imaging of A5C (47) and by uptake of radiolabeled [3H]-ARA (47). FABP1 enhanced initial rate, decreased half-time, and increased maximal binding capacity. Although human FABP1 exhibits the same affinity for ARA as does rat FABP (37), how the structural differences will impact human FABP1’s ability to enhance ARA uptake is not known.
HUMAN AND MURINE FABP1 ROLES AS ENDOCANNABINOID ‘CHAPERONES’
The endogenous ECs [arachidonoylethanolamide (anandamide, AEA), 2-arachidonoylglycerol (2-AG)] derived from ARA-containing phospholipids together with their cannabinoid (CB) receptors constitute a novel system for modulating behavior, pain, inflammation, and satiety (94–100) as well as hepatic lipid accumulation (101–104) by central and/or peripheral mechanisms. FABPs present within brain neuronal and other cells (i.e. FABP3,5,7) have been shown to bind and act as brain cytosolic binding proteins of endocannabinoids (AEA, 2-AG) and cannabinoids (THC, CBD) (105, 106). These brain FABP3,5,7s act as ‘chaperones’ that facilitate re-uptake and targeting of the respective bound ligands to degradative enzymes present in brain organelles (endoplasmic reticulum, mitochondria, lysosomes) or cytosol for metabolism (107–109). Alternately, the brain FABP3,5,7s may also ‘chaperone’ the ECs to the nucleus for regulating nuclear receptors (110). In contrast, until recently the identity of major EC ‘chaperone(s)’ in liver was not clear (89).
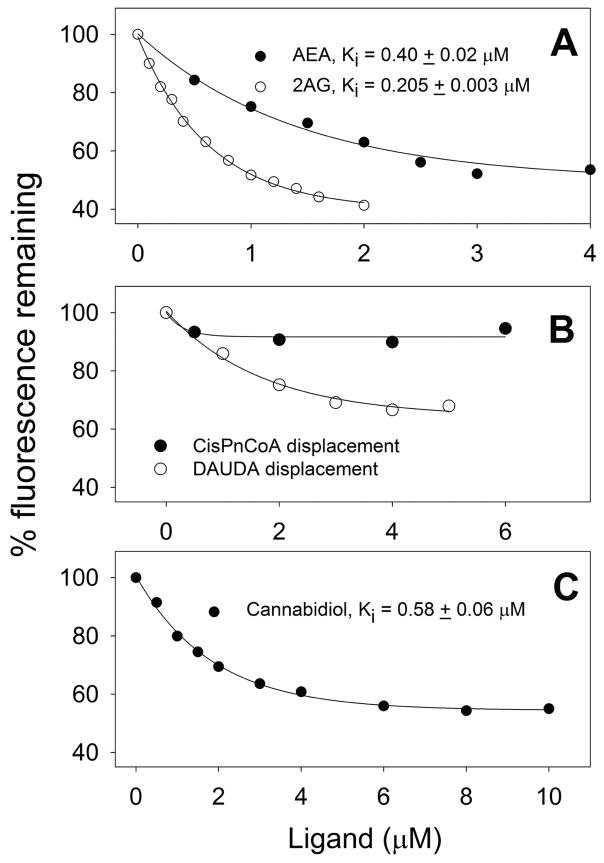
Little is known about how the very hydrophobic, highly-membrane associated endocannabinoids (AEA, 2-AG) traffic within hepatocytes from sites of synthesis for extracellular release, uptake/reuptake, or to intracellular sites for hydrolysis/degradation (102, 111). A cis-parinaroyl-CoA displacement assay developed by our laboratory (112, 113) suggested for the first time that endocannabinoids and phytocannabinoids bind to rat FABP1 (89). The endocannabinoids AEA and 2-AG both displace rat FABP1-bound cis-parinaroyl-CoA (Fig. 1A). Analysis of multiple binding curves yielded Kis of 0.40±0.02 and 0.205±0.003 μM, respectively (89). However, the lack of a suitable fluorescent-labeled AEA and 2-AG assays has been a major limitation in more directly demonstrating AEA and 2-AG binding to FABP1 or other FABPs. This limitation was recently overcome by the development of novel synthetic fluorescent NBD-AEA and NBD-2-AG analogues in collaboration with scientists at Avanti Polar Lipids, Inc. (Fig. 2A). These probes for the first time allowed direct determination of rat FABP1’s binding affinity for these endocannabinoids in a direct binding assay (89). Rat FABP1 affinities for NBD-AEA and NBD-2-AG, Kds of 0.80±0.20 and 0.25±0.05 μM, respectively (Fig. 2B), were in the same range as that for NBD-ARA with Kd of 0.66±0.06 (Fig. 2B). However, the fractional saturation binding curves indicated that each molecule of rat FABP1 protein bound only a single NBD-AEA or NBD-2-AG as compared to two molecules of NBD-ARA (Fig 2B).
Figure 1. Binding of endocannabinoid (AEA, 2AG) and phytocannabinoid (cannabidiol) to rat and human wild-type FABP1: fluorescent ligand displacement assay.
Binding of endocannabinoids (AEA and 2-AG) and a phytocannabinoid (cannabidiol) to rat or human WT FABP1 was measured by displacing bound cis-PnCoA and monitoring cis-PnCoA fluorescence decrease as in (112, 113) and/or by displacing bound 11-(dansylamino)-undecanoic acid (DAUDA) as in (26, 114). Cis-parinaroyl CoA (cisPnCoA) (112, 113) and DAUDA (26, 114) are only weakly fluorescent in buffer, but its fluorescence increased dramatically upon binding to FABP1. The complex of FABP1 (500nM) with the respective fluorescent ligand (500nM) in 10mM phosphate buffer was titrated with displacing ligand: Panel A. Rat FABP1/cis-parinaroyl-CoA with AEA (0–6μM) or 2-AG (0–2μM); Panel B: Human FABP1/cis-parinaroyl-CoA or FABP1/DAUDA with AEA (0–6μM); Panel C: Rat FABP1/cis-parinaroyl-CoA with cannabidiol (0–10μM). CisPnCoA fluorescence (Ex 304nm, Em 425nm) decrease was recorded at 24°C using a Varian Cary Eclipse Fluorescence Spectrophotometer (Varian, Inc., Palo Alto, CA). Fluorescence signals from cis-PnCoA with increasing amount of displacing ligand were used as background and subtracted. EC50 was obtained from curve fitting of the displacement curves. Ki was calculated according to the equation EC50/[cisPnCoA]total = Ki/Kd where [cisPnCoA]total = 500nM and Kd = 228 ± 18nM is the dissociation constant of cis-PnCoA binding to rat L-FABP. DAUDA fluorescence (Ex 330nm, Em 510nm) decrease was recorded at 24°C. Fluorescence signals from DAUDA with increasing amount of AEA were used as background and subtracted. Kd and Ki were determined similarly (data not shown). Kis calculated from multiple displacement curves were presented as mean ± SE (n=3).
In contrast, human FABP1 bound AEA in a different manner from that observed with the rat FABP1. While AEA did not displace human FABP1 bound cis-parinaroyl CoA (Fig. 1B), nevertheless AEA did displace another fluorescent ligand, i.e. 11-(dansylamino)-undecanoic acid (DAUDA), which was bound by human FABP1 albeit more weakly than cis-parinaroyl CoA (Fig 1B). Rat FABP1 also binds DAUDA with affinities in a similar range as does human FABP1 (26, 114). Taken together, these findings indicate that although both human and rat FABP1 bind AEA, they likely differ significantly with regards to affinity and localization of the bound AEA within the respective binding sites.
Similarly, little is known about how the equally hydrophobic, highly-membrane associated phytocannabinoids and synthetic cannabinoids are taken up and traffic within hepatocytes from sites of uptake to intracellular sites for metabolism or secretion at the bile canaliculus. The cis-parinaroyl-CoA displacement assay suggested that FABP1 may also serve this function (89). Cannabidiol displaced rat FABP1-bound cis-parinaroyl-CoA with Kd of 0.58±0.06 μM (Fig. 1C). Rat FABP1 also exhibited high affinity for the psychoactive tetrahydrocannabinol and a variety of synthetic cannabinoid agonists and antagonists (89). Taken together with FABP1’s very high cytosolic concentration (5, 6), these findings suggest FABP1 as a major ‘chaperone’ protein in the liver. Further, the high affinity of FABP1 for cannabidiol suggests that FABP1 may contribute significantly to the very high (90%) first-pass removal of oral cannabinoid (115–119).
FABP1 also binds non-ARA containing potentiating ‘entourage’ (EC*) N-acylethanolamides and 2-monoacylglycerides. Although N-oleoylethanolamide (OEA) and N-palmitoylethanolamide (PEA) do not directly bind/activate CB receptors, they nevertheless act as ‘entourage’ lipids that potentiate AEA (and/or 2-AG) activity by increasing their affinities for CB receptors or decreasing their enzymatic degradation (120–125). Rat FABP1 bound these ‘entourage’ NAE (OEA, PEA) and 2-MG (2-OG, 2-PG) with similar or weaker affinities than for AEA and 2-AG (89). The observation that rat FABP1 binds 2-OG confirms earlier NMR, Lipidex 1000 radioligand competition, and Tyr quenching assays (126, 127). Further, studies with FABP1 gene ablated mice showed that murine FABP1 is the major 3H-2-OG binding protein in mouse liver cytosol (126). In contrast, nothing is known regarding the human FABP1 interaction with such potentiating ‘entourage’ EC* ligands.
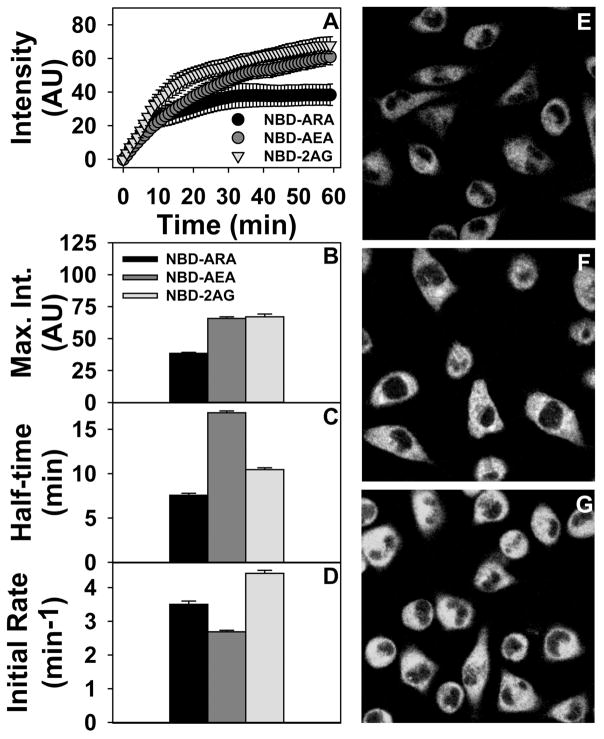
Real-time imaging established that the NBD-labeled AEA and 2-AG as the first potentially useful endocannabinoid analogues for visualizing the uptake, intracellular trafficking, and targeting of these molecules by living cells. L-cells take up NBD-ARA, NBD-AEA and NBD-2AG as shown by representative images (Fig 3E, F, G). Analysis of multiple cells over time revealed showed biphasic uptake curves approaching a maximum for each probe (Fig. 3A). While the initial rates of uptake of these probes was in the order NBD-2-AG > NBD-ARA > NBD-AEA (Fig 3D), the overall half-time of uptake of for both NBD-AEA and NBD-2-AG was longer than that of NBD-ARA (Fig 3C). Furthermore, maximal uptake of NBD-AEA and NBD-2-AG was at least 1.6-fold higher than that of NBD-ARA (Fig. 3B). It should be noted that the half-time of NBD-ARA uptake (Fig. 3C) was in the same range as that of radiolabeled and our earlier A5C fluorescent ARA analogue (47, 128–130)—suggesting that the relative differences in kinetics between NBD-AEA and NBD-2-AG versus NBD-ARA uptake accurately reflect those of unlabeled ARA. These novel analogues now allow real-time determination of the impact of: i) FABP1 overexpression on AEA and 2-AG uptake in murine L-cells; ii) FABP1 gene ablation on hepatic uptake of AEA and 2-AG in vivo or by cultured primary hepatocytes; iii) human T94A variant on uptake of AEA and 2-AG in cultured primary human hepatocytes; iv) FABP1 on both hepatic and brain uptake of ARA, AEA and 2-AG.
Figure 3. Cellular uptake of NBD-ARA, AEA, and 2-AG.
NBD-ARA, NBD-AEA, and NBD-2-AG uptake (A), maximal uptake (B), half-time of uptake (C) and initial rate of uptake (D) by L-cell were measured by confocal imaging in L-cells similarly as for NBD-18:0 (46, 53). Values represent the mean ± SEM, n=20. Panels E, F, and G show representative fluorescent images of NBD-ARA, NBD-AEA, and NBD-2-AG uptake at 60 min, respectively.
HUMAN AND MURINE FABP1 IMPACT LIVER ENDOCANNABINOIDS
The functional significance of endocannabinoids and the CB1 receptor in liver was first established by Kunos et al (101, 131–133). Hepatic CB1 (and CB2) are markedly upregulated in non-alcoholic liver disease (NAFLD) (101–103), while CB1 is upregulated in alcoholic liver disease (AFLD) (102, 104) and in response to high-fat diet-induced obesity (102, 104). Concomitantly, hepatic AEA and 2-AG levels are also elevated in NAFLD, while 2-AG (but not AEA) is elevated in AFLD, and AEA (but not 2-AG) is elevated in response to high-fat diet (101–104). Despite these advances, little is known about hepatic factors contributing to these alterations in the hepatic endocannabinoid system. One possible candidate protein is the hepatic FABP1 which not only binds AEA and 2-AG (Figs 1,2) but loss of FABP1 also elicits hepatic lipid accumulation in vivo (50, 78–81) and in hepatocytes (52–54, 80).
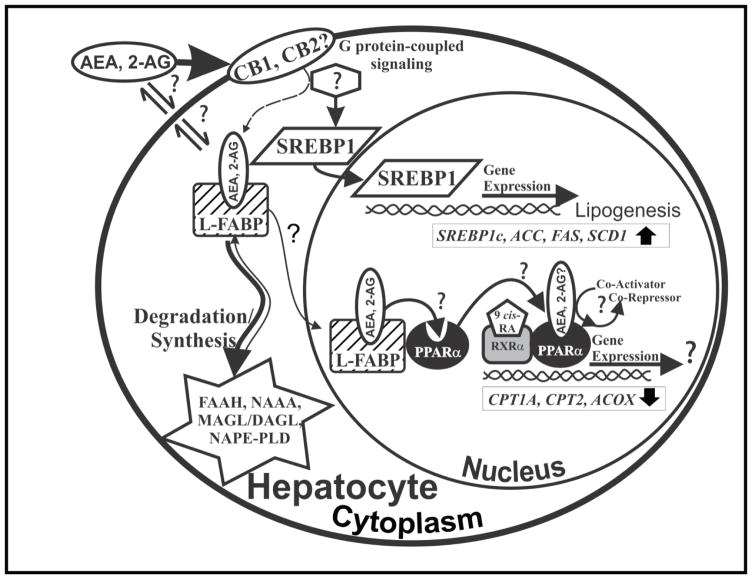
Indeed, FABP1 gene ablation markedly increases hepatic levels of arachidonic acid containing endocannabinoids (EC) such as AEA and 2-AG (Table 1) (89). This increase in hepatic AEA and 2-AG may contribute to hepatic TAG accumulation by a SREBP1 mediated mechanism (Fig 4) (2, 39, 59, 80, 86, 87). CB1 receptor agonists induce SREBP1 that in turn induces transcription of lipogenic enzymes de novo such as acetyl-CoA carboxylase and fatty acid synthase (Fig 4) (134). Concomitantly CB1 agonists reduce LCFA oxidation by inhibiting adenylate cyclase and AMPK activity (134). Alternately loss of FABP1 may reduce transfer of AEA into the nucleus wherein AEA would normally bind to enhances PPARα activation (135) (Fig. 4). It is therefore important to extend these findings toward the human FABP1 and its role regulating the endocannabinoid system and thereby fatty liver disease.
Table 1. Effect of FABP1 gene ablation on N-acylethanolamide and 2-monoacylglycerol levels in male mouse liver.
Male C57BL/6N wild-type (WT) and FABP1 gene ablated mice (8 wk old) placed on a phytol-free, phytoestrogen-free control chow diet for 4 weeks, fasted overnight, and then livers removed/flash frozen and stored at −80°C as in (89).
| Endocannabinoid | Wild-type (WT) | FABP1 Gene Ablated |
|---|---|---|
| N-acylethanolamides (pmol/g Liver) | ||
| AEA | 13 ± 1 | 20 ± 2* |
| OEA | 34 ± 7 | 20 ± 3* |
| PEA | 60 ± 10 | 11 ± 1* |
| 2-monoacylglycerols (nmol/g Liver) | ||
| 2-AG | 0.16 ± 0.02 | 0.37 ± 0.04* |
| 2-OG | 1.4 ± 0.2 | 2.6 ± 0.2* |
| 2-PG | 0.18 ± 0.03 | 0.16 ± 0.01 |
N-acylethanolamides were extracted and analyzed by LC/MS using deuterated internal standards (Cayman Chemical) as in (89, 249) to determine levels of: AEA, arachidonoyl ethanolamide; OEA, oleoyl ethanolamide; PEA, palmitoyl ethanolamide. 2-Monoacylglycerides were extracted similarly, but deuterated internal standards (Cayman Chemical) and LC/MS solvent conditions were modified as in (89, 108) to quantitate liver determine: 2-AG, 2-arachidonoyl monoacylglycerol; 2-OG, 2-oleoyl monoacylglycerol; 2-PG, 2-palmitoyl monoacylglycerol. Values represent the mean ± SEM, n=6–7.
p<0.05, one-way ANOVA for FABP1 gene ablated vs wild-type.
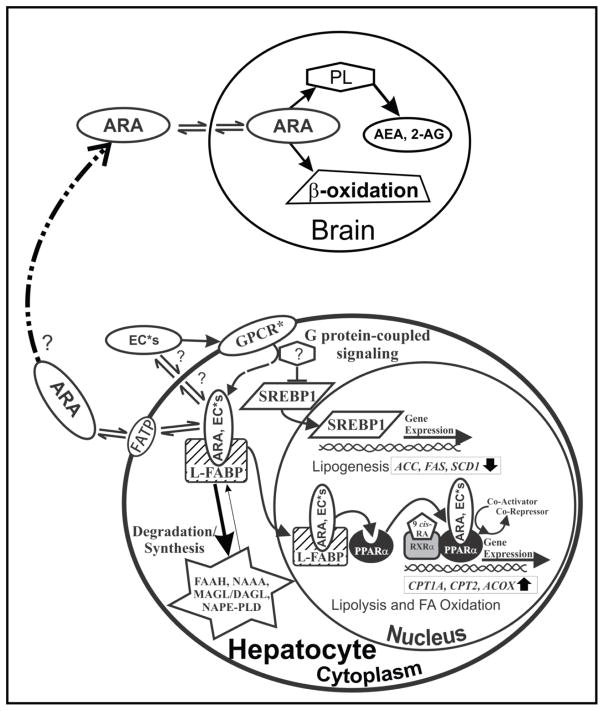
Figure 4. Schematic of FABP1’s role in endocannabinoid (AEA, 2-AG) trafficking and function in primary hepatocytes.
By binding anandamide (AEA) and 2-arachidonoylglycerol (2-AG), FABP1 may influence key aspects of the hepatic endocannabinoid system: A). FABP1 may facilitate AEA and 2-AG release/solubilization into the cytosol after their enzymatic cleavage/synthesis from plasma membrane phospholipids by N-acyl phosphatidylethanolamine phospholipase D (NAPE-PLD) and diacylglycerol lipase α and β (DAGLα and DAGLβ). FABP1 may or may not facilitate bound AEA and 2-AG for transport/efflux across the plasma membrane and activation of CB receptors on the exofacial leaflet and/or intracellular sites for degradation/hydrolysis. FABP1 is known to enhance the cytosolic transport of other bound ligands (5, 46, 52, 141, 251). B). FABP1 may facilitate the re-uptake of AEA and 2-AG from the plasma membrane after these lipophilic ligands cross the plasma membrane by diffusion or via G protein-coupled cannabinoid receptor 1 (CB1). Reuptake of 2-AG may also occur via G protein-coupled endocannabinoid receptor (CB2). CB2 is expressed only in embryonic liver and in diseased conditions such as fatty liver (252, 253). CB1 and CB2 activation has been linked to diet-induced hepatic steatosis, primary biliary cirrhosis, chronic hepatitis, and alcoholic liver (101). CB1 activation in mice enhances lipogenesis through the sterol regulatory element binding protein-1c (SREBP1c) pathway which induces transcription of multiple genes in lipogenesis such as SREBP1c itself, acyl CoA carboxylase (ACC), fatty acid synthase (FAS), and stearoyl CoA desaturase (SCD1) (132). C). Upon FABP1-mediated release of plasma membrane and/or CB receptor bound AEA or 2-AG into the cytosol, FABP1 may transport the bound AEA (and 2-AG?) into the nucleus for PPARα activation. Although anandamide has been shown to bind PPARα and enhance PPARα activation (135), liver fat accumulation in hepatic steatosis results in decreased LCFA oxidation—likely through saturation and/or inhibition of the PPARα pathway by the increasing fat load or damage by inflammation (101). D) FABP1 may also transport the bound AEA to the degradative enzyme fatty acid amide hydrolase (FAAH) localized with smooth endoplasmic reticulum, mitochondria, lipid droplets, and more rarely at the cell membrane (142–144) by a process analogous to that established for other FABP family members (FABP3,5,7) in brain (106–110, 144). In human liver FABP1 may also transport AEA to/from lysosomes, where it is degraded by N-acylethanolamide acid amide hydrolase (NAAA) (143, 145). Finally, FABP1 may transport bound 2-AG for degradation by monoacylglycerol lipase (MAGL), an enzyme found at lower levels in liver than brain or other tissues where it is localized diffusely in cytosol and less so in membranes without overall compartmental preference (142).
FABP1 gene ablation also increases hepatic levels of non-arachidonic acid containing N-acylethanolamides or 2-monoacylglycerols (EC*). Hepatic levels of OEA and PEA (34±7 and 60±10 pmol/g liver, respectively) in male wild-type C57BL/6N mice are about 3–5 fold higher than that of AEA (Table 1) (89). FABP1 gene ablation nearly doubled the hepatic level of the even more highly prevalent (nmol/g vs pmol/g) 2-oleoyl-glycerol (2-OG) which is a finding not compensated for by decreased expression of the much less prevalent OEA and PEA (Table 1) (89). The loss of FABP1 could also contribute to hepatic TAG accumulation by its impact on hepatic EC* levels. By reducing OEA, this would increase the SREBP1 pathway to increase lipogenesis while decreasing lipolysis and fatty acid oxidation through the PPARα pathway, likely through GPR119 (136–140). FABP1 gene ablation would reduce cotransport of EC* ligands (OEA, PEA, 2-OG, 2-PG) into the nucleus for transfer to and activation of PPARα or alternately through G protein-coupled receptors other than CB1/CB2 (GPCR*s) (Fig. 5).
Figure 5. Schematic of FABP1’s role in arachidonic acid (ARA) and EC* (OEA, PEA, 2-OG, 2-PG) targeting/trafficking.
A). FABP1, not detectable in brain, binds and enhances uptake of arachidonic acid (ARA) translocated by fatty acid translocase protein (FATP) in the plasma membrane in cultured cells and likely also in hepatocytes. Within hepatocytes the FABP1 may facilitate transport/targeting of bound ARA to endoplasmic reticulum for incorporation into phospholipids from which AEA and 2-AG are subsequently derived. B) Alternately, FABP1 mediated hepatic uptake may diminish plasma availability for ARA for uptake and conversion into phospholipids from which AEA and 2-AG are derived in brain. Most brain ARA is derived from plasma and rapid hepatic ARA clearance accounts for nearly 50% of ARA removal from the blood. C) By binding other non-ARA containing N-acylethanolamides and 2-monoacylglycerols (EC*s) such as oleoylethanolamide (OEA), palmitoylethanolamide(PEA), 2-oleoylglycerol (2-OG), or 2-palmitoylglycerol (2-PG), FABP1 may influence their synthesis release at the endoplasmic reticulum, transport for efflux at the plasma membrane, re-uptake from the plasma membrane, and targeting for degradation in the endoplasmic reticulum analogous to those of AEA and 2-AG (Fig. 4). D) By binding other EC*s, FABP1 may exert effects on the SREBP1 lipogenic and PPARα oxidative pathways opposite to those of AEA and 2-AG (Fig. 4). For example, the EC* ligands may exert their effects through G protein-coupled receptors other than CB1/CB2 (GPCR*s). For instance, OEA is known to suppress the SREBP1 pathway to reduce lipogenesis while enhancing lipolysis and fatty acid oxidation through the PPARα pathway, likely through activation of GPR119 (136–140, 169). Conversely, FABP1 transports bound ARA and/or EC* into the nucleus for interaction with and induction of PPARα.
Although the non-arachidonic acid containing EC* have no agonist activity at CB1 or CB2 receptors, they are nevertheless bioactive (120). This has led to the suggestion that EC* may directly activate as yet unknown receptor(s) (120). Alternately, it has been proposed that the EC* may act indirectly by enhancing the action of endogenous AEA or 2-AG by increasing their affinity for CB receptors and/or decreasing AEA or 2-AG degradation—possibilities termed ‘entourage’ effects (120, 120–125). However, there is not universal agreement with the ‘entourage’ idea since it is clear that OEA and PEA have alternative targets. For the sake of simplicity in this review, however, the term potentiating ‘entourage’ EC* will be used.
MECHANISM(S) WHEREBY FABP1 IMPACTS LIVER ENDOCANNABINOIDS
While the mechanism(s) whereby FABP1 gene ablation raises hepatic levels of EC (AEA, 2-AG) and some EC* (2-OG) remains to be determined, these increases were not associated with compensatory changes in protein levels of enzymes, receptors, or ‘chaperones’ in the hepatic endocannabinoid system (89). One possibility is based on the fact that FABP1’s high affinity for AEA and 2-AG which suggests potential role(s) for FABP1 in AEA and 2-AG reuptake for ‘chaperoning’ and targeting to degradative enzymes (Fig. 4)—analogous to roles exhibited by brain FABPs 3,5, and 7 (106, 107, 110). Consistent with this possibility increased level of FABP1 enhances (5, 46, 55, 141), while FABP1 gene ablation markedly decreases (52), the cytosolic transport/diffusion of other bound ligands (e.g. NBD-stearic acid). Similar considerations may be proposed for the FABP1 gene ablation-induced increase in hepatic 2-OG (Fig 5) since rat FABP1 has been shown to increase by 7-fold the transfer of a fluorescent 2-OG to model membrane phospholipid vesicles in vitro (126). Thus, loss of FABP1 would reduce AEA, 2-AG, and 2-OG ‘chaperoning’ towards hydrolytic/degradative enzymes such as fatty acid amide hydrolase (FAAH) localized in endoplasmic reticulum, mitochondria, and lipid droplets (142–144), N-acylethanolamide acid amide hydrolase (NAAA) in lysosomes (human but not rodent liver) (143, 145), and monoacylglycerol lipase (MAGL) localized primarily in cytosol at much lower levels in liver than other tissues (142). Less clear is the role of putative as yet to be identified plasma membrane AEA and 2-AG binding/translocase proteins and/or contributions by endocytic reuptake of AEA or 2-AG bound to CB receptors. Finally, it must be noted that the above pathway does not appear account for why FABP1 decreased levels of the more prevalent OEA and PEA or did not change levels of the less prevalent 2-PG. However, a possible explanation for the discrepancy my lie in the finding that other ligands which also bind to FABP1 selectively alter or do not alter its conformation in response to ligand binding (12, 37). In turn ligand-dependent alterations in FABP1 conformation may, depending on the specific ligand, either facilitate or prevent or have no impact on FABP1 intracellular redistribution and/or interaction with other proteins (e.g. PPARα, CPT1) (12, 37, 58, 63).
MURINE FABP IMPACTS BRAIN ENDOCANNABINOID SYSTEM
The presence of cytosolic fatty acid binding proteins (FABP 3, 5, 7), established over 20 years ago (146–153), led to the recent pioneering studies of Deutsch and coworkers identifying these FABPs as endocannabinoid (AEA, 2-AG) ‘chaperones’ for reuptake/intracellular targeting to endoplasmic reticulum for hydrolysis/degradation (105, 106, 108, 110). Ablating or inhibiting FABPs present in brain cytosol (especially FABP3) reduces brain ARA uptake (required for AEA formation)/AEA degradation (154–158). However, it is not completely clear if the impact of ablating or inhibiting these ‘brain’ FABPs is attributable only to their loss/inhibition in brain. For example, FABP3 is also highly prevalent in heart and skeletal muscle, while FABP5 is also found in epidermal cells, mammary gland, liver, kidney, lung, and adipose tissue (3, 159, 160). Likewise, the chemical BMS309403 inhibits the FABP3 and 5 localized in brain and these other tissues as well as FABP4 found in adipose tissue (110). Interestingly, FABP3 gene ablation also diminishes heart uptake of ARA, the precursor of ARA-containing phospholipids from which AEA and 2-AG are synthesized (161). The fact that the liver FABP1 binds the arachidonic acid (ARA) (see above), but is not expressed or detected in brain (156, 162, 163), offers the opportunity to resolve the impact of this extra-CNS FABP on the brain endocannabinoid system (Fig. 5).
LC/MS analysis of brain endocannabinoids of male C57BL/6N mice either expressing or ablated in the liver FABP1 revealed that indeed FABP1 has a role in regulating brain endocannabinoid levels (88). FABP1 gene ablation markedly increased brain levels of both AEA and 2-AG (Table 2). Concomitantly, FABP1 ablation even more markedly increased brain levels of all the potentiating ‘entourage’ N-acylethanolamides (OEA, PEA) and 2-monoacylglycerols (2-OG, 2-PG) (Table 2). Again, these increased levels of endocannabinoids (AEA, 2-AG) and their highly prevalent potentiating ‘entourage’ lipids (OEA, PEA, 2-OG, 2-PG) were not due to altered brain protein levels of brain CB1 receptors or enzymes in endocannabinoid synthesis/degradation.
Table 2.
Effect of FABP1 gene ablation on N-acylethanolamide or 2-monoacylglycerol levels in male mouse brain.
All conditions were as in Table 1, except that LC-MS was used to identify and quantify each N-acylethanolamide or 2-monoacylglycerol as in (88, 250).
| Endocannabinoid | Wild-type (WT) | FABP1 Gene Ablated |
|---|---|---|
| N-acylethanolamides (pmol/g Brain) | ||
| AEA | 13 ± 2 | 25 ± 2* |
| OEA | 70 ± 10 | 200 ± 20* |
| PEA | 74 ± 9 | 130 ± 20* |
| 2-monoacylglycerols (nmol/g Brain) | ||
| 2-AG | 14 ± 1 | 46 ± 3* |
| 2-OG | 4.8 ± 0.4 | 19 ± 1* |
| 2-PG | 6.2 ± 0.8 | 9.5 ± 0.4* |
AEA, arachidonoyl ethanolamide; OEA, oleoyl ethanolamide; PEA, palmitoyl ethanolamide, 2-AG, 2-arachidonoyl monoacylglycerol; 2-OG, 2-oleoyl monoacylglycerol; 2-PG, 2-palmitoyl monoacylglycerol. Results are presented as pmol lipid/g brain for N-acylethanolamides and as nmol lipid/g brain for 2-monoacylglycerols (mean ± SEM, n = 6–7);
p < 0.05 for FABP1 gene ablated vs wild-type (WT).
While the mechanism(s) whereby liver FABP1 gene ablation increases brain AEA and 2-AG levels remains to be resolved, one possibility may lie in the role of FABP1 in hepatic clearance of ARA from plasma to reduce bioavailability for ARA uptake by the brain (Fig. 5) (88). This mechanism is based on the fact that the brain ARA-containing phospholipids (from which AEA and 2-AG are synthesized) are largely derived from ARA taken up from plasma (164, 165). Yet, ARA availability for brain uptake is significantly diminished by high hepatic clearance rate (166–168). Human and rat FABP1 have high affinity for ARA as well as 18:2, n-6 which can be metabolized to ARA in liver, but much less so in brain (18, 37, 90). Overexpressing FABP1 in mouse L-cell fibroblasts increased ARA and ARA analogue uptake (43, 47, 91, 92) more than that of other LCFA (43, 44, 46, 47, 50, 53, 91). Both rodent and human liver cytosolic levels of FABP1 are very high (2–10% of cytosolic protein; 0.1–1.0mM) (2, 5, 6, 37). In fact the hepatic cytosol FABP1 protein concentration is nearly 20 to 100-fold higher than that of all three FABPs (i.e. FABP3, 5, 7) in brain cytosol (146–153). This suggests the liver may very effectively compete with brain for ARA uptake from plasma (Fig. 5). Indeed, nearly half of plasma ARA undergoes hepatic clearance which significantly diminishesavailability for brain uptake (166–168).
Whether a similar explanation may hold for the non-arachidonic acid derived ‘entourage’ EC* (PEA, OEA, 2-OG, 2-PG) is less clear since brain does not need to derive the palmitic acid and oleic acid from plasma for synthesis of palmitic acid and oleic acid containing phospholipids from which the above ‘entourage’ EC* are derived. It is important to note, however, that FABP1 does also bind palmitic and oleic acids, albeit with lower affinity than ARA (18, 47, 90). Furthermore, the uptake of these and other non-ARA fatty acids is increased in FABP1 overexpressing L-cell fibroblasts and correlates directly with FABP1 level in human HepG2 liver cells (5, 43, 44, 46, 48, 92). Conversely, FABP1 gene ablation decreases non-ARA uptake by cultured primary mouse hepatocytes (52–54) and in vivo (50, 79, 80). Full testing of this hypothetical scheme (Fig. 5) and differentiating these possibilities will require future studies with iv injected labeled ARA, palmitic acid, and oleic acid.
Much remains to be done with regards to potential functional consequences of FABP1 on the brain endocannabinoid system. The brain endocannabinoids (AEA, 2-arachidonoylglycerol), the cannabinoid receptors, and the potentiating ‘entourage’ N-acylethanolamides and 2-monoacylglycerols constitute a novel system for modulating behavior, pain and inflammation, food intake, and weight gain (94–100). Since high endocannabinoid levels produce analgesia (108), the FABP1 gene ablation-induced increase in brain AEA level may decrease pain sensitivity (Fig 4). In contrast, the non-ARA containing EC* ligands have opposing influences on food intake and weight gain by differentially impacting LCFA synthesis de novo versus oxidation (Fig 5). For example, increased level of the cannabinoid receptor-1 (CB1) agonist AEA increases food intake and LCFA synthesis de novo by a SREBP1 mediated mechanism (102) (Fig 4), while increased OEA decreases food intake and weight gain by a mechanism involving induction of PPARα transcription of LCFA oxidative genes (134, 137, 169) (Fig. 5). FABP1 gene ablated mice showed unaltered or slightly increased food intake (39, 57, 81, 86, 170, 171)—suggesting the opposing effects of AEA and OEA on food intake were offset since both were increased in parallel by FABP1 gene ablation. The net effect of FABP1 gene ablation mediated changes in brain endocannabinoid levels on other brain functions remains to be elucidated. Since ablation or inhibition of FABPs 3,5, and 7 in brain is known to markedly impact such parameters (108, 172), whether FABP1 gene ablation alters behavior, pain and inflammation remains an intriguing possibility.
FABP1 In Human Health: Impact of the human FABP1 T94A Variant
Increasing evidence points to a role for FABP1 in human health. Hepatic FABP1 level is environmentally responsive, e.g. high fat diet, chronic alcoholism, sex, PPARα agonists. While to date there have been no reports of human genetic variants resulting in complete loss of FABP1, a SNP in the FABP1 gene promoter region is associated with decreased FABP1 and decreased serum TAG (173). Conversely, a SNP in the FABP1 coding region is associated with increased FABP1 level, altered FABP1 conformation/function, human dyslipidemias and non-alcoholic fatty liver disease (NAFLD) (63, 174–176, 176, 177). .
FABP1 is upregulated in both human and rodent models of non-alcoholic fatty liver diseases (NAFLD) (178–182). Upregulation of FABP1 may mitigate the deleterious effects of high LCFA load by: i) preventing LCFA lipotoxicity through binding oxidized and reactive LCFA species (182–189); and ii) partitioning of potentially lipotoxic LCFAs into stable TAG in vivo (190). However, as FABP1 becomes depleted, NAFLD progresses to non-alcoholic steatohepatitis (NASH) (180, 183–187). The human FABP1 directly interacts with human PPARα to facilitate ligand transfer/activation of PPARα transcription of genes in LCFA metabolism, especially oxidation (12, 17, 37, 63, 191). Dysregulation of PPARα is associated with diabetes, cardiovascular disease (CVD), obesity, and NAFLD (182, 191, 192). A human PPARα-V227A variant exacerbates alcohol-induced plasma and liver lipid abnormalities (193, 194).
Interest in the role of the human FABP1 in health has markedly increased since the discovery of several SNPs in the human FABP1 gene. For example, a common polymorphism in the human FABP1 gene promoter region (rs2919872) leads to decreased FABP1 promoter transcriptional activity, decreased FABP1, and decreased plasma TAG in human subjects (173). However, the impact of this single nucleotide polymorphism (SNP) in the human FABP1 promoter region on hepatic TAG accumulation and NAFLD has not been reported. In contrast, a SNP in the coding region of human FABP1 results in a T94A substitution—one of the most prevalent polymorphisms in the FABP family, occurring with 26–38% minor allele frequency and 8.3±1.9% homozygous in the human population (MAF for 1000 genomes in NCBI dbSNP database; ALFRED database) (175–177, 195–198). Impact on the FABP1 T94A variant on whole phenotype, however, is somewhat variable. Several studies correlated T94A variant expression with decreased body mass index (BMI) and waist circumference (174, 175), no change in BMI (195, 196), or increased BMI (176). This variation in whole body phenotype may be associated with the genetic diversity among the different human populations studied. Nevertheless, the expression of the FABP1 T94A variant is associated with clinical dyslipidemias including elevated plasma TAG (174, 175), increased LDL cholesterol (175, 176), and atherothrombotic cerebral infarction (177). With regards to liver phenotype, expression of the human FABP1 T94A variant also elicits non-alcoholic fatty liver disease (NAFLD) (176) and hepatic TAG accumulation concomitant with increased total FABP1 level in cultured primary human hepatocytes (63). Interestingly, the lipid lowering drug fenofibrate binds to both murine and human FABP1 to alter FABP1 conformation and thereby interaction with and activation of PPARα transcriptional activity (12, 17, 41, 42, 63). Fenofibrate, the most commonly prescribed fibrate in the US and Canada (199), lowers serum lipids in both wild-type FABP1 and T94A variant expressing human subjects, but in the FABP1 T94A variant expressers levels are not lowered to baseline (174). Until recently, however, the mechanism(s) whereby this single amino acid substitution in human FABP1 alters its function and responsiveness to fibrates or other drugs remained unclear. While it was initially thought that the T94A substitution results in complete-loss-of-function (i.e. ligand binding ability) analogous to L-FABP gene ablation (49), the following sections demonstrate that the human FABP1 T94A substitution results in an altered structure, structural response to ligand binding, and function rather than loss-of-function.
Molecular Characterization of the Human FABP1 T94A Variant
All previous structures of the recombinant human FABP1 protein were fortuitously derived from cDNAs that each encoded the human WT T94T L-FABP (200–202). In contrast, a commercially available human FABP1 cDNA (OriGene Technologies, Rockville, MD) actually encodes for the human FABP1 T94A variant mutant rather than the wild-type (12). While the number of clones is limited, nevertheless one out of four (i.e. 25%) encoding the FABP1 T94A variant is consistent with the high frequency of the FABP1 T94A variant in the human population (26–38% minor allele freq.; 8.3±1.9% homozygous; MAF for 1000 genomes in NCBI dbSNP database; ALFRED database) (175–177, 195–198). Circular dichroism reveals that the secondary structures of the recombinant human WT FABP1 T94T (obtained by site-directed-mutagenesis of the FABP1 T94A variant cDNA) and the FABP1 T94A variant proteins show key significant differences (12). The non-conservative substitution of a medium sized, uncharged, polar T residue by a smaller, nonpolar, aliphatic A residue at position 94 significantly increases α-helical structure, decreases β-sheet structure, decreases thermal stability, but conversely increases resistance to unfolding by urea. Temperature and chemical denaturation access different aspects of protein stability (12, 203, 204). Thus, the human FABP1 T94A variant represents an altered structure mutation.
While the T94A substitution did not impact the specificity of the human FABP1 protein for a broad variety of ligands, it nevertheless alters affinities for several important ligands (12, 37, 38, 63). T94A substitution does not or only slightly alters FABP1 affinities for long chain fatty acids (saturated, monounsaturated, or polyunsaturated), oleoyl-CoA, lysophosphatidic acid, palmitoyl-oleoyl-phosphatidic acid, n-3 polyunsaturated LCFA (EPA, DHA) PPARα agonists or fibrate PPARα agonists (fenofibrate, fenofibric acid). On the other hand, T94A substitution increases affinity of human FABP1 for cholesterol by 3-fold as demonstrated with a NBD-cholesterol fluorescence binding assay and by cholesterol isothermal titration microcalorimetry (ITC) (38). LCFA binding alters the secondary structure of the human FABP1 WT protein, generally increasing the proportion of α-helical and unordered structures while decreasing that of β-sheet (12, 37, 70). T94A substitution markedly attenuated the ability of the LCFA ligands to alter human FABP1 secondary structure. Likewise, while fenofibric acid (the active metabolite of fenofibrate) also increases the α-helical and unordered structure of human FABP1 WT protein. T94A substitution significantly diminished this response. Fibrate-induced conformational change in human FABP1 is an essential component for human FABP1/PPARα interaction and potentially function (191). Thus, the altered structure of the human FABP1 T94A variant results in an altered ligand-affinity functional mutation rather than loss-of-function.
Functional Impact of the Human FABP1 T94A Variant Expression on Lipidic Ligand Uptake and Metabolism in Cultured Primary Human Hepatocytes
Expression of the human FABP1 T94A variant differentially impacts fatty acid and cholesterol uptake in cultured primary human hepatocytes. FABP1 T94A variant expressing cultured primary human hepatocytes exhibited decreased uptake of poorly metabolizable (fluorescent NBD-stearic acid) and metabolizable ([9,10–3H]-stearic acid) long chain fatty acid (63). Similarly, uptake of radiolabeled palmitic acid by transfected ‘Chang’ liver cells was increased by overexpression of human WT FABP1, but not vector with T94A variant or empty vector (49). Thus, although the affinity of human FABP1 T94A variant for LCFA did not differ from that of the human WT FABP1, nevertheless the T94A substitution decreased LCFA uptake. While the molecular basis for the reduced LCFA uptake exhibited by T94A expressing hepatocytes and transfected cells is not known, it was not attributed to decreased levels of plasma membrane and other intracellular membrane LCFA transport protein. Instead, the finding that mouse FABP1 directly interacts with the plasma membrane fatty acid translocase protein-5 (FATP5) in cultured primary mouse hepatocytes (53) suggests that the altered structure and/or a attenuated conformational response changes FABP1 T94A response to ligand binding (12, 37) may decrease FABP1 T94A interaction with FATP5 and thereby reduce LCFA uptake.
In contrast, human FABP1 T94A substitution oppositely impacts lipoprotein-mediated cholesterol uptake in cultured primary human hepatocytes. Unlike LCFA taken up via membrane fatty acid transport proteins (FATP), lipoprotein cholesterol is taken up via hepatocyte cell surface receptors for LDL and HDL (38). T94A substitution enhances lipoprotein mediated cholesterol uptake which is consistent with its 3-fold higher affinity for cholesterol (38). FABP1 T94A substitution increased cultured primary human hepatocyte uptake of NBD-cholesterol from NBD-cholesterol labeled HDL much more than from LDL (38). Likewise, human FABP1 T94A variant expression in cultured primary human hepatocytes or overexpression of human FABP1 T94A variant (but not human WT FABP1) in cultured ‘Chang’ liver cells increases cholesterol accumulation (49, 63). Consistent with these findings, FABP1 T94A variant expressing human subjects exhibit elevated plasma levels of low-density lipoprotein (LDL) cholesterol (175, 176) concomitant with increased CVD (174, 175), and atherothrombotic cerebral infarction (177).
Expression of the human FABP1 T94A variant elicits lipid accumulation in cultured primary human hepatocytes and livers in vivo. Human subjects expressing the FABP1 T94A variant have increased incidence of non-alcoholic fatty liver disease (NAFLD) as evidenced by ultrasound analysis (176). However, while ultrasound visualizes lipid droplets within liver cytoplasm, it does not actually resolve the types of lipids accumulated therein (205). In contrast, chemical analysis of cultured primary human hepatocyte lipids established that FABP1 T94A variant expression induces accumulation of neutral lipid, especially TAG and cholesteryl esters (CE), a process exacerbated by high LCFA load (63). Accumulation of TAG and CE is consistent with NAFLD in human subjects (206). In contrast, overexpressing human FABP1 T94A variant did not alter TAG mass in human ‘Chang liver’ cells (49). The discrepancy between the impact of FABP1 T94A variant expression in cultured primary human hepatocytes versus ‘Chang liver’ cells may lie in the fact that ‘Chang liver’ cells are derived from human cervical cancer cells rather than human liver (83). Taken together, these data were consistent with cultured primary human hepatocytes providing a useful model for examining the mechanism(s) whereby the human FABP1 T94A variant elicits NAFLD.
FABP1 T94A variant expression increases anabolic mechanism(s) to induce neutral lipid accumulation. Neutral lipid (TAG, CE) accumulation in cultured primary human hepatocytes is associated with upregulation of total liver FABP1. This possibility is supported by earlier studies in vitro showing that WT FABP1 stimulates glycerol-3-phosphate acyltransferase (GPAM), the rate limiting enzyme in lipogenesis (74, 77, 207), acyl-CoA cholesterol acyltransferase (ACAT) (208, 209), as well as increases mRNA expression of downstream enzymes in lipogenesis (GPAM, LPIN2) in heterozygotes, decreases mRNA expression of microsomal triglyceride transfer protein (MTTP), increases secretion of ApoB100 but not TAG. TAG accumulation is not due to increased LCFA uptake, lipogenesis de novo (ACC1, FASN), or the alternate monoacylglycerol acyltransferase (MOGAT) pathway in lipogenesis. Thus, T94A-induced neutral lipid accumulation is associated, at least in part, with increased total FABP1 protein for stimulating neutral lipid synthesis, but less able to load neutral lipids on apoB for secretion (63).
Conversely, FABP1 T94A variant expression impairs catabolic mechanism(s) that would reduce neutral lipid accumulation. Increased neutral lipid accumulation in FABP1 T94A variant expressing human hepatocytes is also attributed at least in part to decreased LCFA β–oxidation (63). T94A substitution decreases β–oxidation of [9,10–3H]-stearic acid by 70% and 40% in heterozygotes and homozygous T94A hepatocytes (63) which is consistent with the decreased β-oxidation in development of NAFLD (205). Impaired LCFA β-oxidation is not due to reduced transcription of LCFA β-oxidative enzymes (CPT1A, CPT2, ACOX1) whose mRNA levels actually increased (63). Instead decreased LCFA β–oxidation was associated with decreased translation of the rate limiting enzyme CPT1A mRNA into CPT1 protein (63). Consistent with this finding, miRNA microarray analysis (Phalanx Biotech Group (San Diego, CA) reveals that T94A increases the level of miR-34a (not shown). miR34a decreases the protein level of CPT1A (rate-limiting enzyme in mitochondrial LCFA β-oxidation) (210) and miR-34a is highly increased in human NAFLD (210, 211).
FABP1 T94A variant expression also impairs ligand-induced PPARα transcription of LCFA β-oxidative enzymes in human hepatocytes (63). While fibrate PPARα agonist efficacy in NAFLD is unclear (212), Very long chain polyunsaturated fatty acids, i.e. VLCn-3PUFA such as eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) reduce lipogenesis de novo (decrease SREBP1c, activate ChREBP) and increase LCFA β-oxidation (activate PPARα) (213–215). T94A impairs fenofibrate and VLCn-3PUFA mediated PPARα transcription of LCFA β-oxidative enzymes (12, 37, 63), suggesting fenofibrate may be less effective in lowering hepatic TAG in T94A subjects. Similarly, fenofibrate is less effective in lowering elevated plasma TAG to basal levels in these T94A variant expressing individuals (174, 175). The decreased ability of the FABP1 T94A variant to mediate ligand activation of PPARα transcriptional activity is attributed at least in part to reduced ability of these ligands to induce redistribution of the FABP1 T94A variant into the nucleus for interaction with and activation of PPARα therein (54). These impaired functions of the T94A variant correlate with FABP1 T94A altered protein structure and reduced protein structural response to ligand binding as noted in the preceding sections.
Does the Human FABP1 T94A Variant Impact the hepatic Endocannabinoid System?
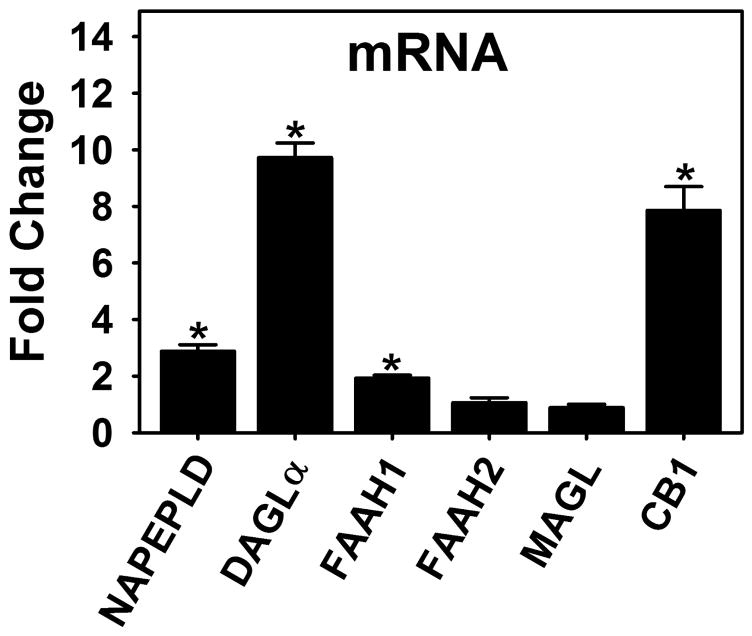
NAFLD in the human FABP1 T94A variant population may at least in part be associated with an altered endocannabinoid system. While underlying causes of NAFLD are unclear (216), genetic variation and environment contribute to the incidence of NAFLD (190, 205, 212, 215, 217–223). Genome wide array studies (GWAS) of NAFLD estimate a 39% heritability of liver lipid accumulation as a continuous trait after controlling for age, gender, race, and BMI (219). The highly prevalent human L-FABP T94A variant (175–177, 195–198) is associated with TAG accumulation in liver (NAFLD) (176), primary hepatocytes (63), and serum (174, 175, 224). Hepatic levels of endocannabinoids and/or receptors (CB1 and/or CB2) of the endocannabinoid system are elevated in NAFLD (101–103), alcoholic liver disease (AFLD) (102, 104), high-fat diet-induced obesity (102, 104), and in response to cannabis with CB1 agonists (e.g. HU-210) (102–104) or CB2 selective agonists (e.g. JWH-133) (102, 104). Expression of the human FABP1 T94A variant markedly induced transcription of key enzymes in AEA and 2-AG synthesis (NAPEPLD, DAGLα) and degradation (FAAH1) as well as their target cannabinoid receptor-1 (CB1) in cultured primary human hepatocytes (Fig 6). These effects are specific since T94A expression has no effect on other AEA and 2-AG degradative enzymes, FAAH2 or MAGL (Fig 6). While the net effect of these opposing influences on AEA and 2-AG levels in the cultured primary human hepatocytes remains to be shown, the 3- and 10-fold increased mRNA levels of enzymes for AEA and 2-AG synthesis (NAPEPLD, DAGLα) concomitant with much smaller or no increase in degradative enzymes (FAAH1, FAAH2, MAGL), suggests increased levels of these endocannabinoids as well as their ‘potentiating’ chaperones. This in turn increases hepatocyte TAG accumulation (63, 176) .
Figure 6. Human FABP1 T94A variant expression induces transcription of enzymes and receptors in the endocannabinoid system.
Primary human hepatocytes as in Fig 6 were cultured as in Fig 6 followed by followed by determination of mRNA levels encoding the human N-acylphosphatidylethanolamide phospholipase-D (NAPE-PLD), diacylglycerol lipase-α (DAGLα), fatty acid amide hydrolase-1 (FARAH1), fatty acid amide hydrolase (FARAH2), monoacylglycerol lipase (MAGL), and cannabinoid receptor-1 (CB1) similarly as for other human mRNA transcripts (213–215). Values are expressed as the fold-change in the ratio of respective mRNA in human FABP1 T94A variant (T94A)/mRNA in wild-type human FABP1. Data are the means ± SEM (n = 7); *p<0.05 for T94A vs WT.
These finding may have important implications for current therapies for NAFLD in human subjects. One approach to reducing TAG levels in NAFLD is to induce PPARα target genes in hepatic fatty acid β-oxidation (63, 176, 225, 226) and lipoprotein metabolism (72, 227–229). In NAFLD individuals, not segregated by T94A or other genotype, fibrate PPARα activators do not uniformly lower TAG and CVD risk (230, 231). Fibrates act by multiple mechanisms, of which many are mediated through PPARα (192, 232–234). For example, fibrates bind and activate PPARα transcription of key genes of LCFA β-oxidation (CPT1, CPT2, ACOX1), LCFA uptake (FATP, L-FABP), and plasma VLDL TAG hydrolysis (LPL) (235–238). PPARα interacts (directly or via cross-talk) with other lipid-regulating genes (HNF4α, LXR, FXR, ANGPTL4); and additional pleiotropic effects. It is important, however, to recognize that fibrates also induce transcription of enzymes involved in LCFA synthesis, desaturation, elongation, and TAG formation de novo (235–238). Fibrates alter endoplasmic reticulum fatty acid composition to enhance cleavage/release of mature SREBP1c which in turn induces nuclear expression of genes involved in LCFA synthesis de novo and TAG formation (235–238). Partitioning of LCFA- or glucose-derived acetyl-CoA toward oxidative versus synthetic pathways will determine the net effect on hepatic TAG and treatment outcome (235, 239, 240). Even if the net effect of fibrate in human FABP1 WT expressers results in more LCFA catabolism than synthesis de novo, however, the available evidence suggests that fibrates may be much less effective in lowering hepatic TAG to treat NAFLD than in lowering serum TAG in T94A expressers (12, 37, 63, 174).
CONCLUSIONS
The discovery of FABP1 nearly forty years ago was followed by elucidation of the rodent FABP1s structure, function in vitro, and more recently physiologically in gene ablated mice. Yet, FABP’s impact on human health is only beginning to be appreciated. Major strides in this regard include the first structural characterizations of the human FABP1, the novel discovery that FABP1 may be the major hepatic endocannabinoid and cannabinoid binding protein, and growing recognition of the highly prevalent human FABP1 T94A variant’s roles in hyperlipidemia and NAFLD. Since NAFLD is also associated with upregulation of hepatic endocannabinoids, it is important to resolve how the T94A variant impacts the endocannabinoid system and transcriptional mechanisms of lipogenesis de novo. This would facilitate development of new nutraceutical approaches to better target elevated TAG in this group, obese subjects and diabetics. One possible candidate is the very long chain n-3 fatty acids (EPA and DHA). While EPA and DHA induce PPARα transcription activity of LCFA oxidative genes, they concomitantly accelerate degradation and/or reduce nuclear distribution of SREBP1c and ChREBP. This decreases SREBP1c (213–215) and ChREBP (241–245) transcription of lipogenic genes which thereby decreases hepatic TAG and NAFLD in human subjects not segregated by FABP1 genotype. Another possibility is suggested by cannabinoid receptor (e.g. CB1) inhibitors that may block the SREBP1c mediated lipogenesis to lower hepatic lipid accumulation (Fig 4). Since FABP1 appears to be involved in the cannabinoid as well as endocannabinoid pathway, it will be important to determine the impact of the FABP1 T94A substitution thereon. In any case, FABP1 (7, 13, 14, 16, 40–42) and the nuclear receptors it impacts, i.e. PPARα (41, 42, 47, 246–248), SREBP1c (213–215), and ChREBP (241–245), continue to be current active therapeutic targets for lipid lowering.
Acknowledgments
This work was supported in part by the US Public Health Service/National Institutes of Health Grant R25 OD016574 (S.M., A.B.K. PD), Merial Veterinary Scholars Program, CVM (S.M., A.B.K. PD), and DA035923 and DA032232 (M.K.).
Abbreviations
- ARA
C20:4n-6 arachidonic acid
- ACC
acetyl-CoA carboxylase
- AEA
N-arachidonoylethanolamide (anandamide)
- ALB
albumin
- ACOX1
acyl-CoA oxidase 1, palmitoyl
- 2-AG
2-arachidonoylglycerol
- CB1
cannabinoid receptor-1
- CB2
cannabinoid receptor-2
- FAS
fatty acid synthase
- cis-PnCoA
cis-parinaroyl-CoA
- CPT1A
carnitine palmitoyl transferase IA, liver
- CPT2
carnitine palmitoyl-CoA transferase II
- DAGLα
diacylglycerol lipase-α
- DAGLβ
diacylglycerol lipase-β
- DAUDA
11-(dansylamino)-undecanoic acid
- DHA
C22:6n-3 docosahexaenoic acid
- EC
arachidonic acid containing endocannabinoids (AEA, 2-AG)
- EC*s
non-ARA containing N-acylethanolamides and 2-monoacylglycerols
- EPA
C20:5n-3 eicosapentaenoic acid
- DGAT2
diacylglycerol O-acyltransferase 2
- FABP1
liver fatty acid binding protein or FABP1
- FABP1 T94T
wild type (WT) human FABP1
- FABP1 T94A
human FABP1 T94A variant
- FABP3
heart fatty acid binding protein-3
- FABP4
adipocyte fatty acid binding protein
- FABP5
epidermal fatty acid binding protein
- FABP7
brain fatty acid binding protein
- FABP KO
FABP1 gene ablated mouse on C57BL/6NCr background
- FAAH
fatty acid amide hydrolase
- FF
fenofibrate
- LCFA
free unesterified long chain fatty acid
- GPAM
glycerol-3-phosphate acyltransferase, mitochondrial
- GPCR*
G protein-coupled receptors other than CB1/CB2
- HDL
high-density lipoprotein
- GPR119
G protein-coupled receptor 119
- HNF4α
hepatocyte nuclear factor-4α
- LCFA
long chain fatty acids, unesterified
- LCFA-CoA
long chain fatty acid-CoA thioester
- LDL
low-density lipoprotein
- LDL-C
low-density lipoprotein-C
- LDLR
Low-Density Lipoprotein (LDL) Receptor
- LPL
lipoprotein lipase
- LSCM
laser scanning confocal microscopy
- MAGL
monoacylglycerol lipase
- MTTP
microsomal triglyceride transfer protein
- NAAA
N-acylethanolamide acid amide hydrolase
- NAFLD
non-alcohol fatty liver disease
- NAPE-PLD
N-acyl phosphatidylethanolamine phospholipase D
- NBD-stearic acid
[12-N-methyl-(7-nitrobenz-2-oxa-1,3-diazo)-aminostearic acid]
- NBD-ARA
NBD-arachidonic acid or [20-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino] arachidonic acid
- NBD-AEA
NBD-N-arachidonoylethanolamide or [20-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino] arachidonoylethanolamide
- NBD-2-AG
NBD-2-arachidonoylglycerol or 2-[20-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino] arachidonoyl glycerol
- NBD-cholesterol
22-(N-(7-nitrobenz-2-oxa-1, 3-diazol-4-yl)-amino)-23, 24-bisnor-5-cholen-3β-ol
- OEA
oleoylethanolamide
- 2-OG
2-oleoylglycerol
- PEA
palmitoylethanolamide
- 2-PG
2-palmitoylglycerol
- PL
phospholipid
- PPARα, -β/δ, or -γ
peroxisome proliferator-activated receptor alpha, beta/delta, or gamma
- SCP-2
sterol carrier protein-2
- SCP-x
sterol carrier protein-X
- SNP
single nucleotide polymorphism
- SRB1
Scavenger receptor class B member 1
- SREBP1c
sterol regulatory element binding protein-1c
- SCD1
stearoyl CoA desaturase
- TAG
triacylglycerol
- VLDL
very low-density lipoprotein
- WT
wild-type C57BL/6NCr mouse.
References
- 1.Ockner RK, Manning JA, Poppenhausen RB, Ho WK. A binding protein for fatty acids in cytosol of intestinal mucosa, liver, myocardium, and other tissues. Science. 1972;177:56–58. doi: 10.1126/science.177.4043.56. [DOI] [PubMed] [Google Scholar]
- 2.Atshaves BP, Martin GG, Hostetler HA, McIntosh AL, Kier AB, Schroeder F. Liver fatty acid binding protein (L-FABP) and Dietary Obesity. Journal of Nutritional Biochemisty. 2010;21:1015–1032. doi: 10.1016/j.jnutbio.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid binding proteins. Annu Rev Nutr. 2008;28:18.1–18.23. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- 4.Thompson J, Reese-Wagoner A, Banaszak L. Liver fatty acid binding protein: species variation and the accomodation of different ligands. Biochim Biophys Acta. 1999;1441:117–130. doi: 10.1016/s1388-1981(99)00146-8. [DOI] [PubMed] [Google Scholar]
- 5.McArthur MJ, Atshaves BP, Frolov A, Foxworth WD, Kier AB, Schroeder F. Cellular uptake and intracellular trafficking of long chain fatty acids. J Lipid Res. 1999;40:1371–1383. [PubMed] [Google Scholar]
- 6.Favretto F, Assfalg M, Gallo M, Cicero DO, D'Onofrio M, Molinari H. Ligand binding promiscuity and human liver fatty acid binding protein: structural and dynamic insights from an interaction study with glycocholate and oleate. ChemBioChem. 2013;14:1807–1819. doi: 10.1002/cbic.201300156. [DOI] [PubMed] [Google Scholar]
- 7.Sharma A, Sharma A. Fatty acid induced remodeling within the human liver fatty acid binding protein. J Bio Chem. 2011;286 doi: 10.1074/jbc.M111.270165.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai J, Lucke C, Chen Z, Qiao Y, Klimtchuk E, Hamilton JA. Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited. Biophys J. 2012;102:2585–2594. doi: 10.1016/j.bpj.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson J, Winter N, Terwey D, Bratt J, Banaszak L. The crystal structure of the liver fatty acid-binding protein. J Biol Chem. 1997;272:7140–7150. doi: 10.1074/jbc.272.11.7140. [DOI] [PubMed] [Google Scholar]
- 10.He Y, Yang X, Wang H, Estephan R, Francis F, Kodukula S, Storch J, Stark RE. Solution-state molecular structure of apo and oleate-liganded liver fatty acid binding protein. Biochemistry. 2007;46:12543–12556. doi: 10.1021/bi701092r. [DOI] [PubMed] [Google Scholar]
- 11.Betts MJ, Russell RB. Amino Acid Properties and Conseaquences of Substitutions. In: Barnes MR, Gray IC, editors. Bioinformatics for Geneticists. 2003. pp. 289–316. [Google Scholar]
- 12.Martin GG, McIntosh AL, Huang H, Gupta S, Atshaves BP, Kier AB, Schroeder F. Human liver fatty acid binding protein (L-FABP) T94A variant alters structure, stability, and interaction with fibrates. Biochemistry. 2013;52:9347–9357. doi: 10.1021/bi401014k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long D, Yang D. Millisecond timescale dynamics of human liver fatty acid binding protein: testing of its relevance to the ligand entry process. Biophys J. 2011;98:3054–3061. doi: 10.1016/j.bpj.2010.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long D, Yang D. Buffer interference with protein dynamics: a case study on human liver fatty acid binding protein. Biophys J. 2009;96:1482–1488. doi: 10.1016/j.bpj.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long D, Yang D. Millisecond timescale dynamics of human liver fatty acid binding protein: testing of its relevance to the ligand entry process. Biophys J. 2010;98:3054–3061. doi: 10.1016/j.bpj.2010.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai J, Lucke C, Qiao Y, Klimtchuk E, Hamilton JA. Solution structure and backbone dynamics of human liver fatty acid binding protein. Biophys J. 2010;98:238a. doi: 10.1016/j.bpj.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velkov T. Interactions between human liver fatty acid binding protein and peroxisome proliferator activated receptor drugs. PPAR Research. 2013;2013:1–14. doi: 10.1155/2013/938401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolov A, Cho TH, Murphy EJ, Schroeder F. Isoforms of rat liver fatty acid binding protein differ in structure and affinity for fatty acids and fatty acyl CoAs. Biochemistry. 1997;36:6545–6555. doi: 10.1021/bi970205t. [DOI] [PubMed] [Google Scholar]
- 19.Frolov A, Miller K, Billheimer JT, Cho TC, Schroeder F. Lipid specificity and location of the sterol carrier protein-2 fatty acid binding site: A fluorescence displacement and energy transfer study. Lipids. 1997;32:1201–1209. doi: 10.1007/s11745-997-0154-5. [DOI] [PubMed] [Google Scholar]
- 20.Wolfrum C, Ellinghaus P, Fobker M, Seedorf U, Assmann G, Borchers T, Spener F. Phytanic acid is ligand and transcriptional activator of murine liver fatty acid binding protein. J Lipid Res. 1999;40:708–714. [PubMed] [Google Scholar]
- 21.Hanhoff T, Benjamin S, Borchers T, Spener F. Branched-chain fatty acids as activators of peroxisome proliferators. Eur J Lip Sci Technol. 2005;107:716–729. [Google Scholar]
- 22.Paulussen RJA, Veerkamp JH. Intracellular fatty acid-binding proteins characteristics and function. In: Hilderson HJ, editor. Subcellular Biochemistry. Vol. 16. Plenum Press; New York: 1990. pp. 175–226. [DOI] [PubMed] [Google Scholar]
- 23.Banaszak L, Winter N, Xu Z, Bernlohr DA, Cowan S, Jones TA. Lipid-binding proteins: A family of fatty acid and retinoid transport proteins. Adv Protein Chem. 1994;45:89–151. doi: 10.1016/s0065-3233(08)60639-7. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J, Ory J, Reese-Wagoner A, Banaszak L. The liver fatty acid binding protein-comparison of cavity properties of intracellular lipid binding proteins. Mol Cell Biochem. 1999;192:9–16. [PubMed] [Google Scholar]
- 25.Thumser AE, Wilton DC. The binding of natural and fluorescent lysophospholipids to wild- type and mutant rat liver fatty acid-binding protein and albumin. Biochem J. 1995;307:305–311. doi: 10.1042/bj3070305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thumser AE, Voysey JE, Wilton DC. The binding of lysophospholipids to rat liver fatty acid-binding protein and albumin. Biochem J. 1994;301:801–806. doi: 10.1042/bj3010801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maatman RG, van Moerkerk HT, Nooren IM, van Zoelen EJ, Veerkamp JH. Expression of human liver fatty acid-binding protein in Escherichia coli and comparative analysis of its binding characteristics with muscle fatty acid-binding protein. Biochim Biophys Acta. 1994;1214:1–10. doi: 10.1016/0005-2760(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 28.Hagan RM, Worner-Gibbs J, Wilton DC. Tryptophan insertion mutagenesis of liver fatty acid binding protein. J Biol Chem. 2005;280:1782–1789. doi: 10.1074/jbc.M407131200. [DOI] [PubMed] [Google Scholar]
- 29.Di Pietro SM, Santome JA. Isolation, characterization, and binding properties of two rat liver fatty acid binding protein isoforms. Biochim Biophys Acta. 2000;1478:186–200. doi: 10.1016/s0167-4838(00)00042-x. [DOI] [PubMed] [Google Scholar]
- 30.Dietrich A, Dieminger W, Fuchte K, Stoll GH, Schlitz E, Gerok W, Kurz G. Functional signficance of interaction of hepatic FABP with sulfated and nonsulfated taurine-conjugated bile salts in rat liver. J Lipid Res. 1995;36:1745–1755. [PubMed] [Google Scholar]
- 31.Dietrich A, Dieminger W, Nelly SM, Gerok W, Kurz G. Synthesis and applicability of a photolabile 7, 7-azi analogue of 3-sulfated taurine-conjugated bile acids. J Lipid Res. 1995;36:1729–1744. [PubMed] [Google Scholar]
- 32.Thumser AE, Wilton DC. The binding of cholesterol and bile salts to recombinant rat liver fatty acid-binding protein. Biochem J. 1996;320:729–733. doi: 10.1042/bj3200729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaikaus RM, Bass NM, Ockner RK. Functions of fatty acid binding proteins. Experientia. 1990;46:617–630. doi: 10.1007/BF01939701. [DOI] [PubMed] [Google Scholar]
- 34.Martin GG, Atshaves BP, Landrock KK, Landrock D, Storey SM, Howles PN, Kier AB, Schroeder F. Ablating L-FABP in SCP-2/SCP-x null mice impairs bile acid metabolism and biliary HDL-cholesterol secretion. Am J Physiol Gastrointest and Liver Phys. 2014;307:G1130–G1143. doi: 10.1152/ajpgi.00209.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maatman RG, Van Kuppevelt TH, Veerkamp JH. Two types of fatty acid-binding protein in human kidney. Isolation, characterization and localization. Biochem J. 1991;273:759–766. doi: 10.1042/bj2730759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carbone V, Velkov T. Interaction of phthalates and phonxy acid herbicide environmental pollutants with intestinal intracellular lipid binding proteins. Chem Res in Tox. 2013;26:1240–1250. doi: 10.1021/tx400170t. [DOI] [PubMed] [Google Scholar]
- 37.Huang H, McIntosh AL, Martin GG, Landrock K, Landrock D, Gupta S, Atshaves BP, Kier AB, Schroeder F. Structural and functional interaction of fatty acids with human liver fatty acid binding protein (L-FABP) T94A variant. FEBS J. 2014;281:2266–2283. doi: 10.1111/febs.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H, McIntosh AL, Martin GG, Landrock KK, Landrock D, Storey SM, Gupta S, Atshaves BP, Kier AB, Schroeder F. Human L-FABP T94A variant enhances cholesterol uptake. Biochim Biophys Acta. 2015;1851:946–955. doi: 10.1016/j.bbalip.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin GG, Atshaves BP, Huang H, McIntosh AL, Williams BW, Pai PJ, Russell DH, Kier AB, Schroeder F. Hepatic phenotype of liver fatty acid binding protein (L-FABP) gene ablated mice. Am J Physiol. 2009;297:G1053–G1065. doi: 10.1152/ajpgi.00116.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai J, Lucker C, Chen Z, Klimtchuk E, Qiao Y, Hamilton JA. Human liver fatty acid binding protein: Solutoiin structure and ligand binding. Biophys J. 2009;96:600a. doi: 10.1016/j.bpj.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuang S, Velkov T, Horne J, Wielens J, Chalmers DK, Porter CJH, Scanlon MJ. Probing fibrate binding specificity of rat liver fatty acid binding protein. J Med Chem. 2009;52:5344–5355. doi: 10.1021/jm801349e. [DOI] [PubMed] [Google Scholar]
- 42.Chuang S, Velkov T, Horne J, Porter CJH, Scanlon MJ. Characterization of the drug binding specificity of rat liver fatty acid binding protein. J Med Chem. 2008;51:3755–3764. doi: 10.1021/jm701192w. [DOI] [PubMed] [Google Scholar]
- 43.Murphy EJ, Prows DR, Jefferson JR, Schroeder F. Liver fatty acid binding protein expression in transfected fibroblasts stimulates fatty acid uptake and metabolism. Biochim Biophys Acta. 1996;1301:191–198. doi: 10.1016/0005-2760(96)00024-0. [DOI] [PubMed] [Google Scholar]
- 44.Prows DR, Murphy EJ, Schroeder F. Intestinal and liver fatty acid binding proteins differentially affect fatty acid uptake and esterification in L-Cells. Lipids. 1995;30:907–910. doi: 10.1007/BF02537481. [DOI] [PubMed] [Google Scholar]
- 45.Prows DR, Murphy EJ, Moncecchi D, Schroeder F. Intestinal fatty acid-binding protein expression stimulates fibroblast fatty acid esterification. Chem Phys Lipids. 1996;84:47–56. doi: 10.1016/s0009-3084(96)02619-9. [DOI] [PubMed] [Google Scholar]
- 46.Murphy EJ. L-FABP and I-FABP expression increase NBD-stearate uptake and cytoplasmic diffusion in L-cells. Am J Physiol. 1998;275:G244–G249. doi: 10.1152/ajpgi.1998.275.2.G244. [DOI] [PubMed] [Google Scholar]
- 47.McIntosh AL, Huang H, Atshaves BP, Wellburg E, Kuklev DV, Smith WL, Kier AB, Schroeder F. Fluorescent n-3 and n-6 very long chain polyunsaturated fatty acids: three photon imaging and metabolism in living cells overexpressing liver fatty acid binding protein. J Biol Chem. 2010;285:18693–18708. doi: 10.1074/jbc.M109.079897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfrum C, Buhlman C, Rolf B, Borchers T, Spener F. Variation of liver fatty acid binding protein content in the human hepatoma cell line HepG2 by peroxisome proliferators and antisense RNA affects the rate of fatty acid uptake. Biochim Biophys Acta. 1999;1437:194–201. doi: 10.1016/s1388-1981(99)00008-6. [DOI] [PubMed] [Google Scholar]
- 49.Gao N, Qu X, Yan J, Huang Q, Yuan HY, Ouyang DS. L-FABP T94A decreased fatty acid uptake and altered hepatic triglyceride and cholesterol accumulation in Chang liver cells stably transfected with L-FABP. Mol Cell Biochem. 2010;345:207–214. doi: 10.1007/s11010-010-0574-7. [DOI] [PubMed] [Google Scholar]
- 50.Martin GG, Danneberg H, Kumar LS, Atshaves BP, Erol E, Bader M, Schroeder F, Binas B. Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid binding protein (L-FABP) gene. J Biol Chem. 2003;278:21429–21438. doi: 10.1074/jbc.M300287200. [DOI] [PubMed] [Google Scholar]
- 51.Atshaves BP, Foxworth WB, Frolov AA, Roths JB, Kier AB, Oetama BK, Piedrahita JA, Schroeder F. Cellular differentiation and I-FABP protein expression modulate fatty acid uptake and diffusion. Am J Physiol. 1998;274:C633–C644. doi: 10.1152/ajpcell.1998.274.3.C633. [DOI] [PubMed] [Google Scholar]
- 52.Atshaves BP, McIntosh AL, Lyuksyutova OI, Zipfel WR, Webb WW, Schroeder F. Liver fatty acid binding protein gene ablation inhibits branched-chain fatty acid metabolism in cultured primary hepatocytes. J Biol Chem. 2004;279:30954–30965. doi: 10.1074/jbc.M313571200. [DOI] [PubMed] [Google Scholar]
- 53.Storey SM, McIntosh AL, Huang H, Martin GG, Landrock KK, Landrock D, Payne HR, Kier AB, Schroeder F. Loss of intracellular lipid binding proteins differentially impacts saturated fatty acid uptake and nuclear targeting in mouse hepatocytes. Am J Physiol Gastrointest and Liver Phys. 2012;303:G837–G850. doi: 10.1152/ajpgi.00489.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McIntosh AL, Atshaves BP, Hostetler HA, Huang H, Davis J, Lyuksyutova OI, Landrock D, Kier AB, Schroeder F. Liver type fatty acid binding protein (L-FABP) gene ablation reduces nuclear ligand distribution and peroxisome proliferator activated receptor-alpha activity in cultured primary hepatocytes. Arch Biochem Biophys. 2009;485:160–173. doi: 10.1016/j.abb.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weisiger RA. Cytosolic fatty acid binding proteins catalyze two distinct steps in intracellular transport of their ligands. Mol Cell Biochem. 2005;239:35–42. [PubMed] [Google Scholar]
- 56.Atshaves BP, McIntosh AL, Payne HR, Gallegos AM, Landrock K, Maeda N, Kier AB, Schroeder F. Sterol carrier protein-2/sterol carrier protein-x gene ablation alters lipid raft domains in primary cultured mouse hepatocytes. J Lipid Res. 2007;48:2193–2211. doi: 10.1194/jlr.M700102-JLR200. [DOI] [PubMed] [Google Scholar]
- 57.Atshaves BP, McIntosh AL, Payne HR, Mackie J, Kier AB, Schroeder F. Effect of branched-chain fatty acid on lipid dynamics in mice lacking liver fatty acid binding protein gene. Am J Physiol. 2005;288:C543–C558. doi: 10.1152/ajpcell.00359.2004. [DOI] [PubMed] [Google Scholar]
- 58.Hostetler HA, Lupas D, Tan Y, Dai J, Kelzer MS, Martin GG, Woldegiorgis G, Kier AB, Schroeder F. Acyl-CoA binding proteins interact with the acyl-CoA binding domain of mitochondrial carnitine palmitoyltransferase I. Mol Cell Biochem. 2011;355:135–148. doi: 10.1007/s11010-011-0847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atshaves BP, McIntosh AL, Kier AB, Schroeder F. High dietary fat exacerbates weight gain and obesity in female liver fatty acid binding protein gene ablated mice. Lipids. 2010;45:97–110. doi: 10.1007/s11745-009-3379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woldegiorgis G, Bremer J, Shrago E. Substrate inhibition of carnitine palmitoyltransferase by palmitoyl-CoA and activation by phospholipids and proteins. Biochim Biophys Acta. 1985;837:135–140. doi: 10.1016/0005-2760(85)90236-x. [DOI] [PubMed] [Google Scholar]
- 61.Petrescu AD, McIntosh AL, Storey SM, Huang H, Martin GG, Landrock D, Kier AB, Schroeder F. High glucose potentiates liver fatty acid binding protein (L-FABP) mediated fibrate induction of PPARa in mouse hepatocytes. Biochim Biophys Acta. 2013;1831:1412–1425. doi: 10.1016/j.bbalip.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petrescu AD, Huang H, Martin GG, McIntosh AL, Storey SM, Landrock D, Kier AB, Schroeder F. Impact of L-FABP and glucose on polyunsaturated fatty acid induction of PPARa regulated b-oxidative enzymes. Am J Physiol Gastrointest and Liver Phys. 2013;304:G241–G256. doi: 10.1152/ajpgi.00334.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McIntosh AL, Huang H, Storey SM, Landrock K, Landrock D, Petrescu AD, Gupta S, Atshaves BP, Kier AB, Schroeder F. Human FABP1 T94A variant impacts fatty acid metabolism and PPARa activation in cultured human female hepatocytes. Am J Physiol Gastrointest and Liver Phys. 2014;307:G164–G176. doi: 10.1152/ajpgi.00369.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hostetler HA, McIntosh AL, Atshaves BP, Storey SM, Payne HR, Kier AB, Schroeder F. Liver type Fatty Acid Binding Protein (L-FABP) interacts with peroxisome proliferator activated receptor-α in cultured primary hepatocytes. J Lipid Res. 2009;50:1663–1675. doi: 10.1194/jlr.M900058-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hostetler HA, Balanarasimha M, Huang H, Kelzer MS, Kaliappan A, Kier AB, Schroeder F. Glucose regulates fatty acid binding protein interaction with lipids and PPARα. J Lipid Res. 2010;51:3103–3116. doi: 10.1194/jlr.M005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hostetler HA, McIntosh AL, Petrescu AD, Huang H, Atshaves BP, Murphy EJ, Kier AB, Schroeder F. Fluorescence methods to assess the impact of lipid binding proteins on ligand activated gene expression. In: Murphy EJ, Rosenberger TA, editors. Methods in Lipid-Mediated Signaling. CRC Press-Taylor and Francis Group, LLC; Boca Raton, FL: 2010. pp. 299–348. [Google Scholar]
- 67.Huang H, McIntosh AL, Martin GG, Petrescu AD, Landrock K, Landrock D, Kier AB, Schroeder F. Inhibitors of fatty acid synthesis induce PPARa-regulated fatty acid b-oxidative enzymes: synergistic roles of L-FABP and glucose. PPAR Research. 2013;2013:1–22. doi: 10.1155/2013/865604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Atshaves BP, Payne HR, McIntosh AL, Tichy SE, Russell D, Kier AB, Schroeder F. Sexually dimorphic metabolism of branched chain lipids in C57BL/6J mice. J Lipid Res. 2004;45:812–830. doi: 10.1194/jlr.M300408-JLR200. [DOI] [PubMed] [Google Scholar]
- 69.Atshaves BP, Storey SM, Petrescu AD, Greenberg CC, Lyuksyutova OI, Smith R, Schroeder F. Expression of fatty acid binding proteins inhibits lipid accumulation and alters toxicity in L-cell fibroblasts. Am J Physiol. 2002;283:C688–C703. doi: 10.1152/ajpcell.00586.2001. [DOI] [PubMed] [Google Scholar]
- 70.Atshaves BP, Storey S, Huang H, Schroeder F. Liver fatty acid binding protein expression enhances branched-chain fatty acid metabolism. Mol Cell Biochem. 2004;259:115–129. doi: 10.1023/b:mcbi.0000021357.97765.f2. [DOI] [PubMed] [Google Scholar]
- 71.Richert L, Lamboley C, Viollon-Abadie C, Grass P, Hartmann N, Laurent S, Heyd B, Mantion G, Chibout SD, Staedtler F. Effects of clofibric acid on mRNA expression profiles in primary cultures of rat, mouse, and human hepatocytes. Toxicol Appl Pharmacol. 2003;191:130–146. doi: 10.1016/s0041-008x(03)00231-x. [DOI] [PubMed] [Google Scholar]
- 72.Rakhshandehroo M, Hooiveld G, Muller M, Kersten S. Comparative analysis of gene regulation by the transcription factor PPARα between mouse and human. PLoS ONE. 2009;4:e6796. doi: 10.1371/journal.pone.0006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McIntosh AL, Petrescu AD, Hostetler HA, Kier AB, Schroeder F. Liver-type fatty acid binding protein interacts with hepatocyte nuclear factor 4alpha. FEBS Lett. 2013;587:3787–3791. doi: 10.1016/j.febslet.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bordewick U, Heese M, Borchers T, Robenek H, Spener F. Compartmentation of hepatic fatty-acid-binding protein in liver cells and its effect on microsomal phosphatidic acid biosynthesis. Biol Chem Hoppe-Seyler. 1989;370:229–238. doi: 10.1515/bchm3.1989.370.1.229. [DOI] [PubMed] [Google Scholar]
- 75.Jolly CA, Hubbell T, Behnke WD, Schroeder F. Fatty acid binding protein: stimulation of microsomal phosphatidic acid formation. Arch Biochem Biophys. 1997;341:112–121. doi: 10.1006/abbi.1997.9957. [DOI] [PubMed] [Google Scholar]
- 76.Jolly CA, Murphy EJ, Schroeder F. Differential influence of rat liver fatty acid binding protein isoforms on phospholipid fatty acid composition: phosphatidic acid biosynthesis and phospholipid fatty acid remodeling. Biochim Biophys Acta. 1998;1390:258–268. doi: 10.1016/s0005-2760(97)00186-0. [DOI] [PubMed] [Google Scholar]
- 77.Schroeder F, Jolly CA, Cho TH, Frolov AA. Fatty acid binding protein isoforms: structure and function. Chem Phys Lipids. 1998;92:1–25. doi: 10.1016/s0009-3084(98)00003-6. [DOI] [PubMed] [Google Scholar]
- 78.Martin GG, Huang H, Atshaves BP, Binas B, Schroeder F. Ablation of the liver fatty acid binding protein gene decreases fatty acyl CoA binding capacity and alters fatty acyl CoA pool distribution in mouse liver. Biochem. 2003;42:11520–11532. doi: 10.1021/bi0346749. [DOI] [PubMed] [Google Scholar]
- 79.Erol E, Kumar LS, Cline GW, Shulman GI, Kelly DP, Binas B. Liver fatty acid-binding protein is required for high rates of hepatic fatty acid oxidation but not for the action of PPAR-a in fasting mice. FASEB J. 2004;18:347–349. doi: 10.1096/fj.03-0330fje. [DOI] [PubMed] [Google Scholar]
- 80.Newberry EP, Xie Y, Kennedy S, Buhman KK, Luo J, Gross RW, Davidson NO. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid binding protein gene. J Biol Chem. 2003;278:51664–51672. doi: 10.1074/jbc.M309377200. [DOI] [PubMed] [Google Scholar]
- 81.Lagakos WS, Gajda AM, Agellon LB, Binas B, Choi V, Mandap B, Russnak T, Zhou YX, Storch J. Different functions of intestinal and liver-type fatty acid binding proteins in intestine and in whole body energy homeostasis. Am J Physiol Gastrointest and Liver Phys. 2011;300:G803–G814. doi: 10.1152/ajpgi.00229.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Newberry EP, Xie Y, Kennedy SM, Luo J, Davidson NO. Protection against western diet-induced obesity and hepatic steatosis in liver fatty acid binding protein knockout mice. Hepatology. 2006;44:1191–1205. doi: 10.1002/hep.21369. [DOI] [PubMed] [Google Scholar]
- 83.Masters JR. Cell line misidentification: the beginning of the end. Nature Rev Cancer. 2010;10:441–448. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- 84.Martin GG, Atshaves BP, Landrock KK, Landrock D, Schroeder F, Kier AB. Loss of L-FABP, SCP-2/SCP-x, or both Induces Hepatic Lipid Accumulation in Female Mice. Arch Biochem Biophys. 2015;580:41–49. doi: 10.1016/j.abb.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martin GG, Landrock D, Landrock KK, Howles PN, Atshaves BP, Kier AB, Schroeder F. Relative contributions of L-FABP, SCP-2/SCP-x, or both to hepatic biliary phenotype of female mice. Arch Biochem Biophys. 2015;588:25–32. doi: 10.1016/j.abb.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin GG, Atshaves BP, McIntosh AL, Mackie JT, Kier AB, Schroeder F. Liver fatty acid binding protein gene ablation enhances age-dependent weight gain in male mice. Mol Cell Biochem. 2009;324:101–115. doi: 10.1007/s11010-008-9989-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martin GG, Atshaves BP, McIntosh AL, Mackie JT, Kier AB, Schroeder F. Liver Fatty Acid Binding Protein GeneAblated Female Mice Exhibit Increased AgeDependent Obesity. J Nutr. 2008;138:1859–1865. doi: 10.1093/jn/138.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martin GG, Chung S, Landrock D, Landrock KK, Huang H, Dangott LJ, Peng X, Kaczocha M, Murphy EJ, Kier AB, Schroeder F. Fatty acid binding protein-1 ablation differentially impacts the brain endocannabinoid system of male vs female mice. J Neurochem Revision pending 2015 [Google Scholar]
- 89.Huang H, Martin GG, Landrock D, Chung S, McIntosh AL, Dangott LJ, Li S, Kier AB, Schroeder F. FABP1: a novel hepatic endocannabinoid binding protein. Am J Physiol Gastrointest and Liver Phys. 2016 doi: 10.1021/acs.biochem.6b00446. In review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Murphy EJ, Edmondson RD, Russell DH, Colles SM, Schroeder F. Isolation and characterization of two distinct forms of liver fatty acid binding protein from the rat. Biochim Biophys Acta. 1999;1436:413–425. doi: 10.1016/s0005-2760(98)00150-7. [DOI] [PubMed] [Google Scholar]
- 91.Murphy EJ, Prows DR, Jefferson JR, Incerpi S, Hertelendy ZI, Heiliger CE, Schroeder F. Cis-parinaric acid uptake in L-cells. Arch Biochem Biophys. 1996;335:267–272. doi: 10.1006/abbi.1996.0507. [DOI] [PubMed] [Google Scholar]
- 92.Schroeder F, Jefferson JR, Powell D, Incerpi S, Woodford JK, Colles SM, Myers-Payne S, Emge T, Hubbell T, Moncecchi D, Prows DR, Heyliger CE. Expression of rat L-FABP in mouse fibroblasts: role in fat absorption. Mol Cell Biochem. 1993;123:73–83. doi: 10.1007/BF01076477. [DOI] [PubMed] [Google Scholar]
- 93.Sklar LA, Hudson BS, Simoni RD. Conjugated polyene fatty acids as fluorescent probes: binding to bovine serum albumin. Biochemistry. 1977;16:5100–5108. doi: 10.1021/bi00642a024. [DOI] [PubMed] [Google Scholar]
- 94.Sarchielli P, Pini LA, Coppola F, et al. Endocannabinoids in chronic migraine: CSF findings suggest a system failure. Neuropsychopharmacology. 2007;32:1384–1390. doi: 10.1038/sj.npp.1301246. [DOI] [PubMed] [Google Scholar]
- 95.Christie MJ, Mallet C. Endocannabinoids can open the pain gate. Sciencesignaling. 2009;2:pe57. doi: 10.1126/scisignal.288pe57. [DOI] [PubMed] [Google Scholar]
- 96.Sagar DR, Burston JJ, Woodhams SG, Chapman V. Dynamic changes to the endocannabinoid system in models of chronic pain. Phil Trans R Soc B. 2012;367:3300–3311. doi: 10.1098/rstb.2011.0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fine PG, Rosenfeld MJ. The endocannabinoid system, cannabinoids, and pain. Rambam Maimonides Medical Journal. 2013;4:e0022. doi: 10.5041/RMMJ.10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guasti L, Richardson D, Jhaveri M, et al. Minocycline treatment inhibits microglial activation and alters spinal levels of endocannabinoids in a rat model of neuropathic pain. Molecular Pain. 2009;5 doi: 10.1186/1744-8069-5-35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.De Petrocellis L, Melck D, Bisogno T, Di Marzo V. Endocannabinoids and fatty acid amides in cancer, inflammation, and related disorders. Chem Phys Lip. 2000;108:191–209. doi: 10.1016/s0009-3084(00)00196-1. [DOI] [PubMed] [Google Scholar]
- 100.Luongo L, Malcangio M, Salvemini D, Starowicz K. Chronic pain: New insights in molecular and cellular mechanisms. BioMed Res Int. 2015:1–2. doi: 10.1155/2015/676725. org/10.1155/2015/676725. [DOI] [PMC free article] [PubMed]
- 101.Tam J, Liu J, Mukhopadhyay B, Cinar R, Godlewski G, Kunos G. Endocannabinoids in liver disease. Hepatology. 2011;53:346–355. doi: 10.1002/hep.24077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alswat KA. The role of endocannabinoid system in fatty liver disease and therapeutic potential. Saudi Journal of Gastroenterology. 2015;19:144–151. doi: 10.4103/1319-3767.114505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Regnell SE. Cannabinoid 1 receptor in fatty liver. Hepatol Res. 2013;43:131–138. doi: 10.1111/j.1872-034X.2012.01085.x. [DOI] [PubMed] [Google Scholar]
- 104.Purohit V, Rapaka R, Shurtleff D. Role of cannabinoids in the development of fatty liver (steatosis) AAPS Journal. 2010;12:233–237. doi: 10.1208/s12248-010-9178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Elmes MW, Kaczocha M, Berger WT, Leung KN, Ralph BP, Wang L, Sweeney JM, Miyauchi JT, Tsirka SE, Ojima I, Deutsch DG. Fatty acid binding proteins are intracellular carriers for delta-9-tetrahydrocannabinol (THC) and cannabidiol (CBD) J Biol Chem. 2015;290:8711–8721. doi: 10.1074/jbc.M114.618447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaczocha LM, Glaser ST, Deutsch DG. Identification of intracellular carriers for the endocannabinoid anandamide. Proc Natl Acad Sci U S A. 2009;106:6375–6380. doi: 10.1073/pnas.0901515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kaczocha M. Ph D Thesis. Stony Brook University; 2009. Role of fatty acid binding proteins and FAAH-2 in endocannabinoid uptake and inactivation. [Google Scholar]
- 108.Kaczocha M, Rebecchi MJ, Ralph BP, Teng YHG, Berger WT, Galbavy W, Elmes MW, Glaser ST, Wang L, Rizzo RC, Deutsch DG, Ojima I. Inhibition of fatty acid binding protein elevates brain anandamide levels and produces analgesia. PLoS ONE. 2014;9:e94200. doi: 10.1371/journal.pone.0094200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leung K, Elmes MW, Glaser ST, Deutsch DG, Kaczocha M. Role of FAAH-like anandamide transporter in anandamide inactivation. PLoS ONE. 2013;8:e79355. doi: 10.1371/journal.pone.0079355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kaczocha M, Vivieca S, Sun J, Glaser ST, Deutsch DG. Fatty acid binding proteins transport N-acylethanolamines to nuclear receptors and are targets of endocannabinoid transport inhibitors. J Biol Chem. 2012;287:3415–3424. doi: 10.1074/jbc.M111.304907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fowler CJ. Transport of endocannabinoids across plasma membrane and within the cell. FEBS J. 2013;280:1895–1904. doi: 10.1111/febs.12212. [DOI] [PubMed] [Google Scholar]
- 112.Frolov A, Cho TH, Billheimer JT, Schroeder F. Sterol carrier protein-2, a new fatty acyl coenzyme A-binding protein. J Biol Chem. 1996;271:31878–31884. doi: 10.1074/jbc.271.50.31878. [DOI] [PubMed] [Google Scholar]
- 113.Huang H, Atshaves BP, Frolov A, Kier AB, Schroeder F. Acyl-coenzyme A binding protein expression alters liver fatty acyl coenzyme A metabolism. Biochemistry. 2005;44:10282–10297. doi: 10.1021/bi0477891. [DOI] [PubMed] [Google Scholar]
- 114.Wilkinson TC, Wilton DC. Studies on fatty acid-binding proteins. The detection and quantification of the protein from rat liver by using a fluorescent fatty acid analogue. Biochem J. 1986;238:419–424. doi: 10.1042/bj2380419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huestis MA. Human cannabinoid pharmacokinetics. Chem Biodivers. 2007;4:1770–1804. doi: 10.1002/cbdv.200790152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mattes RD, Shaw LM, Edling-Owens J, Engelman K, Elsohly MA. Bypassing the first-pass effect for the therapeutic use of cannabinoids. Pharm Biochem and Behavior. 1993;44:745–747. doi: 10.1016/0091-3057(93)90194-x. [DOI] [PubMed] [Google Scholar]
- 117.Trevaskis NL, Shackleford DM, Charman WN, Edwards GA, et al. Intestinal lymphatic transport enhances the post-prandial oral bioavailability of a novel cannabinoid receptor agonist via avoidance of first-pass metabolism. Pharm Res. 2009;26:1486–1495. doi: 10.1007/s11095-009-9860-z. [DOI] [PubMed] [Google Scholar]
- 118.Grotenhermen F. Clin Pharmacokinet. 2003;42:327–360. doi: 10.2165/00003088-200342040-00003. [DOI] [PubMed] [Google Scholar]
- 119.Ashton CH. Pharmacology and effects of cannabis: a brief review. Br J Psychiatry. 2001;178:101–106. doi: 10.1192/bjp.178.2.101. [DOI] [PubMed] [Google Scholar]
- 120.Ho WSV, Barrett DAR. Entourage effects of N-palmitoylethanolamide and N-oleoylethanolamide on vasorelaxaton to anandamide occur through TRPV1 receptors. Br J Pharmacol. 2008;155:837–846. doi: 10.1038/bjp.2008.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Smart D, Jonsson KO, Vanvoorde S, Lambert DM, Fowler CJ. Entourage effects of N-acyl ethanolamines at human vanilloid receptors. Comparison of effects upon anandamide-induced vanilloid receptor activation and upon anandamide metabolism. Br J Pharmacol. 2002;136:452–458. doi: 10.1038/sj.bjp.0704732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Piomelli S, Seaman C. Mechanism of red blood cell aging: relationship of cell density and cell age [Review] Am J Hematology. 1993;42:46–52. doi: 10.1002/ajh.2830420110. [DOI] [PubMed] [Google Scholar]
- 123.Franklin A, Parmentier-Batteur S, Walter L, Greenbert DA, Stella N. Palmitoylethanolamide increases after focal cerebral ischemia and potentiates microglial cell motility. J Neuroscience. 2003;23:7767–7775. doi: 10.1523/JNEUROSCI.23-21-07767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ben-Shabat S, Fride E, Sheskin T, et al. An entoruage effects: inactive endogenous fatty acid glycerol esters enhance 2-arachidonoyl-glycerol cannabinoid activity. Eur J Pharm. 1998;353:23–31. doi: 10.1016/s0014-2999(98)00392-6. [DOI] [PubMed] [Google Scholar]
- 125.Mechoulam R, Fride E, Hanus L, et al. Anandamide may mediate sleep induction. Nature. 1997;389:25–26. doi: 10.1038/37891. [DOI] [PubMed] [Google Scholar]
- 126.Lagakos WS, Guan X, Ho SY, Sawicki LR, Corsico B, Murota K, Stark RE, Storch J. L-FABP binds monoacylglycerol in vitro and in mouse liver cytosol. J Biol Chem. 2013;288:19805–19815. doi: 10.1074/jbc.M113.473579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Storch J. Diversity of fatty acid-binding protein structure and function: studies with fluorescent ligands [Review] Mol Cell Biochem. 1993;123:45–53. doi: 10.1007/BF01076474. [DOI] [PubMed] [Google Scholar]
- 128.Strokin ML, Sergeeva MG, Mevkh AT. The influence of serum fatty acid binding proteins on arachidonic acid uptake by macrophages. Applied Biochemistry and Biotechnology. 2000;88:195–200. [Google Scholar]
- 129.Strokin ML, Sergeva MG, Mevkh AT. Differences in the uptake of low and high concentrations of arachidonic acid into murine peritoneal macrophages. Biocatalysis-2000: Fundamentals and Applications. 2000;41:91–94. [Google Scholar]
- 130.Campbell FM, Clohessy AM, Gordon MJ, Page KR, Roy-Dutta AK. Uptake of long chain fatty acids by human placental choriocarcinoma (BeWo) cells: role of plasma membrane fatty acid-binding protein. Journal of Lipid Research. 1997;38:25582568. [PubMed] [Google Scholar]
- 131.Osei-Hyiaman D, Liu J, Zhou L, Godlewski G, Harvey-White J, Jeong W, Batkai S, Marsicano G, Lutz B, Buettner C, Kunos G. Hepatic CB1 receptor is required for development of diet-induced steatosis, dyslipidemia, and insulin and leptin resistance in mice. J Clin Inv. 2008;118:3160–3169. doi: 10.1172/JCI34827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Osei-Hyiaman D, DePetrillo M, Pacher P, Kunos G. Endocannabinoid activation at hepatic CB1 receptors stimulates fatty acid synthesis and contributes to diet-induced obesity. J Clin Inv. 2005;115:1298–1305. doi: 10.1172/JCI23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pacher P, Batkai S, Kunos G. The endocannabinoid system as an emerging target of pharmacotherapy. Pharmacological Reviews. 2006;58:389–462. doi: 10.1124/pr.58.3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Maccarrone M, Gasperi V, Catani MV, Diep TI, Dainese E, Hansen HS, Avigliano L. The endocannabinoid system and its relevance to nutrition. Annu Rev Nutr. 2010;30:423–440. doi: 10.1146/annurev.nutr.012809.104701. [DOI] [PubMed] [Google Scholar]
- 135.Sun Y, Alexander SPH, Garle MJ, et al. Cannabinoid activation of PPARa: a novel neuroprotective mechanism. Br J Pharmacol. 2007;152:734–743. doi: 10.1038/sj.bjp.0707478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fu J, Gaetani S, Ovelsi F, Verme JL, Serrano A, de Fonseca FR, Rosengarth A, Luecke H, Di Giacomo B, Tarzia G, Plomelli D. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPARα. Nature. 2003;425:90–93. doi: 10.1038/nature01921. [DOI] [PubMed] [Google Scholar]
- 137.Naughton SS, Mathai ML, Hryciw DH, McAinch AJ. Fatty acid modulation of the endocannabinoid system and the effect on food intake and metabolism. Int J Endocrinol ID. 2013;361895:1–11. doi: 10.1155/2013/361895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Syed SK, Bui HH, Beavers LS, et al. Regulation of GPR119 receptor activity with endocannabinoid-like lipids. Am J Physiol Endocrinol Metab. 2012;303:E1469–E1478. doi: 10.1152/ajpendo.00269.2012. [DOI] [PubMed] [Google Scholar]
- 139.Long L, Li L, Chen LLX, et al. Effect of oleoylethanolamide on diet-induced non-alcoholic fatty liver in rats. J Pharm Sci. 2015;127:244–250. doi: 10.1016/j.jphs.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 140.Yang JW, Kim HS, Im JH, et al. GPR119: a promising target for non-alcoholic fatty liver disease. FASEB J. 2016;30:324–335. doi: 10.1096/fj.15-273771. [DOI] [PubMed] [Google Scholar]
- 141.Weisiger RA. Cytoplasmic transport of lipids: Role of binding proteins. Comp Biochem Physiol. 1996;115B:319–331. [Google Scholar]
- 142.Labar G, Wouters J, Lambert DM. A review of the monoacylglycerol lipase: at teh interface between fat and endocannabinoid signaliling. Current Medicinal Chemistry. 2010;17:2588–2607. doi: 10.2174/092986710791859414. [DOI] [PubMed] [Google Scholar]
- 143.Tsuboi K, Takezaki N, Ueda N. The N-acylethanolamine-hydrolyzing acid amidase (NAAA) Chemistry and Biodiversity. 2007;4:1914–1925. doi: 10.1002/cbdv.200790159. [DOI] [PubMed] [Google Scholar]
- 144.Kaczocha M, Glaser ST, Chae J, Brown DA, Deutsch DG. Lipid droplets are novel sites fo N-acylethanolamine inactivation by FAAH2. J Biol Chem. 2010;285:2796–2806. doi: 10.1074/jbc.M109.058461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ueda N, Tsuboi K, Uyama T. N-acylethanolamine metabolism with special reference to N-acylethanolamine-hydrolyzing acid amidase (NAAA) Prog Lipid Res. 2010;49:299–315. doi: 10.1016/j.plipres.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 146.Myers-Payne SC, Hubbell T, Pu L, Schnutgen F, Borchers T, Wood WG, Spener F, Schroeder F. Isolation and characterization of two fatty acid binding proteins from mouse brain. J Neurochem. 1996;66:1648–1656. doi: 10.1046/j.1471-4159.1996.66041648.x. [DOI] [PubMed] [Google Scholar]
- 147.Pu L, Igbavboa U, Wood WG, Roths JB, Kier AB, Spener F, Schroeder F. Expression of Fatty Acid Binding Proteins Is Altered in Aged Mouse Brain. Molecular and Cellular Biochemistry. 1999;198:69–78. doi: 10.1023/a:1006946027619. [DOI] [PubMed] [Google Scholar]
- 148.Pu L, Annan RS, Carr SA, Frolov A, Wood WG, Spener F, Schroeder F. Isolation and identification of a native fatty acid binding protein form mouse brain. Lipids. 1999;34:363–373. doi: 10.1007/s11745-999-0374-8. [DOI] [PubMed] [Google Scholar]
- 149.Owada Y, Yoshimoto T, Kondo H. Spatio-temporally differential expression of genes for three members of fatty acid binding proteins in developing and mature rat brains. Journal of Chemical Neuroanatomy. 1996;12:113–122. doi: 10.1016/s0891-0618(96)00192-5. [DOI] [PubMed] [Google Scholar]
- 150.Schnutgen F, Borchers T, Muller T, Spener F. Heterologous expression and characterization of mouse brain fatty acid binding protein. Biol Chem Hoppe-Seyler. 1996;377:211–215. doi: 10.1515/bchm3.1996.377.3.211. [DOI] [PubMed] [Google Scholar]
- 151.Bennett E, Stenvers KL, Lund PK, Popko B. Cloning and characterization of a cDNA encoding a novel fatty acid binding protein from rat brain. J Neurochem. 1994;63:1616–1624. doi: 10.1046/j.1471-4159.1994.63051616.x. [DOI] [PubMed] [Google Scholar]
- 152.Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 153.Kurtz A, Zimmer A, Schnutgen F, Bruning G, Spener F, Muller T. The expression pattern of a novel gene encoding brain-fatty acid binding protein correlates with neuronal and glial cell development. Development. 1994;120:2637–2649. doi: 10.1242/dev.120.9.2637. [DOI] [PubMed] [Google Scholar]
- 154.Yu S, Levi L, Casadesus G, Kunos G, Noy N. Fatty acid binding protein 5 (FABP5) regulates cognitive function both by decreasing anandamide levels and by activating the nuclear receptor peroxisome proliferator activated receptor β/δ (PPARβ/δ) in the brain. J Biol Chem. 2014;289:12748–12758. doi: 10.1074/jbc.M114.559062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Murphy EJ, Owada Y, Kitanaka N, Kondo H, Glatz JFC. Brain arachidonic acid incorportion is decreased in heart FABP gene ablated mice. Biochem. 2005;44:6350–6360. doi: 10.1021/bi047292r. [DOI] [PubMed] [Google Scholar]
- 156.Owada Y, Abdelwahab SA, et al. Altered emotional behavioral responses in mice lacking brain type FABP gene. Eur J Neurosci. 2006;24:175–187. doi: 10.1111/j.1460-9568.2006.04855.x. [DOI] [PubMed] [Google Scholar]
- 157.Owada Y. Fatty acid binding protein: localization and functional significance in brain. Tohoku J Exp Med. 2008;214:213–220. doi: 10.1620/tjem.214.213. [DOI] [PubMed] [Google Scholar]
- 158.Moulle VSF, Cansell C, Luquet S, Cruciani-Guglielmacci C. Multiple roles of fatty acid handling proteins in brain. Frontiers in Physiology. 2012;3:1–6. doi: 10.3389/fphys.2012.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Haunerland NH, Spener F. Fatty acid binding proteins--insights from genetic manipulations. Prog Lipid Res. 2004;43:328–349. doi: 10.1016/j.plipres.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 160.Storch J, Thumser AE. The fatty acid transport functions of fatty acid binding proteins. Biochim Biophys Acta. 2000;1486:28–44. doi: 10.1016/s1388-1981(00)00046-9. [DOI] [PubMed] [Google Scholar]
- 161.Murphy EJ, Barcelo-Coblijn G, Binas B, Glatz JFC. Heart fatty acid uptake is decreased in heart fatty acid binding protein gene-ablated mice. J Biol Chem. 2004;279:34481–34488. doi: 10.1074/jbc.M314263200. [DOI] [PubMed] [Google Scholar]
- 162.Myers-Payne S, Fontaine RN, Loeffler AL, Pu L, Rao AM, Kier AB, Wood WG, Schroeder F. Effects of chronic ethanol consumption on sterol transfer protein in mouse brain. J Neurochem. 1996;66:313–320. doi: 10.1046/j.1471-4159.1996.66010313.x. [DOI] [PubMed] [Google Scholar]
- 163.Avdulov NA, Chochina SV, Myers-Payne S, Hubbell T, Igbavboa U, Schroeder F, Wood WG. Expression and lipid binding of sterol carrier protein-2 and liver fatty acid binding proteins: differential effects of ethanol in vivo and in vitro. In: Riemersma WR, editor. Essential Fatty Acids and Eicosanoids: Invited Papers from the Fourth International Congress. American Oil Chemists Society Press; Champaign, IL: 1998. pp. 324–327. [Google Scholar]
- 164.Mitchell RW, Hatch GM. Fatty acid transport into the brain: of fatty acid fables and lipid tails. Prost Leukot Essen Fatty Acids. 2011;85:293–302. doi: 10.1016/j.plefa.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 165.Bazinet RP, Laye S. PUFA and their metabolites in brain function and disease. Nature Reviews Neuroscience. 2014;15:771–785. doi: 10.1038/nrn3820. [DOI] [PubMed] [Google Scholar]
- 166.Mashek DG. Hepatic fatty acid trafficking: multiple forks in the road. Adv Nutr. 2013;4:697–710. doi: 10.3945/an.113.004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Havel RJ, Felts JM, Van Duyne CM. Formation and fate of endogenous triglycerides in blood plasma of rabbits. J Lipid Res. 1962;3:297–308. [Google Scholar]
- 168.Kohout M, Kohoutova B, Heimberg M. The regulaton of hepatic triglyceride metabolism by free fatty acids. J Biol Chem. 1971;246:5067–5074. [PubMed] [Google Scholar]
- 169.Fu J, Oveisi F, Gaetani S, Lin E, Piomelli D. Oleoylethanolamine, an endogenous PPAR-a agonist, lowers body weight and hyperlipidemia in obese rats. Neuropharmacology. 2005;48:1153. doi: 10.1016/j.neuropharm.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 170.Martin GG, Atshaves BP, McIntosh AL, Mackie JT, Kier AB, Schroeder F. Liver fatty acid binding protein (L-FABP) gene ablation alters liver bile acid metabolism in male mice. Biochem J. 2005;391:549–560. doi: 10.1042/BJ20050296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gajda AM, Storch J. Enterocyte fatty acid binding proteins (FABPs): different functions of liver and intestinal FABPs in the intestine. Prost Leukot Essen Fatty Acids. 2015;93:9–15. doi: 10.1016/j.plefa.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kaczocha M, Glaser ST, Maher T, Clavin B, Hamilton J, O'Rourke J, Rebecchi M, Puopolo M, Owada Y, Thanos PK. Fatty acid binding protein deletion suppresses inflammatory pain through endocannabinoid/N-acylethanolamine-dependent mechanisms. Molecular Pain. 2015;11:52. doi: 10.1186/s12990-015-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Peng X-E, Wu Y-L, Zhu Y, Huang R-D, Lu Q-Q, Lin X. Association of a human FABP1 gene promoter region polymorphism with altered serum triglyceride levels. PLoS ONE. 2015:1–13. doi: 10.137/journal.pone..0139417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Brouillette C, Bose Y, Perusse L, Gaudet D, Vohl MC. Effect of liver fatty acid binding protein (FABP) T94A missense mutation on plasma lipoprotein responsiveness to treatment with fenofibrate. J Hum Gen. 2004;49:424–432. doi: 10.1007/s10038-004-0171-2. [DOI] [PubMed] [Google Scholar]
- 175.Fisher E, Weikert C, Klapper M, Lindner I, Mohlig M, Spranger J, Boeing H, Schrezenmeir J, Doring F. L-FABP T94A is associated with fasting triglycerides and LDL-cholesterol in women. Mol Gen and Metab. 2007;91:278–284. doi: 10.1016/j.ymgme.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 176.Peng X-E, Wu YL, Lu Q-Q, Ju Z-J, Lin X. Two genertic variants in FABP1 and susceptibility to non-alcoholic fatty liver disease in a Chinese population. Gene. 2012 doi: 10.1016/j.gene.2012.03.050. in press. [DOI] [PubMed] [Google Scholar]
- 177.Yamada Y, Kato K, Oguri M, Yoshida T, Yokoi K, Watanabe S, Metoki N, Yoshida H, Satoh K, Ichihara S, Aoyagi Y, Yasunaga A, Park H, Tanaka M, Nozawa Y. Association of genetic variants with atherothrombotic cerebral infarction in Japanese individuals with metabolic syndrome. Int J Mol Med. 2008;21:801–808. [PubMed] [Google Scholar]
- 178.Yang SY, He XY, Schulz H. Fatty acid oxidation in rat brain is limited by the low activity of 3-ketoacyl-coenzyme A thiolase. J Biol Chem. 1987;262:13027–13032. [PubMed] [Google Scholar]
- 179.Higuchi N, Kato M, Tanaka M, et al. Effects of insulin resistance and hepatic lipid accumulation on hepatic mRNA expression levels of apoB, MTP, and L-FABP in non-alcoholic fatty liver disease. Exp and Ther Med. 2011;2:1077–1081. doi: 10.3892/etm.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Charlton M, Viker K, Krishnan A, Sanderson S, Veldt B, Kaalsbeek AJ, Kendrick M, Thompson G, Que F, Swain J, Sarr M. Differential expression of lumican and fatty acid binding protein-1: new insights into the histologic spectrum of nonalcoholic fatty liver disease. Hepatology. 2009;49:1375–1384. doi: 10.1002/hep.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Baumgardner JN, Shankar K, Hennings L, Badger TM, Ronis MJJ. A new model for nonalcoholic steatohepatitis in the rat utilizing total enteral nutrition to overfeed a high-polyunsaturated fat diet. Am J Physiol Gastrointest and Liver Phys. 2007;294:G27–G38. doi: 10.1152/ajpgi.00296.2007. [DOI] [PubMed] [Google Scholar]
- 182.Guzman C, Benet M, Pisonero-Vaquero S, Moya M, Garcia-Mediavilla MV, Martinez-Chantar ML, Gonzalez-Gallego J, Castell JV, Sanchez-Campos S, Jover R. The human liver fatty acid binding protein (FABP1) gene is activated by FOXA1 and PPARa; and repressed by C/EBPa: implicaiton in FABP1 down-regulation in nonalcoholic liver disease. Biochim Biophys Acta. 2013;1831:803–818. doi: 10.1016/j.bbalip.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 183.Rajaraman G, Wang GQ, Yan J, Jiang P, Gong Y, Burczynski FJ. Role of cytosolic liver fatty acid binding protein in hepatocellular oxidative stress: effect of dexamethasone and clofibrate treatment. Mol Cell Biochem. 2007;295:27–34. doi: 10.1007/s11010-006-9268-6. [DOI] [PubMed] [Google Scholar]
- 184.Wang G, Gong Y, Anderson J, Sun D, Minuk G, Robertes MS, Burczynski FJ. Antioxidative function of L-FABP in L-FABP stably transfected Chang liver cells. Hepatology. 2005;42:871–879. doi: 10.1002/hep.20857. [DOI] [PubMed] [Google Scholar]
- 185.Wang G, Shen H, Rajaraman G, Roberts MS, Gong Y, Jiang P, Burczynski FJ. Expression and antioxidant function of liver fatty acid binding protein in normal and bile-duct ligated rats. Eur J Pharm. 2007;560:61–68. doi: 10.1016/j.ejphar.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 186.Yan J, Gong Y, She YM, Wang G, Roberts MS, Burczynski FJ. Molecular mechanism of recombinant liver fatty acid binding protein's antioxidant activity. J Lipid Res. 2009;50:2445–2454. doi: 10.1194/jlr.M900177-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yan J, Gong Y, She YM, Wang G, Robertes MS, Burczynski FJ. Molecular mechanism of recombinant L-FABP's antioxidant activity. J Lipid Res. 2010;50:2445–2454. doi: 10.1194/jlr.M900177-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Smathers RL, Fritz KS, Galligan JJ, Shearn CT, Reigan P, Marks MJ, Petersen DR. Characterizationof 4-HNE modified L-FABP reveals alterations in structural and functional dynamics. PPAR Research. 2012;7:e38459. doi: 10.1371/journal.pone.0038459. doi:10/1371/journal.pone.0038459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fan W, Chen K, Zheng G, Wang W, Teng A, Liu A, Ming D, Yan P. Role of liver fatty acid binding protein in hepatocellular injury: Effect of CrPic treatment. J Inorganic Biochem. 2013;124:46–53. doi: 10.1016/j.jinorgbio.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 190.Anstee QM, Daly AK, Day CP. Genetic modifiers of non-alcoholic fatty liver disease progression. Biochim Biophys Acta. 2011;1812:1557–1566. doi: 10.1016/j.bbadis.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 191.Velkov T, Rimmer KA, Headey SJ. Ligand enhanced expression and in-cell assay of human peroxisome proliferator activated receptor alpha ligand binding domain. Protein Exp Purif. 2010;70:260–269. doi: 10.1016/j.pep.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 192.Desvergne B, Michalik L, Wahli W. Be fit or be sick: peroxisome proliferator-activated receptors are down the road. Mol Endocrinology. 2004;18:1321–1332. doi: 10.1210/me.2004-0088. [DOI] [PubMed] [Google Scholar]
- 193.Naito H, Kamijima M, Yamanoshita O, Nakahara A, Katoh T, Tanaka N, Aoyama T, Gonzalez FJ, Nakajima T. Differential effects of aging, drinking, and exercise on serum cholesterol levels dependent on the PPARA-V277A polymorphism. J Occup Health. 2007;49:353–362. doi: 10.1539/joh.49.353. [DOI] [PubMed] [Google Scholar]
- 194.Galli A, Pinaire J, Fischer M, Dorris R, Crabb DW. The transcriptional and DNA binding activity of peroxisome proliferator activated receptor-α is inhibited by ethanol metabolism. J Biol Chem. 2001;276:68–75. doi: 10.1074/jbc.M008791200. [DOI] [PubMed] [Google Scholar]
- 195.Robitaille J, Brouillette C, Lemieux S, Perusse L, Gaudet D, Vohl MC. Plasma concentrations of apolipoprotein B are modulated by a gene-diet interaction effect between the L-FABP T94A polymorphism and dietary fat intake in French-Canadian men. Mol Gen and Metab. 2004;82:296–303. doi: 10.1016/j.ymgme.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 196.Weikert MO, Loeffelholz Cv, Roden M, Chandramouli V, Brehm A, Nowotny P, Osterhoff MA, Isken F, Spranger J, Landau BR, Pfeiffer A, Mohlig M. A Thr94Ala mutation in human liver fatty acid binding protein contributes to reduced hepatic glycogenolysis and blunted elevation of plasma glucsoe levels in lipid-exposed subjects. Am J Physiol Endocrinol Metab. 2007;293:E1078–E1084. doi: 10.1152/ajpendo.00337.2007. [DOI] [PubMed] [Google Scholar]
- 197.Bu L, Salto LM, De Leon KJ, De Leon M. Polymorphisms in fatty acid binding protein 5 show association with type 2 diabetes. Diabetes Res Clin Prac. 2011;92:82–91. doi: 10.1016/j.diabres.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mansego ML, Martinez F, Martinez-Larrad MT, Zabena C, Rojo G, Morcillo S, Soriguer F, Martin-Escudero JC, Serrano-Rios M, Redon J, Chaves FJ. Common variants of the liver fatty acid binding protein gene influence the risk of Type 2 Diabetes and insulin resistance in Spanish population. PLoS ONE. 2012;7:e31853. doi: 10.1371/journal.pone.0031853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jackevicius CA, Tu JV, Ross JS, Ko DT, Carreon D, Krumholz HM. Use of fibrates in the United States and Canada. JAMA. 2012;305:1217–1224. doi: 10.1001/jama.2011.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lowe JB, Boguski MS, Sweetser DA, Elshourbagy N, Taylor JM, Gordon JI. Human liver fatty acid binding protein: Isolation of a full length cDNA and comparative sequence analyses of orthologous and paralogous proteins. J Biol Chem. 1985;260:3417. [PubMed] [Google Scholar]
- 201.Chan L, Wei CF, Li WH, Yang CY, Ratner P, Pownall H, Gotto AM, Jr, Smith LC. Human liver fatty acid binding protein cDNA and amino acid sequence. Functional and evolutionary implications. J Biol Chem. 1985;260:2629–2632. [PubMed] [Google Scholar]
- 202.Maatman RG, van de Westerlo EM, Van Kuppevelt TH, Veerkamp JH. Molecular identification of the liver- and the heart-type fatty acid-binding proteins in human and rat kidney. Use of the reverse transcriptase polymerase chain reaction. Biochem J. 1992;288:285–290. doi: 10.1042/bj2880285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Farruggia B, Pico GA. Thermodynamic features of the chemical and thermal denaturations of human serum albumin. Int J Biol Macromolecules. 1999;26:317–323. doi: 10.1016/s0141-8130(99)00054-9. [DOI] [PubMed] [Google Scholar]
- 204.Wang Q, Christiansen A, Samiotakis A, Wittung-Stafshede P, Cheung MS. Comparison of chemical and thermal protein denaturation by combination of computational and experimental approaches. II. J Chem Phys. 2011;135:175102. doi: 10.1063/1.3656692. [DOI] [PubMed] [Google Scholar]
- 205.Gariani K, Philippe J, Jornayvaz FR. Non-alcoholic liver disease and insulin resistance: from bench to bedside. Diabetes and Metabolism. 2013;39:16–26. doi: 10.1016/j.diabet.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 206.Puri P, Baillie RA, Wiest MM, Mirshahi F, Choudhury J, Cheung O, Sergeant C, Contos MJ, Sanyal AJ. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46:1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 207.Jolly CA, Hubbell T, Behnke WD, Schroeder F. Fatty acid binding protein: Stimulation of microsomal phosphatidic acid formation. Arch Biochem Biophys. 1997;341:112–121. doi: 10.1006/abbi.1997.9957. [DOI] [PubMed] [Google Scholar]
- 208.Chao H, Zhou M, McIntosh A, Schroeder F, Kier AB. Acyl CoA binding protein and cholesterol differentially alter fatty acyl CoA utilization by microsomal acyl CoA: cholesterol transferase. J Lipid Res. 2003;44:72–83. doi: 10.1194/jlr.m200191-jlr200. [DOI] [PubMed] [Google Scholar]
- 209.Nemecz G, Schroeder F. Selective binding of cholesterol by recombinant fatty acid-binding proteins. J Biol Chem. 1991;266:17180–17186. [PubMed] [Google Scholar]
- 210.Fu T, Choi S-E, Kim D-H, Seok S, Suino-Powell KM, Xu HE, Kemper JK. Aberrantly elevated microRNA-34a in obesity attenuates hepatic responses to FGF19 by targeting a membrane coreceptor b-Klotho. PNAS Early Edition. 2012:1–6. doi: 10.1073/pnas.1205951109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ceccarelli S, Panera N, Gnani D, Nobili V. Dual role of microRNAs in NAFLD. Int J Mol Sci. 2013;14:8437–8455. doi: 10.3390/ijms14048437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Tailleux A, Wouters K, Staels B. Role of PPARs in NAFLD: potential therapeutic targets. Biochim Biophys Acta. 2012;1821:809–818. doi: 10.1016/j.bbalip.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 213.Shearer GC, Savinova OV, Harris WS. Fish oil--How does it reduce plasma triglycerides. Biochim Biophys Acta. 2012;1821:843–851. doi: 10.1016/j.bbalip.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chamouton J, Latruffe N. PPARa/HNF4a interplay on diversified response elements. Relevance in the regulation of liver peroxisomal fatty acid catabolism. Curr Drug Metabolism. 2012;13:1436–1453. doi: 10.2174/138920012803762738. [DOI] [PubMed] [Google Scholar]
- 215.Scorletti E, Byrne CD. Omega-3 fatty acids, hepatic lipid metabolism, and NAFLD. Annu Rev Nutr. 2013;33:231–248. doi: 10.1146/annurev-nutr-071812-161230. [DOI] [PubMed] [Google Scholar]
- 216.Pagliassotti MJ. ER stress in NAFLD. Annu Rev Nutr. 2012;32:17–33. doi: 10.1146/annurev-nutr-071811-150644. [DOI] [PubMed] [Google Scholar]
- 217.Johansen CT, Hegele RA. Allelic and phenotypic spectrum of plasma triglycerides. Biochim Biophys Acta. 2012;1821:833–842. doi: 10.1016/j.bbalip.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 218.Kazantzis M, Stahl A. Fatty acid transport proteins, implications in physiology and disease. Biochim Biophys Acta. 2012;1821:852–857. doi: 10.1016/j.bbalip.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chalasani N, Guo X, Loomba R, Goodarzi MO, et al. Genome wide association study identifies variants associated with histologic features of nonalcoholic fatty liver disease. Gastroenterology. 2010;139:1567–1576. doi: 10.1053/j.gastro.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, et al. Genome wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Greco D, Kotronen A, Westerbacka J, Puig O, et al. Gene expression in human NAFLD. Am J Physiol Gastrointest and Liver Phys. 2008;294:G1281–G1287. doi: 10.1152/ajpgi.00074.2008. [DOI] [PubMed] [Google Scholar]
- 222.Tiniakos DG, Vos MB, Brunt EM. Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu Rev Pathol Mech Dis. 2010;5:145–171. doi: 10.1146/annurev-pathol-121808-102132. [DOI] [PubMed] [Google Scholar]
- 223.Anderson N, Borlak J. Molecular mechanisms and therapeutic targets in steatosis and steatohepatitis. Pharm Rev. 2008;60:311–357. doi: 10.1124/pr.108.00001. [DOI] [PubMed] [Google Scholar]
- 224.Tian Y, Li H, Wang S, Chen Z, Li Z, Zhou H, Ouyang D-S. Association of L-FABP T94A, MTP I128T polymorphisms and hyperlipidemia in Chinese subjects. Lipids. 2014 doi: 10.1007/s11745-015-3990-3. in press. [DOI] [PubMed] [Google Scholar]
- 225.Kawano Y, Cohen DE. Mechanisms of hepatic triglyceride accumulation in non-alcoholic liver disease. J Gastroenterology. 2013;48:434–441. doi: 10.1007/s00535-013-0758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Green CJ, Pramfalk C, Moren KJ, Hodson L. From whole body to cellular models of hepatic triglyceride metabolism: man has got to know his limitations. Am J Physiol Endocrinol Metab. 2015;308:E1–E20. doi: 10.1152/ajpendo.00192.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Sanderson LM, Boekschoten MV, Desvergne B, Muller M, Kersten S. Transcriptional profiling reeals divergent roles of PPARα and PPARβ/δ in regulatoin of gene expression in mouse liver. Physiol Genomics. 2010;41:42–52. doi: 10.1152/physiolgenomics.00127.2009. [DOI] [PubMed] [Google Scholar]
- 228.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 229.Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142:4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- 230.Robins SJ. Fibrates and coronary heart disease reduction. Curr Op in Endocrin & Diabetes. 2002;9:312–322. [Google Scholar]
- 231.Saha SA, Arora RR. Fibrates in the prevention of cardiovascular disease in pateints with type 2 diabetes mellitus--A pooled meta-analysis of randomized placebo-controlled clinical trials. Int J Cardiol. 2010;141:157–166. doi: 10.1016/j.ijcard.2008.11.211. [DOI] [PubMed] [Google Scholar]
- 232.Chapman MJ. Fibrates in 2003: therapeutic action in atherogenic dyslipidemia and future perspectives. Atherosclerosis. 2003;171:1–13. doi: 10.1016/s0021-9150(03)00156-4. [DOI] [PubMed] [Google Scholar]
- 233.Dayspring T, Pokrywka G. Fibrate therapy in patients with metabolic syndrome and diabetes mellitus. Current Atherosclerosis Reports. 2006;8:356–364. doi: 10.1007/s11883-006-0032-x. [DOI] [PubMed] [Google Scholar]
- 234.Staels B, Maes M, Zambon A. Fibrates and future PPARα agonists in the treatment of cardiovascular disease. Nature Clin Pract Cardiovasc Med. 2008;5:542–553. doi: 10.1038/ncpcardio1278. [DOI] [PubMed] [Google Scholar]
- 235.Oosterveer MH, Grefhourst A, van Dijk TH, Havinga R, Staels B, Kuipers F, Groen AK, Reijngoud DJ. Fenofibrate simultaneously induces hepatic fatty acid oxidation, synthesis, and elongation in mice. J Biol Chem. 2009;284:34036–34044. doi: 10.1074/jbc.M109.051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sekiya M, Yhagi N, Matsuzaka T, Najima Y, Nakakuki M, Nagai R, Ishibashi S, Osuga JI, Yamada N, Shimano H. Polyunsaturated fatty acids ameliorate hepatic steatosis in obese mice by SREBP-1 suppression. Hepatology. 2003;38:1529–1539. doi: 10.1016/j.hep.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 237.Froyland L, Madsen L, Vaagenes H, Totland GK, Auwerx J, Kryvi H, Staels B, Berge RK. Mitochondrion is the principal target for nutritional and pharmacological control of triglyceride metabolism. J Lipid Res. 1997;38:1851–1858. [PubMed] [Google Scholar]
- 238.Bijland S, Pieterman EJ, Maas ACE, van der Hoorn JWA, van Erk MJ, van Klinken JB, Havekes LM, van Dijk KW, Princen HM, Rensen PCN. Fenofibrate increases VLDL triglyceride production despite reducing plasma triglyceride levels in APOE3-Leiden. CETP mice. J Biol Chem. 2010;285:25168–25175. doi: 10.1074/jbc.M110.123992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Rioux V, Catheline D, Legrand P. In rat hepatocytes, myristic acid occurs through lipogenesis, palmitic acid shortening and lauric acid elongation. Animal. 2007;1:820–826. doi: 10.1017/S1751731107000122. [DOI] [PubMed] [Google Scholar]
- 240.Jacobson TA. Role of n-3 fatty acids in the treatment of hypertriglyceridemia and cardiovascular disease. Am J Clin Nutr. 2008;87:1981S–1990S. doi: 10.1093/ajcn/87.6.1981S. [DOI] [PubMed] [Google Scholar]
- 241.de Castro GS, Cardoso JFR, Calder PC, Joarao AA, Vannucchi H. Fish oil decreases hepatic lipogenic genes in rats fasted and refed on a high fructose diet. Nutrients. 2015;7:1644–1656. doi: 10.3390/nu7031644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Sun H, Jiang R, Wang S, He B, Zhang Y, Piao D, Yu C, Wu N, Han P. The effect of LXRa, ChREBP and Elovl6 in liver and white adipose tissue on medium- and long-chain fatty acid diet induced insulin resistance. Diab Res and Clin Practice. 2013;102:183–192. doi: 10.1016/j.diabres.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 243.Jump DB, Botolin D, Wang Y, Xu J, Demeure O, Christian B. Docosahexaenoic acid (DHA) and hepatic gene expression. Chem Phys Lip. 2008;153:3–13. doi: 10.1016/j.chemphyslip.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metabolism. 2006;4:107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 245.Nakagawa T, Ge Q, Pawlosky R, Wynn RM, Veech RL, Uyeda K. Metabolite regulation of nucleo-cytosolic trafficking of ChREBP. J Biol Chem. 2013;288:28358–28367. doi: 10.1074/jbc.M113.498550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hostetler HA, Syler LR, Hall LN, Zhu G, Schroeder F, Kier AB. A novel high-throughput screening assay for putative anti-diabetic agents through PPARa interactions. J Biomol Screening. 2008;13:855–861. doi: 10.1177/1087057108323127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hostetler HA, Kier AB, Schroeder F. Very-long-chain and branched-chain fatty acyl CoAs are high affinity ligands for the peroxisome proliferator-activated receptor alpha (PPARalpha) Biochemistry. 2006;45:7669–7681. doi: 10.1021/bi060198l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Staels B. Fibrates in CVD: a step towards personalized medicine. Lancet. 2011;375:1847–1848. doi: 10.1016/S0140-6736(10)60758-1. [DOI] [PubMed] [Google Scholar]
- 249.Wang L, Liu J, Harvey-White J, Zimmer A, Kunos G. Endocannabinoid signaling via cannabinoid receptor 1 is invovled in ethanol prference and its age-dpendent decline in mice. Proc Natl Acad Sci U S A. 2003;100:1393–1398. doi: 10.1073/pnas.0336351100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Jian W, Edom R, Weng N, Zannikos P, Zhang Z, Wang H. Validation and application of an LC-MS/MS method for quantitation of three fatty acid ethanolamides as biomarkers for fatty acid hydrolase inhibitionin human placenta. J Chrom B. 2010;878:1687–1699. doi: 10.1016/j.jchromb.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 251.Luxon BA, Weisiger RA. Sex differences in intracellular fatty acid transport: role of cytoplasmic binding proteins. Am J Physiol. 1993;265:G831–41. doi: 10.1152/ajpgi.1993.265.5.G831. [DOI] [PubMed] [Google Scholar]
- 252.Lotersztain S, Mallat A. The endocannabinoid system as a therapeutic target for liver diseases. CMReJournal. 2008;1:36–40. [Google Scholar]
- 253.Mendez-Sanchez N, Zamora-Valdes D, Pichardo-Bahena R, Barredo-Prieto B, et al. Endocannabinoid receptor CB2 in nonalcoholic fatty liver disease. Liver International. 2006;27:215–219. doi: 10.1111/j.1478-3231.2006.01401.x. [DOI] [PubMed] [Google Scholar]