Abstract
Cloned DNA fragments from 14 characterized maize genes and 91 random fragments used for genetic mapping in maize were tested for their ability to hybridize and detect restriction fragment length polymorphisms in sorghum and other related crop species. Most DNA fragments tested hybridized strongly to DNA from sorghum, foxtail millet, Johnsongrass, and sugarcane. Hybridization to pearl millet DNA was generally weaker, and only a few probes hybridized to barley DNA under the conditions used. Patterns of hybridization of low-copy sequences to maize and sorghum DNA indicated that the two genomes are very similar. Most probes detected two loci in maize; these usually detected two loci in sorghum. Probes that detected one locus in maize generally detected a single locus in sorghum. However, maize repetitive DNA sequences present on some of the genomic clones did not hybridize to sorghum DNA. Most of the probes tested detected polymorphisms among a group of seven diverse sorghum lines tested; over one-third of the probes detected polymorphism in a single F2 population from two of these lines. Cosegregation analysis of 55 F2 individuals enabled several linkage groups to be constructed and compared with the linkage relationships of the same loci in maize. The linkage relationships of the polymorphic loci in the two species were usually conserved, but several rearrangements were detected.
Full text
PDF
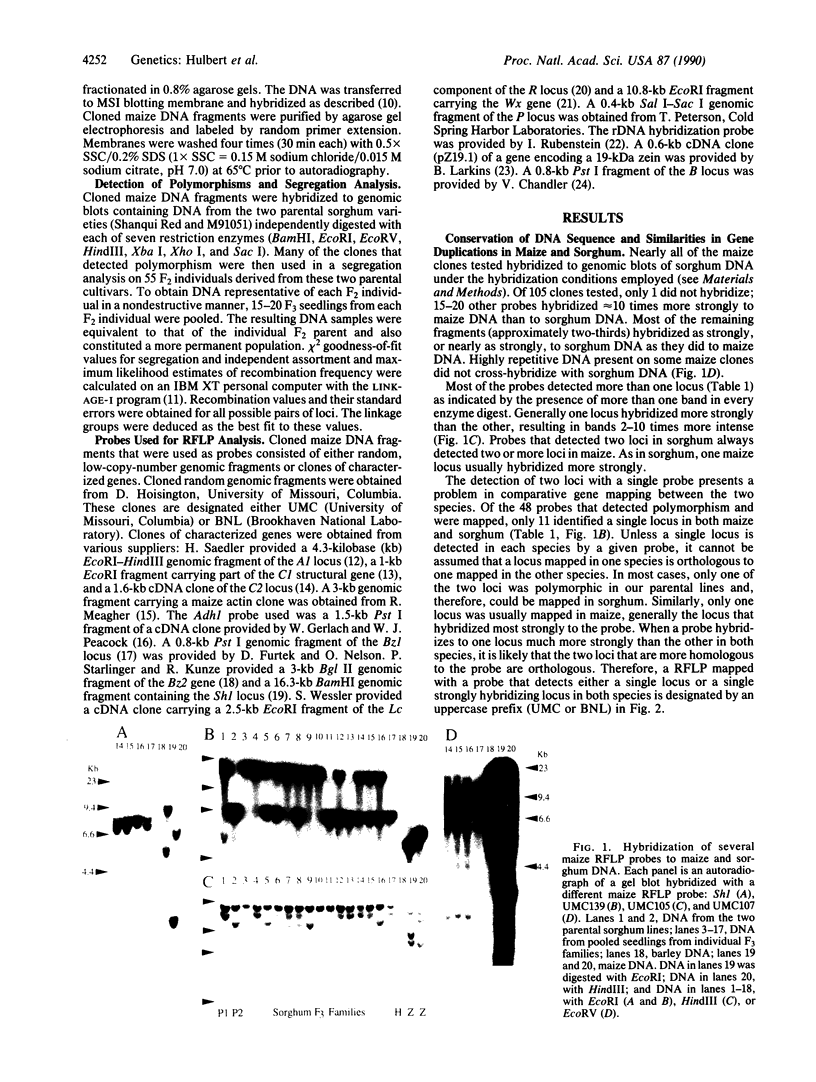
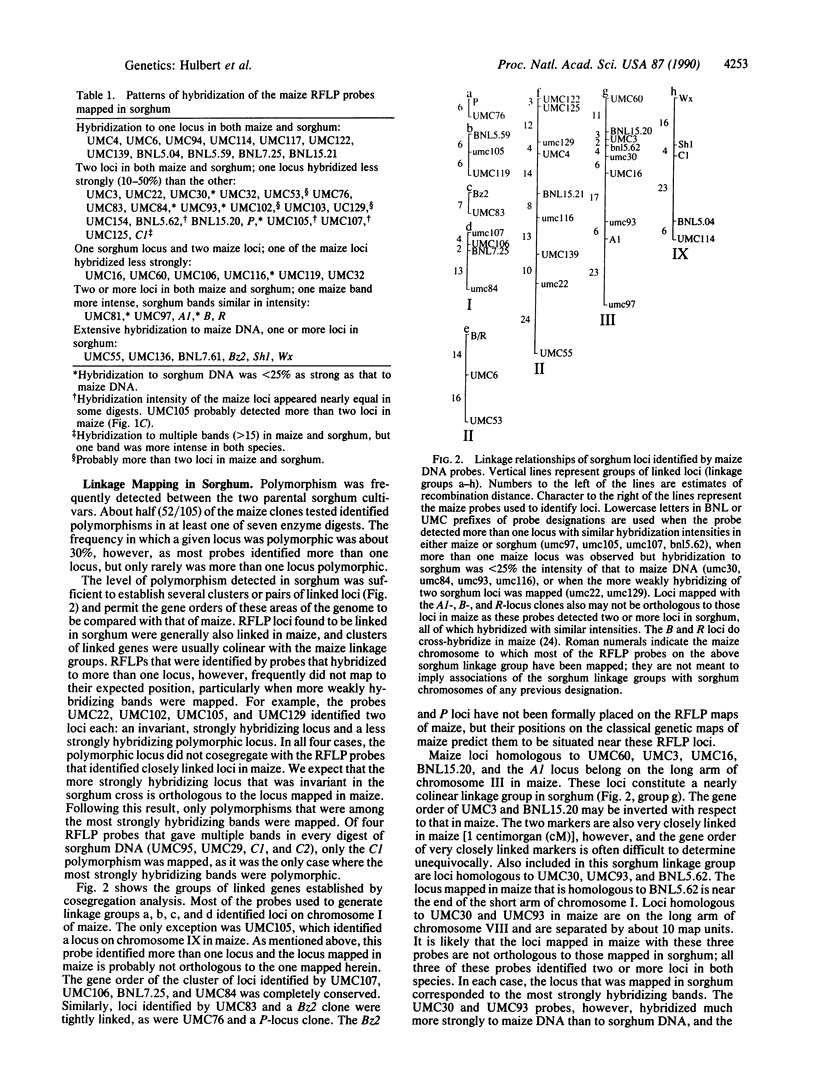


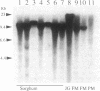
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L. Transposable element Mu1 is found in multiple copies only in Robertson's Mutator maize lines. J Mol Appl Genet. 1984;2(6):519–524. [PubMed] [Google Scholar]
- Bonierbale M. W., Plaisted R. L., Tanksley S. D. RFLP Maps Based on a Common Set of Clones Reveal Modes of Chromosomal Evolution in Potato and Tomato. Genetics. 1988 Dec;120(4):1095–1103. doi: 10.1093/genetics/120.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr B., Burr F. A., Thompson K. H., Albertson M. C., Stuber C. W. Gene mapping with recombinant inbreds in maize. Genetics. 1988 Mar;118(3):519–526. doi: 10.1093/genetics/118.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler V. L., Radicella J. P., Robbins T. P., Chen J., Turks D. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell. 1989 Dec;1(12):1175–1183. doi: 10.1105/tpc.1.12.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. V., Furtek D. B., Nelson O. E. Cloning of the bronze locus in maize by a simple and generalizable procedure using the transposable controlling element Activator (Ac). Proc Natl Acad Sci U S A. 1984 Jun;81(12):3825–3829. doi: 10.1073/pnas.81.12.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser M., Weck E., Döring H. P., Werr W., Courage-Tebbe U., Tillmann E., Starlinger P. Genomic clones of a wild-type allele and a transposable element-induced mutant allele of the sucrose synthase gene of Zea mays L. EMBO J. 1982;1(11):1455–1460. doi: 10.1002/j.1460-2075.1982.tb01337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach W. L., Pryor A. J., Dennis E. S., Ferl R. J., Sachs M. M., Peacock W. J. cDNA cloning and induction of the alcohol dehydrogenase gene (Adh1) of maize. Proc Natl Acad Sci U S A. 1982 May;79(9):2981–2985. doi: 10.1073/pnas.79.9.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helentjaris T., Weber D., Wright S. Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics. 1988 Feb;118(2):353–363. doi: 10.1093/genetics/118.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig S. R., Habera L. F., Dellaporta S. L., Wessler S. R. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M. D., Hunter B., Phillips R. L., Rubenstein I. The structure of the maize ribosomal DNA spacer region. Nucleic Acids Res. 1986 Jun 25;14(12):4953–4968. doi: 10.1093/nar/14.12.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly C., Shepherd N. S., Pereira A., Schwarz-Sommer Z., Bertram I., Robertson D. S., Peterson P. A., Saedler H. Molecular cloning of the a1 locus of Zea mays using the transposable elements En and Mu1. EMBO J. 1985 Apr;4(4):877–882. doi: 10.1002/j.1460-2075.1985.tb03713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J., Wienand U., Peterson P. A., Saedler H. Molecular cloning of the c locus of Zea mays: a locus regulating the anthocyanin pathway. EMBO J. 1986 May;5(5):829–833. doi: 10.1002/j.1460-2075.1986.tb04291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K., Devereux J., Wilson D. R., Sheldon E., Larkins B. A. Cloning and sequence analysis reveal structural variation among related zein genes in maize. Cell. 1982 Jul;29(3):1015–1026. doi: 10.1016/0092-8674(82)90465-2. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., Allard R. W. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984 Dec;81(24):8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D. M., Hightower R. C., Meagher R. B. Genes encoding actin in higher plants: intron positions are highly conserved but the coding sequences are not. J Mol Appl Genet. 1983;2(1):111–126. [PubMed] [Google Scholar]
- Shure M., Wessler S., Fedoroff N. Molecular identification and isolation of the Waxy locus in maize. Cell. 1983 Nov;35(1):225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- Suiter K. A., Wendel J. F., Case J. S. LINKAGE-1: a PASCAL computer program for the detection and analysis of genetic linkage. J Hered. 1983 May-Jun;74(3):203–204. doi: 10.1093/oxfordjournals.jhered.a109766. [DOI] [PubMed] [Google Scholar]
- Tanksley S. D., Bernatzky R., Lapitan N. L., Prince J. P. Conservation of gene repertoire but not gene order in pepper and tomato. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6419–6423. doi: 10.1073/pnas.85.17.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theres N, Scheele T, Starlinger P. Cloning of the Bz2 locus of Zea mays using the transposable element Ds as a gene tag. Mol Gen Genet. 1987 Aug;209(1):193–197. doi: 10.1007/BF00329858. [DOI] [PubMed] [Google Scholar]