Abstract
Fluorescent lipids are important tools for live imaging in cell culture and animal models, yet their metabolism has not been well-characterized. Here we describe a novel combined HPLC and LC-MS/MS method developed to characterize both total lipid profiles and the products of fluorescently labeled lipids. Using this approach, we found that lipids labeled with the fluorescent tags, 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene (BODIPY FL), 4,4-difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-s-indacene [BODIPY(558/568)], and dipyrrometheneboron difluoride undecanoic acid (TopFluor) are all metabolized into varying arrays of polar and nonpolar fluorescent lipid products when they are fed to larval zebrafish. Quantitative metabolic labeling experiments performed in this system revealed significant effects of total dietary lipid composition on fluorescent lipid partitioning. We provide evidence that cholesterol metabolism in the intestine is important in determining the metabolic fates of dietary FAs. Using this method, we found that inhibitors of dietary cholesterol absorption and esterification both decreased incorporation of dietary fluorescent FAs into cholesterol esters (CEs), suggesting that CE synthesis in enterocytes is primarily responsive to the availability of dietary cholesterol. These results are the first to comprehensively characterize fluorescent FA metabolism and to demonstrate their utility as metabolic labeling reagents, effectively coupling quantitative biochemistry with live imaging studies.
Keywords: diet and dietary lipids, fatty acid/metabolism, lipids/chemistry, mass spectrometry, high-performance liquid chromatography, cholesterol/metabolism, zebrafish, isotopic tracers, BODIPY, charged aerosol detection
When dietary FAs are absorbed by intestinal enterocytes, those that are not oxidized are incorporated into complex lipids (lipid molecules containing more than one acyl chain and/or sterol group), including phospholipids, triglycerides (TGs), and cholesterol esters (CEs). This process involves a network of transporters, enzymes, and cellular components, the roles and regulation of which are still being characterized (1–3). The processes by which dietary FAs are distributed among various complex lipid classes and how assembly of complex lipids from dietary FAs may be influenced by other nutrients, including dietary cholesterol, are not fully understood.
To investigate dietary FA metabolism in a quantitative manner, it was necessary to develop an animal model wherein individual FAs could be studied in the context of the digestion and absorption of a physiologically relevant mixed-lipid diet. While cell culture is a well-established model system for the study of lipid metabolism, it cannot replicate the digestive and metabolic physiology of a whole vertebrate animal with a functional liver, intestine, pancreas, and microbiota, such as the larval zebrafish (4–11). An additional strength of this model is that live imaging and biochemical experiments may be performed in parallel using the same fluorescent lipid reagents and experimental system (12–21). Many publications using fluorescent lipid reagents did not examine the metabolism of these labels [with some asserting that some or all fluorescent FAs are not metabolizable (22–24)], but prior work has shown that 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene (BODIPY® FL)-labeled FAs may be esterified and incorporated into products, including phospholipids, TGs, and CEs (15, 25–29). However, with the exception of two HPLC-fluorescence profiles of the phospholipid products of BODIPY FL-C12 (BODIPY FL-dodecanoic acid) (25, 26), all published characterization of the metabolic products of fluorescent lipids has been performed with TLC, which, unlike HPLC, does not resolve individual lipid species within each class. For this study, we developed a novel total-lipid HPLC method that allowed the complex lipid product profiles of BODIPY FL-labeled FAs and other fluorescent lipids commonly used in imaging experiments to be described in detail and facilitated investigation of the role of nutritional context in how fluorescent lipids are metabolized.
Emulsification of fluorescent FAs into chicken egg yolk [a high-fat/high-cholesterol (HF/HC) meal relative to standard larval fish diets] has been shown to promote their metabolism when they are fed to larval zebrafish, but how the nutritional context in which fluorescent lipids are delivered regulates their partitioning has not been characterized (15). There is evidence from mammalian models for interaction between the dietary cholesterol and FA uptake and metabolism pathways: CEs are synthesized in enterocytes and dietary long-chain FAs enhance dietary cholesterol uptake in rats, but the mechanisms for this interaction remain unknown (30, 31). We applied our novel fluorescent lipid profiling method to examine the effect of the metabolic availability of dietary cholesterol on intestinal CE synthesis in order to better understand the role of cholesterol regulation mechanisms in dietary FA metabolism.
In this study, we describe total lipid profiles of the 6 days post-fertilization (dpf) larval zebrafish after both low-fat/low-cholesterol (LF/LC) and HF/HC meals, and the fluorescent product profiles synthesized from the most commonly used commercially available fluorescent lipids when they are fed to larval zebrafish in several different diets. This thorough characterization of the metabolism of fluorescent lipids will greatly expand the depth of physiological information that may be gained from using these reagents in well-established live fluorescent imaging methods. We also demonstrate a novel application of fluorescent FAs as metabolic labeling reagents by showing that pharmaceutical manipulations of intestinal cholesterol metabolism result in significant changes in dietary fluorescent FA partitioning, suggesting coupling between dietary cholesterol uptake and esterification.
MATERIALS AND METHODS
Zebrafish
Embryos were collected from group matings of wild-type (AB) Danio rerio (zebrafish) and raised to the 6 dpf larval stage at 28°C on a 14:10 on:off light cycle in embryo media. All experiments with zebrafish (protocol #139) were approved by the Carnegie Institution Department of Embryology IACUC.
Food preparation and feeding
The fluorescent FAs, 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoic acid (BODIPY® FL-C5) (Thermo Fisher Scientific), 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoic acid (BODIPY® FL-C12) (Thermo Fisher Scientific), 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-hexadecanoic acid (BODIPY® FL-C16) (Thermo Fisher Scientific), 4,4-difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-s-indacene-3-dodecanoic acid [BODIPY®(558/568)-C12] (Thermo Fisher Scientific), and dipyrrometheneboron difluoride undecanoic acid (TopFluor®-C11) (Avanti Polar Lipids) were administered to larval zebrafish in HF/HC meals: Solutions of 5% chicken egg yolk liposomes containing 4 μg/ml fluorescent FA were prepared as previously described (15). Larval zebrafish (6 dpf) were immersed in these solutions for 2 h, protected from light in a shaking incubator at 30°C and 30 rpm. After feeding, larvae were rinsed in fresh embryo media and screened for full intestines under a stereomicroscope. Larvae that had eaten were maintained in embryo media at room temperature for a 4–24 h chase period (measured from the start of feeding) depending on the experiment. Samples of 10–100 pooled larvae were collected for lipid extraction and stored dry at −80°C.
To compare the metabolism of BODIPY FL-C12 and palmitate, larvae were fed equimolar amounts of BODIPY FL-C12 (9.56 μM) and palmitic acid (0.85 μM 3H-palmitic acid and 8.71 μM unlabeled palmitic acid) in a HF/HC meal for 2 h at 25°C and 30 rpm. Larvae were maintained at 25°C in embryo media for 18 h, and then samples of 25 larvae each were taken for lipid extraction.
For experiments in which lipids were extracted from larval zebrafish intestines, larvae were anesthetized in tricaine (3-amino benzoic acid ethyl ester) and mounted in 3% methylcellulose. Intestines were removed using tungsten wire dissecting tools prepared according to published methods (32) and stored in lipid extraction buffer (1 mM EDTA, 20 mM Tris-Cl) at −80°C. Alternately, larvae could be fixed 24–72 h at 4°C in 4% paraformaldehyde, transferred to PBS, and then dissected. No differences in lipid profiles, as analyzed by HPLC-charged aerosol detection (CAD), were observed between fixed and unfixed samples. All data shown are from lipid extracts of unfixed larvae.
Preparation of LF/LC and low-fat/high-cholesterol (LF/HC) foods with fluorescent lipids was adapted from published methods (21). For the LF/LC diet, 2.5 nmol of fluorescent lipid {BODIPY FL-C12, BODIPY(558/568)-C12, cholesterol BODIPY FL-C12 (Thermo Fisher Scientific), or cholesterol cholesterol 4,4-difluoro-5-(2-pyrrolyl)-4-bora-3a,4a-diaza-s-indacene-3-undecanoate [BODIPY(576/589)-C11]; Molecular Probes, discontinued} were diluted in 0.5 ml diethyl ether, combined with 0.1 g Sera® Micron larval fish food, and solvents were allowed to evaporate. For LF/HC diets, 4 mg of cholesterol (4% w/w) were added as well. Larval zebrafish (5 dpf) were fed LF/LC or LF/HC meals twice per day for 2 days at 25°C, and then LF/LC or LF/HC meals with fluorescent lipid for 3 days at 25°C protected from light. Food was then withdrawn and samples of 20–75 larvae were taken after an overnight chase to allow larvae to excrete any unabsorbed food remaining in the intestinal lumen. Larger numbers of larvae per sample were required for experimental groups fed red fluorescent lipids.
Sample preparation, HPLC, and data analysis
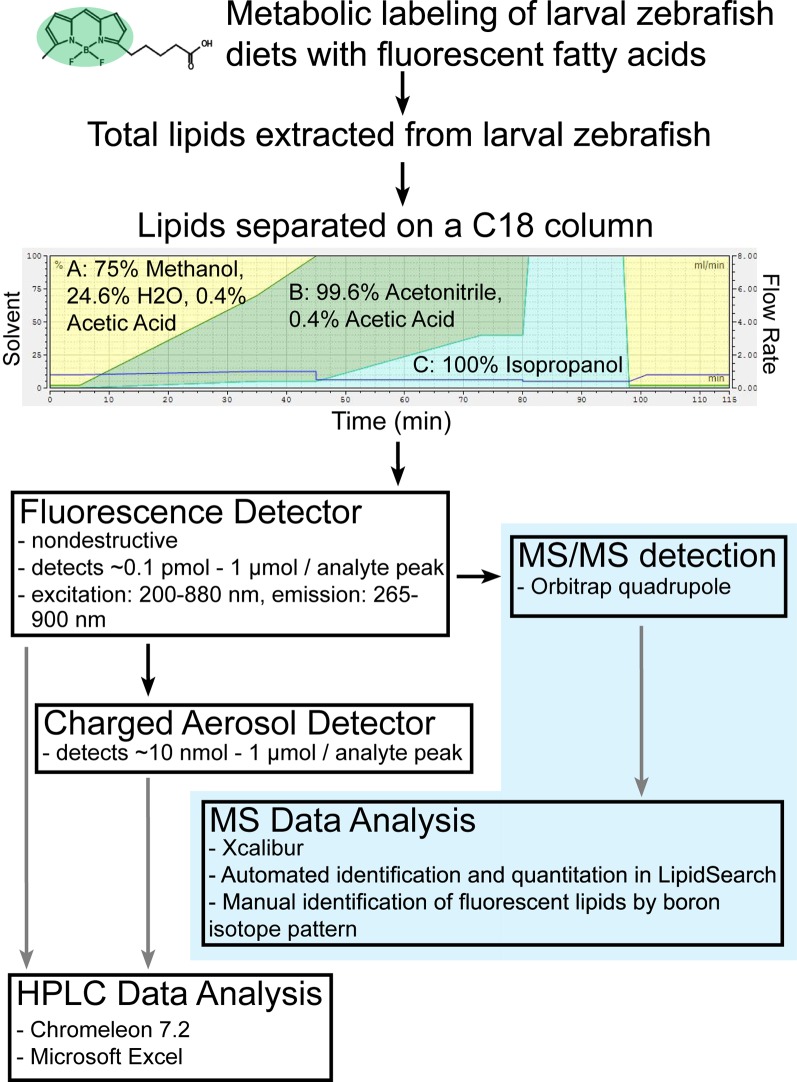
Lipids were extracted from frozen larvae samples by a Bligh-Dyer procedure, dried under vacuum, and resuspended in 100 μl HPLC-grade isopropanol per sample as the HPLC injection solvent. The components of each sample were separated and detected by an HPLC system using a LPG-3400RS quaternary pump, WPS-3000TRS autosampler (maintained at 20°C), TCC-3000RS column oven (maintained at 40°C), Accucore C18 column (150 × 3.0 mm, 2.6 μm particle size), FLD-3100 fluorescence detector (8 μl flow cell maintained at 45°C), and a Dionex Corona Veo charged aerosol detector (all from Thermo Fisher Scientific). Component peaks were resolved over an 80 min time range in a multistep mobile phase gradient as follows: 0–5 min = 0.8 ml/min in 98% mobile phase A (methanol-water-acetic acid, 750:250:4) and 2% mobile phase B (acetonitrile-acetic acid, 1,000:4); 5–35 min = 0.8–1.0 ml/min, 98–30% A, 2–65% B, and 0–5% mobile phase C (2-propanol); 35–45 min = 1.0 ml/min, 30–0% A, 65–95% B, and 5% C; 45–73 min = 1.0 ml/min, 95–60% B and 5–40% C; and 73–80 min = 1.0 ml/min, 60% B, and 40% C. (HPLC-grade acetic acid and 2-propanol were purchased from Fisher Scientific and HPLC-grade methanol and acetonitrile were purchased from Sigma-Aldrich.) The following adjustments were made to the solvent gradients for better separation of nonpolar products of BODIPY FL-C5 and BODIPY FL-C12: In the “B-C5” instrument method, the flow rate was decreased to 0.5 ml/min at 40–80 min, and in the “B-C12” method, the flow rate was decreased to 0.5 ml/min at 45–80 min. Between 5 and 99 μl were injected per sample to produce peak shapes suitable for quantitation. For HPLC-fluorescence, the following excitation and detection wavelengths were used: 576/596 nm for products of cholesterol BODIPY(576/598)-C11, 558/578 nm for products of BODIPY(558/568)-C12, and 488/512 nm for products of BODIPY FL(503/512) and TopFluor-labeled lipids. It was not possible to use excitation and detection settings that matched the maxima for each BODIPY-lipid because, for the FLD-3100 fluorescent HPLC detector, these settings must be at least 20 nm apart. Optimal fluorescent detector settings were determined empirically (data not shown).
Chromatographic peaks produced by cholesterol, as well as several fluorescent and nonfluorescent CEs, were identified by comparison with standards (Sigma-Aldrich and Thermo Fisher Scientific). Non-cholesterol-containing lipids were identified and quantitated by MS as described below. Additionally, acetone precipitation [adapted from (33)] was performed on some samples prior to HPLC so that fluorescent analytes that could not be precisely identified by MS could be classified as polar or nonpolar lipids. Quantitation of lipid species was performed using Chromeleon 7.2 (Thermo Fisher Scientific, Germering, Germany). Peak baselines were drawn manually and areas were determined automatically. Further data analysis (where applicable) was performed using Microsoft Excel with the StatPlus package.
To compare the metabolic products of BODIPY-C12 and 3H-palmitic acid, lipids extracted from larval zebrafish fed these FAs simultaneously, as described above, were subjected to HPLC-fluorescence analysis using the B-C12 method, after which the eluent was collected in 1 min increments (Model 2110 Fraction Collector; Bio-Rad Laboratories) and radioactivity was quantitated using a liquid scintillation counter (Tri-Carb 2810 TR liquid scintillation analyzer, Perkin-Elmer; 3a70b scintillation fluid, Research Products International Corp.). Fluorescent chromatograms were quantitated in Chromeleon 7.2 by simulated fraction collection: a flat baseline was drawn from 0 to 80 min and each 1 min area was assigned as a “peak” by placing a delimiter perpendicular to the baseline.
LC-MS and data analysis
For LC-MS experiments, chromatography was performed using the procedure and equipment described above, with eluent directed to a mass spectrometer in place of the charged aerosol detector after exiting the fluorescence detector. High-resolution accurate-mass data were recorded on a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a heated electrospray-ionization probe (HESI-II). Data were acquired in positive and negative polarity with the following source parameters: spray voltage, 3.5 kV for positive polarity and 3.2 kV for negative polarity; sheath gas flow, 70 (arbitrary units); auxiliary gas flow, 25 (arbitrary units); sweep gas flow, 1 (arbitrary units); auxiliary gas heater temperature, 400°C; capillary temperature, 285°C; S-Lens RF level, 45%. The instrument was operated with external mass calibration without use of a lock mass. Mass spectra were recorded in data-dependent MS/MS mode as a Top-12 experiment with the following acquisition settings: mass resolution, 140,000 for full-MS and 17,500 for MS/MS; full-MS scan range, 200–1,600 Da; quadrupole isolation window, 1.2 Da; normalized collision energy, stepped from 20% to 30% to 40%; exclusion mass list, 200 entries (created individually for each polarity mode from injection solvent blank runs). Data acquisition and quantitation was performed using Xcalibur (version 3.0.63; Thermo Fisher Scientific), and identification of nonfluorescent lipid analytes was performed with LipidSearch (version 4.0; MKI, Tokyo, Japan). Fluorescent lipids were identified manually by using Xcalibur to search for analyte peaks containing boron, as this element lends a unique isotopic signature to the BODIPY FL fluorescent tag. The two most common boron isotopes, 10B and 11B, exist naturally and in BODIPY FL-FAs (verified experimentally using standards; data not shown) in a 1:10 to 1:4 ratio and have a Δm/z of 0.9964 amu. Pairs of analytes that fit this signature, eluted at the same retention time and specific to a single fluorescent HPLC peak, were identified as specific BODIPY-lipids by comparing the experimental m/z to the expected m/z of each of a range of potential products of BODIPY FL-C5, -C12, and -C16. Expected m/z was calculated from molecular formulas using the Lipid MAPS® exact mass tool (http://www.lipidmaps.org/tools/structuredrawing/masscalc.php).
Confocal imaging
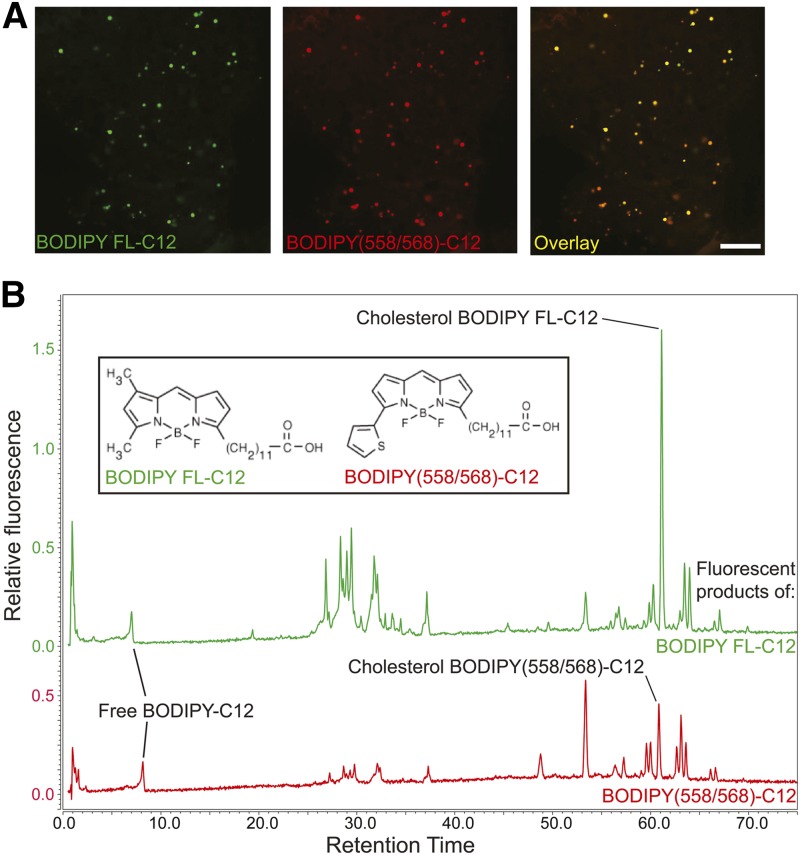
Larval zebrafish were fed both BODIPY FL-C12 and BODIPY(558/568)-C12 emulsified in 5% chicken egg yolk at 4 μg/ml for 2 h at 30°C, as described above, and then transferred to fresh embryo media and incubated at room temperature. Live confocal imaging was performed 6 h after removal of larvae from the HF/HC food (Fig. 4). Larvae were anesthetized with tricaine and live mounted for imaging in 3% methylcellulose under a coverslip at room temperature. Fluorescent images were acquired with a Leica TCS SP5 II confocal microscope equipped with an Argon laser, Leica 63×/1.4 oil-immersion objective, and Leica Application Suite Advanced Fluorescence 2.7.3.9723 image acquisition software. Images were adjusted for brightness and contrast using Fiji image analysis software (National Institutes of Health).
Fig. 4.
BODIPY FL-C12 and BODIPY(558/568)-C12 are processed similarly to each other when fed to larval zebrafish in a HF/HC meal. A: When BODIPY FL-C12 and BODIPY(558/568)-C12 are fed simultaneously in a HF/HC meal, they label the same subcellular structures in the larval zebrafish liver 8 h postfeeding. Scale bar: 25 μm. B: Similar arrays of phospholipid, TG, and CE products of BODIPY FL-C12 and BODIPY(558/568)-C12 are observed 8 h postfeeding, though the products form in different proportions to each other. Results are representative of six samples per group.
Pharmaceutical treatments
Larval zebrafish were treated with ezetimibe using a protocol adapted from published methods, which demonstrated that ezetimibe blocks dietary cholesterol uptake in larval zebrafish (34, 35): at 5 dpf, larvae were immersed in a 5 μM ezetimibe (SCH58053; Santa Cruz Biotech) and 0.1% ethanol (vehicle) solution and maintained at 25°C for 20 h. While being fed experimental diets, larvae were treated with 10 μM ezetimibe. Following meals, larvae were returned to a 5 μM ezetimibe solution until samples were taken for lipid extraction.
A similar protocol for treatment of larval zebrafish with an ACAT inhibitor (CAY10486, N-[3-(4-hydroxyphenyl)-1-oxo-2-propenyl]L-phenylalanine methyl ester; Cayman Chemical) was developed from a published method for ACAT inhibition in embryos (17). Beginning at 5 dpf, larvae were immersed in a 100 μM ACAT inhibitor and 0.5% DMSO (vehicle) solution and maintained at 25°C for 20 h. This ACAT inhibitor concentration was maintained during and after feeding, until samples were taken.
RESULTS
A comprehensive total lipid HPLC method facilitated detection and quantitation of all classes of lipids in a single sample
In order to obtain the greatest possible depth of information from metabolic labeling with dietary fluorescent FAs, it was crucial to understand the lipidomic context surrounding these metabolic tracers. Lipid profiling by HPLC with various detection methods is a well-established practice (36–42) that allows for relatively high-throughput and low-cost experiments (when compared with LC-MS). However, precise identification of complex lipids resolved by HPLC using standards is often impractical, as there are thousands of possible unique phospholipid species and tens of thousands of possible TGs. For this reason lipid researchers often turn to MS-based lipidomics, which can provide precise identification (if high resolution accurate mass detection methods are used) and quantitation of complex lipids, but often at the cost of limiting the number of samples and replicates that may be analyzed. Here we describe a combined HPLC-CAD and LC-MS workflow in which a lipidomic data set obtained from a short series of LC-MS experiments is used to interpret results from a large number of HPLC experiments, retaining the high-throughput/low-cost advantage of HPLC, while enhancing the type and quality of data that may be obtained from HPLC lipidomics. A multistep gradient HPLC procedure for analysis of the larval zebrafish lipidome was designed to resolve the components of a mixed sample consisting of lipids of a wide range of molecular weights, degrees of polarity, and degrees of saturation. A single LC-MS/MS experiment (n = 3 biological replicates) was then performed using the chromatography component of our established HPLC method in order to identify the specific lipid contents of each analyte peak detected by HPLC-CAD/fluorescence (Fig. 1). The lipidomic data set this experiment yielded (supplemental Table S1) could then be used to interpret the results of any HPLC experiment performed with the same instrument method. Cholesterol and most CEs were not detected by the MS method we employed, but were detected by HPLC-CAD and identified by comparison with purchased standards (Sigma-Aldrich). The composition of each lipid peak detected by HPLC-CAD is summarized in supplemental Table S2. The total FA composition of larval zebrafish was similar in groups fed a single LF/LC or HF/HC meal, despite differences in dietary FA composition. FA composition analysis of only the TGs showed that oleic acid was enriched 2.3-fold and linoleic acid was enriched 1.9-fold in the TGs of larvae fed a HF/HC meal (vs. LF/LC), and that DHA was enriched 6.2-fold in larvae fed a LF/LC meal (vs. HF/HC) (supplemental Table S3). This was consistent with differences in dietary FA composition, where the HF/HC diet was 35% oleic acid (vs. 14% in LF/LC) and 16.5% linoleic acid (vs. 9% in LF/LC), and the LF/LC diet was 8.6% DHA (vs. 0.5% in HF/HC). Less prevalent species enriched in the TGs of LF/LC-fed larvae (vs. larvae fed the HF/HC meal) included 20:5, 16:1, and 20:4. No other changes in FA composition greater than 1.5-fold were observed between the HF/HC and LF/LC groups in FA species accounting for >0.3% of the total in TGs.
Fig. 1.
A combined HPLC-CAD and LC-MS/MS workflow for total lipid profiling of larval zebrafish. Once an HPLC method was established that resolved lipids of all classes present in larval zebrafish, the instrument was temporarily reconfigured to route sample flow to a quadrupole-Orbitrap mass spectrometer instead of the charged aerosol detector. Data obtained from a series of LC-MS/MS experiments with exact mass detection was then used to identify the components of peaks observed by HPLC-CAD/fluorescence. Through this combined workflow, peak composition data from a single LC-MS/MS experiment greatly enhances the depth of information that may be obtained from all future HPLC experiments performed using the same instrument method.
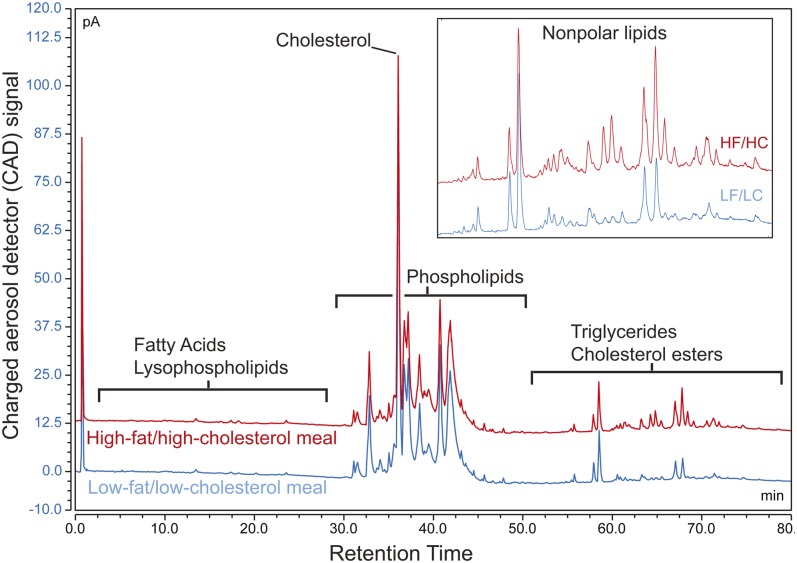
HPLC-CAD data revealed that the whole-body lipid content of larval zebrafish was enriched in a variety of TGs following a single HF/HC meal, while other lipid species were unchanged (Fig. 2). The TG content of larval zebrafish fed a HF/HC meal was increased 10-fold (on average) over larval zebrafish fed a LF/LC meal when calculated from the LC-MS/MS data (n = 3). A similar change in TG levels plus an increase in CE levels was observed when HPLC-CAD data were processed through the combined HPLC-CAD/LC-MS data analysis workflow (Table 1). The combined workflow was advantageous over simple quantitation of HPLC-CAD data because it allowed for analysis of lipid species that were not identified by HPLC-CAD (e.g., ceramides, monoglycerides, and diglycerides), and also allowed quantitation of free cholesterol and two more CE species than what could be detected in larval zebrafish samples analyzed with MS/MS detection (Table 1; supplemental Tables S1, S2).
Fig. 2.
Multistep gradient HPLC with CAD facilitates resolution and quantitation of all classes of lipids in a single underivatized sample. Several TG species are increased in larval zebrafish following a single HF/HC meal, while other lipid classes remain unchanged. Similar results are obtained when lipid class distribution is quantitated directly from the MS dataset using LipidSearch, and when quantitation of HPLC-CAD analyte peaks (identified previously using LC-MS/MS) is performed in Chromeleon (Table 1). The HF/HC (red) trace is offset by 12.5 pA (picoamperes). This chromatogram represents results from three independent experiments.
TABLE 1.
Lipid class composition of larval zebrafish fed LF/LC or HF/HC diets
| MS (% of total lipid) | HPLC-CAD (% of total lipid) | Combined Workflow (% of total lipid) | ||||
| Class | LF/LC | HF/HC | LF/LC | HF/HC | LF/LC | HF/HC |
| FA | 0.9 ± 0.3 | 0.8 ± 0.2 | 1.1 ± 0.8 | 0.6 ± 0.2 | 0.2 ± 0.1 | 0.2 ± 0.1 |
| Phospholipid | 94.3 ± 29.9 | 90.5 ± 30.5 | 62.1 ± 36.0 | 62.9 ± 20.8 | 62.9 ± 34.5 | 67.5 ± 18.5 |
| TG | 0.5 ± 0.1 | 4.8 ± 1.1 | 6.1 ± 3.6 | 12.6 ± 3.5 | 0.7 ± 0.3 | 7.4 ± 1.9 |
| Diglyceride | 0.1 ± 0.1 | 0.1 ± 0.0 | —a | — | 0.8 ± 0.4 | 0.6 ± 0.2 |
| Monoglyceride | 0.2 ± 0.1 | 0.1 ± 0.1 | — | — | 13.3 ± 10.5 | 4.9 ± 2.8 |
| Lysophospholipid | 0.6 ± 0.2 | 0.6 ± 0.2 | 1.4 ± 1.1 | 0.4 ± 0.3 | 1.2 ± 0.7 | 0.8 ± 0.3 |
| Ceramide | 0.7 ± 0.2 | 0.6 ± 0.2 | — | — | 0.7 ± 0.3 | 0.6 ± 0.2 |
| CE | 0.0 ± 0.0 | 0.0 ± 0.0 | 3.1 ± 1.2 | 5.4 ± 1.4 | 1.7 ± 0.5 | 2.8 ± 0.8 |
| Cholesterol | — | — | 26.3 ± 6.3 | 17.9 ± 2.4 | 16.9 ± 1.1 | 13.9 ± 1.4 |
| Other | 2.7 ± 1.0 | 2.6 ± 0.7 | — | — | 11.6 ± 0.9 | 1.4 ± 0.4 |
Lipid class composition was determined by LC-MS/MS, HPLC-CAD, or the combined workflow described in Fig. 1. Lipids are quantitated by class as a percentage of the total signal ± standard deviation (n = 3).
Dashes indicate lipid class could not be detected and/or identified by this method.
BODIPY-labeled FAs were metabolized by larval zebrafish into different arrays of complex lipids depending on FA chain length
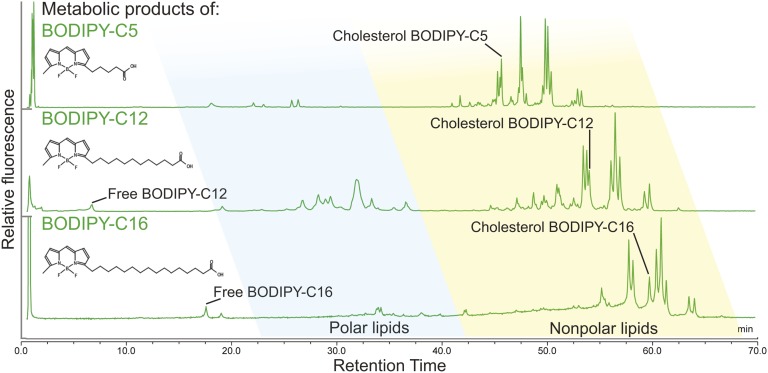
TLC assays have shown that larval zebrafish fed BODIPY FL-C5, -C12, and -C16 in a HF/HC meal incorporate these fluorescent FAs into the three major classes of complex lipids (phospholipids, TGs, and CEs) in different proportions (15). Because HPLC resolves individual lipid species within each class, where TLC does not, we repeated the experiments of Carten, Bradford, and Farber (15) in order to investigate the fluorescent lipid product profile of each of these BODIPY FL-FAs at a higher chromatographic resolution. Larval zebrafish were fed BODIPY FL-FAs in a HF/HC meal for 2 h, and total lipids were extracted after a 16 h chase period. The products of each BODIPY FL-FA were analyzed by HPLC with fluorescence detection, which revealed that carbon chain length of fluorescent saturated FAs influenced their channeling into complex lipids: while BODIPY FL-C12 was incorporated into a wide array of nonpolar lipids and phospholipids, BODIPY FL-C16 and BODIPY FL-C5 were largely incorporated into nonpolar lipids and a smaller amount of phospholipid. A single CE product (confirmed through the use of standards and an ACAT inhibitor, data not shown) of each BODIPY FL-FA was observed. No oxidation intermediates of BODIPY FL-FAs were found (Fig. 3, Table 2).
Fig. 3.
The chain length of BODIPY FL-labeled saturated FAs affects their incorporation into complex lipids. HPLC-fluorescence analysis resolves products of BODIPY FL-C5, -C12, and -C16 synthesized by larval zebrafish that have been given these fluorescent FAs in a HF/HC meal. Results are representative of >10 independent HPLC-fluorescence analyses of total lipid extracts taken from 6 dpf larval zebrafish sampled 16–20 h postfeeding.
TABLE 2.
Distribution of fluorescent products of BODIPY FL-FAs among the major lipid classes 16 h postfeeding
| FA | PLs | CE | TGs | Total Polar:Total Nonpolar Lipids | |
| BODIPY FL-C5 | —a | 7.5 ± 1.3 | 9.6 ± 0.6 | 82.9 ± 1.9 | 0.08 |
| BODIPY FL-C12 | 1.5 ± 0.7 | 39.5 ± 1.4 | 11.0 ± 1.3 | 48.0 ± 2.0 | 0.67 |
| BODIPY FL-C16 | 5.5 ± 1.6 | 5.6 ± 0.5 | 3.0 ± 0.5 | 85.9 ± 2.0 | 0.06 |
Fluorescent products of BODIPY FL-FAs fed to larval zebrafish in a HF/HC meal are quantitated by class as percentage of total fluorescence ± standard deviation (n = 3 for BODIPY FL-C5, n = 6 for BODIPY FL-C12 and -C16). PL, polar lipid.
Free BODIPY FL-C5 could not be quantitated in this assay as it runs in the solvent front.
Though resolution and quantitation of individual complex lipid products of BODIPY FL-FAs were achieved by HPLC-fluorescence, it was not practical to use standards to identify each putative fluorescent phospholipid and TG peak, due to the number of possibilities. MS detection identified 31 unlabeled FAs in larval zebrafish that could combine with BODIPY FL-FAs to form fluorescent complex lipids with the observed retention times, which gave a total of 273 possible phospholipid products and 496 possible TG products of each BODIPY FL-FA (assuming that none were double-labeled, which was unlikely due to the low ratio of labeled to unlabeled dietary FAs in these experiments).
Based on retention times of purchased standards and nonfluorescent peaks identified by LC-MS/MS, we hypothesized that fluorescent peaks eluting between 15 and 45 min on the HPLC-fluorescence chromatograms in Fig. 3 were phospholipids and fluorescent peaks eluting between 45 and 80 min were nonpolar lipids (e.g., TGs and sterol esters). To test this hypothesis, total lipid extracts from larval zebrafish fed BODIPY FL-FAs in a HF/HC meal were subjected to acetone precipitation to separate polar and nonpolar fractions, each of which was analyzed by HPLC with fluorescent detection (supplemental Fig. S1). These data confirmed that the predicted class of each peak was as expected. For more precise identification of specific fluorescent complex lipids, the products of each BODIPY FL-FA were also analyzed by LC-MS/MS as described above, and the unique isotopic signature of boron was used to identify putative BODIPY-lipids. Five BODIPY FL-C12 phospholipids, two BODIPY FL-C5 phospholipids, a BODIPY FL-C16 TG, and the CEs of BODIPY FL-C5 and BODIPY FL-C12 were identified by this method (supplemental Table S4).
Fluorescent FAs are especially useful metabolic labeling reagents in the larval zebrafish model because biochemical and live imaging assays may be performed in parallel. The fluorescent lipid feeding procedures used in this study are well-established as methods to investigate dietary lipid transport and deposition in tissues, cells, and subcellular structures in the larval zebrafish, as well as other models (26, 43, 44). At 4–8 h from the start of feeding BODIPY FL-C5 in a HF/HC meal, fluorescence is seen by live confocal microscopy in a variety of subcellular structures and membranes in the larval zebrafish liver, pancreas, and intestinal enterocytes, while BODIPY FL-C16-derived fluorescence in these organs is concentrated in lipid droplets (15). Based on this previously published imaging data, we expected to detect a larger amount of phospholipid products of BODIPY FL-C5 when compared with BODIPY FL-C16. This hypothesis was supported by HPLC-fluorescence lipid profiles, in which the products of BODIPY FL-C5 in samples taken 8 h postfeeding were 6.6 ± 1.3% phospholipid (n = 13) and products of BODIPY FL-C16 in samples taken 6–10 h postfeeding were 2.1 ± 0.5% phospholipid (n = 4, P = 0.000012; Student’s t-test).
As all variants of the BODIPY fluorophore are large compared with the labeled FAs, we hypothesized that the metabolism of BODIPY-FAs would not exactly replicate that of their unlabeled counterparts, but would resemble the metabolism of larger FAs. Specifically, as the BODIPY FL fluorophore is approximately the length of a four-carbon chain, we tested the hypothesis that BODIPY FL-C12 and 3H-palmitate would yield similar product profiles. BODIPY FL-C12 and 3H-palmitate were fed to larval zebrafish in equimolar amounts in a HF/HC meal, and products were analyzed 18 h postfeeding by HPLC. Following fluorescent detection, HPLC eluent was collected in 1 min fractions, which were assayed for radioactivity so that the metabolism of both radioactive and fluorescently labeled FAs could be characterized simultaneously. The complex lipid product profiles of BODIPY FL-C12 and 3H-palmitate, while not identical, contained similarly high ratios of labeled phospholipid to labeled nonpolar lipid when compared with the products of BODIPY FL-C5 and BODIPY FL-C16 (Tables 2, 3). Full product profiles of BODIPY FL-C12 and 3H-palmitate present in larval zebrafish 18 h postfeeding are shown in supplemental Fig. S2.
TABLE 3.
Distribution of labeled products of BODIPY FL-C12 and 3H-C16 FAs among the major lipid classes
| FA | PLs | CE | TGs | Total Polar:Total Nonpolar Lipids | |
| BODIPY FL-C12 | 22.5 ± 6.2 | 43.4 ± 7.4 | 9.9 ± 0.4 | 24.2 ± 1.3 | 1.27 |
| 3H-C16 | 4.3 ± 0.7 | 76.8 ± 4.0 | 2.4 ± 0.6 | 16.5 ± 2.9 | 4.06 |
Larvae were fed and incubated postfeeding at room temperature. For all other experiments, feeding was conducted at 30°C. Products of labeled FAs fed to larval zebrafish in a HF/HC meal and analyzed 18 h postfeeding are quantitated by class as percentage of total signal ± standard deviation (n = 4).
Fluorescent lipid analogs of similar chain lengths and different fluorescent tags were metabolized by larval zebrafish into different arrays of complex lipid products
As BODIPY FL-C12 is especially physiologically relevant due to its similarity to palmitate and forms a wider array of polar and nonpolar products than the other two BODIPY FL-FAs examined, we focused on this chain length for the remaining experiments, which addressed the effects of dietary context and variants of the BODIPY fluorophore on the metabolism of fluorescent FAs. In addition to BODIPY FL, lipids labeled with many other variants of BODIPY are commonly used in live fluorescent microscopy. As BODIPY-labeled FAs are metabolizable, the fluorescent signal in live imaging experiments is derived not only from the BODIPY-FA, but also from its metabolic products. To better understand how differences in BODIPY fluorophores impact the metabolism of labeled FAs and possibly the subcellular localization of their products, we set out to explore the effect of varying fluorophore chemistry with constant FA chain length.
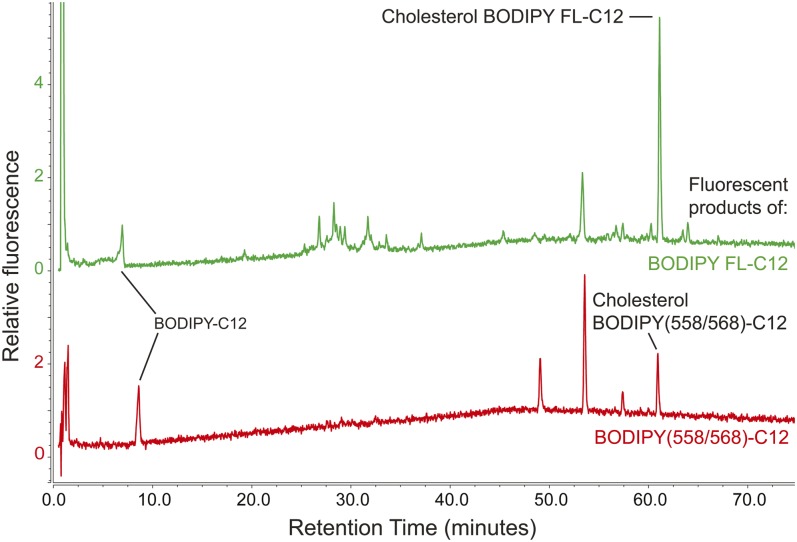
BODIPY(558/568)-C12 fluoresces red and is therefore especially useful in fluorescent microscopy experiments involving cells or animals expressing GFP. This red fluorescent tag, at 272 g/mol, is substantially larger than BODIPY FL (218 g/mol) and carries an additional five-member thienyl ring structure (Fig. 4, inset). Previous work comparing the metabolism of the red and green BODIPY-C12 variants by mouse primary hepatocytes showed, by qualitative TLC assays, that both of these fluorescent FAs are incorporated into both nonpolar complex lipids and phospholipids (29). Because the BODIPY tag accounts for over half of the molecular weight and over 25% of the length of the BODIPY-C12 molecule, we hypothesized that structural differences between BODIPY variants would correlate with differences in the metabolism of BODIPY-lipids that could be quantitated by HPLC-fluorescence.
To test this hypothesis, a mixture of BODIPY FL-C12 and BODIPY(558/568)-C12 in a HF/HC meal was fed to larval zebrafish, which were imaged by live confocal microscopy 8 h postfeeding. Red and green fluorescence, derived from the products of both BODIPY-C12 variants, appeared in the same subcellular structures in the larval zebrafish liver (Fig. 4A). Additionally, similar arrays of complex lipid products were observed when each of these fluorescent FAs was fed individually (also in a HF/HC meal) and HPLC-fluorescence analysis was performed on samples taken 8 h postfeeding (Fig. 4B). However, the relative amounts of each BODIPY-C12 product varied depending on the type of BODIPY tag: BODIPY FL-C12 was more readily metabolized into complex lipid products than BODIPY(558/568)-C12, and was incorporated mostly into polar lipids, while BODIPY(558/568)-C12 was incorporated mostly into TGs (Table 4).
TABLE 4.
BODIPY FL-C12 and BODIPY(558/568)-C12 are incorporated into similar products, but in different proportions
| FAs | PLs | CEs | TGs | |
| BODIPY FL-C12 | 2.3 ± 0.1 | 59.7 ± 0.8 | 14.2 ± 1.5 | 23.7 ± 0.8 |
| BODIPY(558/568)-C12 | 6.6 ± 0.2 | 23.8 ± 1.0 | 11.9 ± 1.4 | 60.0 ± 0.8 |
Fluorescent products of BODIPY FL-FAs fed to larval zebrafish in a HF/HC meal and analyzed 8 h postfeeding are quantitated by class as the percentage of total fluorescence ± standard deviation (n = 2).
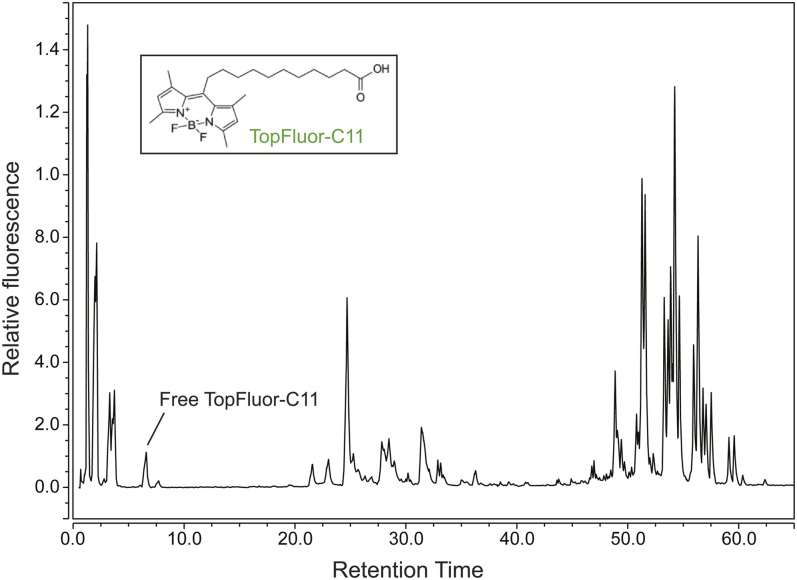
To further examine the effect of BODIPY variant on BODIPY-FA partitioning in the larval zebrafish, we carried out similar feeding and HPLC-fluorescence experiments using the fluorescent FA, TopFluor®-C11. TopFluor is a green variant of BODIPY that carries two additional methyl groups and is bonded to its lipid conjugate at carbon 1 of its central ring, forming a T-shaped molecule in contrast with the more linear BODIPY FL-FAs (Fig. 5, inset). We hypothesized that, though the fluorophores were chemically similar, the position at which the labeled lipid was attached might result in variations in metabolism. When fed to larval zebrafish in a HF/HC meal, TopFluor-C11 was processed into an array of products similar in class to the products of BODIPY FL-C12, though the specific fluorescent HPLC peaks that appeared were different (compare Figs. 3, 5).
Fig. 5.
HPLC-fluorescence product profile of the BODIPY-lipid, TopFluor-C11. Larval zebrafish synthesize a similar array of polar and nonpolar lipids from BODIPY FL-C12 (Figs. 3, 6) and TopFluor-C11 when these FA analogs are delivered in a HF/HC meal. Products of TopFluor-C11 20 h postfeeding are 1.6 ± 0.1% free TopFluor-C11, 22.1 ± 1.7% polar lipids, and 76.0 ± 1.5% nonpolar lipids (n = 3). Unlike BODIPY FL-C12, some oxidation intermediates of TopFluor-C11 (eluting before 5 min) may be present. No CE of TopFluor-C11 was identified.
Fluorescent lipid analogs of similar chain lengths were metabolized by larval zebrafish into different arrays of complex lipid products when dietary lipid content was varied
We then set out to explore the effect of dietary lipid content on fluorescent lipid metabolism, again using BODIPY FL-C12, the most widely metabolized BODIPY FL-FA. LF/LC and LF/HC diets were investigated as BODIPY-lipid carriers. BODIPY FL-C12 fed in a LF/LC or LF/HC meal was used to make an array of complex lipid products similar to those synthesized when this fluorescent FA was fed in a HF/HC meal, but in different proportions [Fig. 6 (upper trace), Table 5]. [Chromatograms in Fig. 6 show the fluorescent lipid products of BODIPY FL-C12 and BODIPY(558/568)-C12 delivered to larval zebrafish in LF/LC meals. Results were similar for these fluorescent FAs fed in LF/HC meals (chromatograms not shown; quantitative data is summarized in Table 4).] Specifically, similar amounts of fluorescent phospholipid were synthesized when BODIPY FL-C12 was fed in all three diets, but with the HF/HC diet, less fluorescent CE (HF/HC-fed larvae: CE = 11.0 ± 1.3% of total) and more fluorescent TG were made when compared with the LF/LC (CE = 42.9 ± 3.6% of total) and LF/HC meals (CE = 51.2 ± 2.0% of total) (Tables 4, 5). We hypothesized that the larger amount of FA in the HF/HC diet generally increased TG synthesis and, therefore, caused more fluorescent FA to be channeled into TGs when compared with the LF/LC and LF/HC diets. Addition of cholesterol to a low-fat meal also correlated with increased CE synthesis from BODIPY FL-C12, at the expense of incorporation into phospholipid (Table 5, P < 0.05).
Fig. 6.
The array of complex lipid products formed when BODIPY-C12 is fed to larval zebrafish in a LF/LC diet varies depending on the type of BODIPY label. BODIPY FL-C12 fed in a LF/LC meal is used to make an array of polar and nonpolar lipid products similar to those synthesized when this fluorescent FA is fed in a HF/HC meal (Fig. 4), but in different proportions. In contrast, BODIPY(558/568)-C12 is no longer incorporated into a detectable amount of phospholipid when delivered in a LF/LC diet. Results are representative of five samples per group.
TABLE 5.
BODIPY-lipids with similar FA chain lengths are metabolized differently depending on the BODIPY variant, BODIPY-lipid class, and cholesterol content of the diet
| Products of BODIPY-Lipid Fed in LF/LC Diet | Products of BODIPY-Lipid Fed in LF/HC Diet | |||||||
| BODIPY-Lipid | FA | PL | TG | CE | FA | PL | TG | CE |
| BODIPY FL-C12 | No signal | 40.1 ± 5.2a | 16.9 ± 1.7 | 42.9 ± 3.6a | No signal | 29.3 ± 0.2a | 19.5 ± 2.2 | 51.2 ± 2.0a |
| BODIPY(558/568)-C12 | 15.8 ± 3.0 | No signal | 52.5 ± 1.2 | 31.7 ± 3.0 | 45.9 ± 17.2 | No signal | 34.1 ± 11.8 | 20.0 ± 8.9 |
| Cholesterol BODIPY FL-C12 | No signalb | 27.2 ± 0.6b | 16.8 ± 9.9b | 56.0 ± 10.5b | No signal | 30.0 ± 7.7 | 26.2 ± 7.3 | 43.7 ± 7.6 |
| Cholesterol BODIPY(576/589)-C11 | No signalb | No signalb | No signalb | No signalb | No signal | No signal | 58.9 ± 3.8 | 41.1 ± 3.8 |
Fluorescent products of BODIPY-lipids fed to larval zebrafish in a HF/HC meal are quantitated by class as percentage of total fluorescence ± standard deviation.
P < 0.05 when LF/LC and LF/HC groups are compared by Student’s t-test.
n = 2. For all other groups, n = 3.
We then sought to determine whether high dietary cholesterol had a similar effect on the metabolism of the red fluorescent FA, BODIPY(558/568)-C12. BODIPY(558/568)-C12 contributed fluorescence to all four lipid classes that were measured when delivered in a HF/HC meal (Table 4), but was no longer metabolized into a detectable amount of phospholipid when delivered in a LF/LC or LF/HC meal (Table 5). No significant differences were observed between the fluorescent product profiles of BODIPY(558/568)-C12 delivered in a LF/LC or LF/HC diet (Table 5). In general, BODIPY(558/568)-C12 appeared to be metabolized into complex lipid products to a lesser extent than BODIPY FL-C12.
Metabolism of BODIPY-CEs by larval zebrafish was responsive to the cholesterol content of the diet and the BODIPY variant
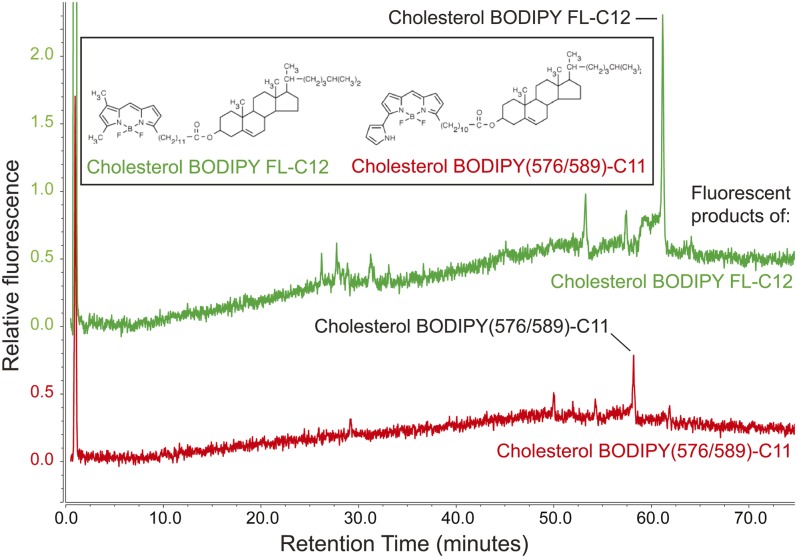
Stoletov et al. (21) established the larval zebrafish as a model for hypercholesterolemia in which they observed fluorescent punctae in blood vessels of larval zebrafish fed a red fluorescent CE analog [cholesterol BODIPY(576/589)-C11] for several days in a high-cholesterol diet. The chemical composition of the BODIPY-lipids labeling these punctae, however, was not known. As the fluorescent label of cholesterol BODIPY(576/589)-C11 is located on the acyl chain, we hypothesized that the fluorescent acyl chain would be cleaved from the cholesterol molecule in the intestine by luminal lipases, and that it would subsequently be metabolized like a BODIPY-FA. To test this hypothesis and gain a better understanding of the larval zebrafish as a model of nascent hypercholesterolemia, we repeated the experiments of Stoletov et al. (21) with HPLC-CAD/fluorescence analysis, with the addition of larval zebrafish fed cholesterol BODIPY FL-C12 for comparison. Each BODIPY-CE was added to LF/LC or LF/HC food, and total lipids were extracted from larval zebrafish after 3 days of feeding. No fluorescent products of BODIPY(576/589)-C11 were recovered from larval zebrafish fed this fluorescent lipid in the LF/LC diet, suggesting that cholesterol BODIPY(576/589)-C11 is excreted without being digested or absorbed (Table 5, supplemental Fig. S3A). A separate series of HPLC-fluorescence experiments confirmed that cholesterol BODIPY(576/589)-C11 was present in the food given to this experimental group and in larvae that still had visible food particles in their intestines (data not shown). When cholesterol BODIPY(576/589)-C11 was delivered in a LF/HC diet, however, fluorescent TGs and cholesterol BODIPY(576/589)-C11 were detected. In contrast, fluorescence derived from cholesterol BODIPY FL-C12 delivered in a LF/LC or LF/HC meal appeared in an array of products similar to those produced by feeding the FA, BODIPY FL-C12 (Fig. 7, supplemental Fig. S3, Table 5). These results suggest that the fluorescent lipid punctae observed in larval zebrafish blood vessels may be labeled by a mixture of fluorescent TGs and CEs, and that cholesterol BODIPY FL-C12 is more readily metabolized by larval zebrafish than its red counterpart, cholesterol BODIPY(576/589)-C11.
Fig. 7.
When the fluorescent CE cholesterol BODIPY FL-C12 is fed to larval zebrafish in a LF/HC diet, fluorescent phospholipid, TG, and CE products are observed. Cholesterol BODIPY(576/589)-C11 yields fluorescent TG (supplemental Fig. S3B) and CE products when delivered in a LF/HC diet. Results are representative of four samples per group.
Incorporation of dietary BODIPY-labeled FA analogs into CEs was responsive to the availability of dietary cholesterol for uptake by enterocytes
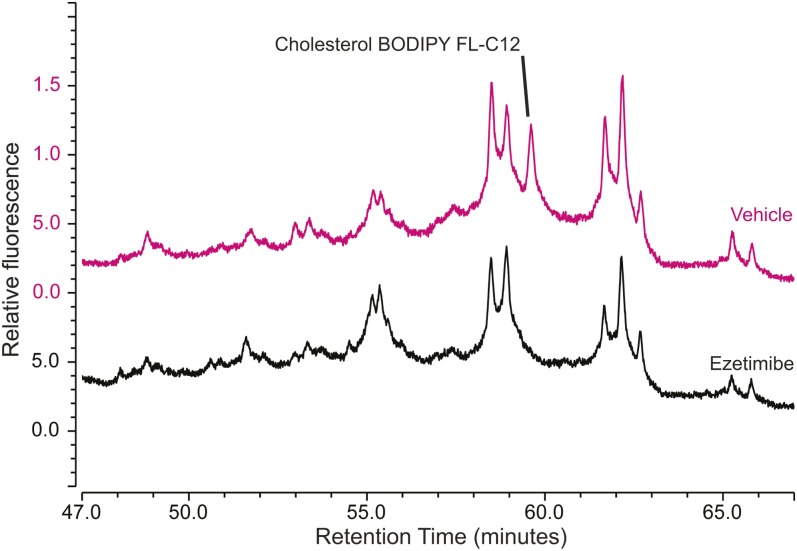
To further investigate the role of cholesterol in regulating the metabolism of dietary FAs, we sought to understand the effect of the metabolic availability of dietary cholesterol on the partitioning of dietary fluorescent FAs among complex lipid classes. Using two pharmaceutical inhibitors of cholesterol absorption and metabolism, we set out to explore the degree to which we could track cholesterol metabolism and its influence on dietary FA partitioning by monitoring fluorescent CE synthesis. Ezetimibe is an inhibitor of dietary cholesterol absorption by enterocytes that acts by interfering with the npc1l1 cholesterol uptake pathway. It has been shown to decrease absorption of dietary radioactive cholesterol or fluorescent cholesterol analogs by 70–80% in humans, other mammals, and larval zebrafish (14, 45–47). The 4-hydroxycinnamic acid (L-phenylalanine methyl ester) amide (CAY10486; Cayman Chemical) is an ACAT inhibitor that has been shown to reduce CE synthesis by 70% in mammalian cell culture and by 40% in 3 dpf larval zebrafish (17, 48). We hypothesized that because a high-cholesterol diet increased CE synthesis from dietary fluorescent FAs, limiting cholesterol absorption would have the opposite effect. Treatment of larval zebrafish with ezetimibe or an ACAT inhibitor before, during, and after a HF/HC meal containing BODIPY-labeled FAs resulted in a statistically significant decrease in fluorescent CE synthesis (Table 6). The respective decreases in CE synthesis resulting from treatment with ezetimibe and the ACAT inhibitor were within one standard deviation of each other in experiments with both BODIPY FL-C12 and BODIPY FL-C16. Neither ezetimibe nor the ACAT inhibitor altered the total cholesterol content of larval zebrafish in a 48 h drug treatment period with a single HF/HC meal (supplemental Fig. S4B). Changes in CE synthesis from fluorescent dietary FAs observed in these experiments were determined by dissection to take place in the intestine (Fig. 8, supplemental Fig. S5), suggesting that CE synthesis from newly absorbed dietary FAs depends primarily on availability of newly absorbed or synthesized cholesterol.
TABLE 6.
CE synthesis from dietary FAs is decreased by blocking dietary cholesterol absorption
| Ezetimibe (5 μM) | ACAT Inhibitor (100 μM) | |||
| BODIPY-FA | CE (treated:control) | P | CE (treated:control) | P |
| BODIPY FL-C5 | 0.73 ± 0.08 | 0.02a | 0.64 ± 0.07 | 0.005a |
| BODIPY FL-C12 | 0.52 ± 0.16 | 0.004b | 0.44 ± 0.05 | 0.0007a |
| BODIPY(558/568)-C12 | 0.35 ± 0.02 | 0.002a | 0.61 ± 0.12 | 0.018a |
| BODIPY FL-C16 | 0.25 ± 0.10 | 0.0003b | 0.19 ± 0.03 | 0.00005a |
Fold changes are given as the treated:control group ratio of the average CE peak area per larval equivalent in samples taken 18 h postfeeding ±standard deviation. All sample groups pass Levene’s test for equal variance.
Student’s t-test, unpaired, equal variance, n = 3.
Nested ANOVA, df = 11 (two experiments; two to three technical replicates per group per experiment).
Fig. 8.
Ezetimibe reduces CE synthesis from dietary FAs in the intestine. Larval zebrafish (6 dpf) were treated with ezetimibe (or 0.1% ethanol vehicle) and then fed BODIPY FL-C12 in the HF/HC diet. Intestines were dissected from 35 larvae per group 20 h postfeeding and pooled for lipid extraction. The cholesterol BODIPY FL-C12 peak is present in the HPLC-fluorescence trace from intestines of larvae in the vehicle control group (upper chromatogram), and absent in the trace from intestines of ezetimibe-treated larvae (lower chromatogram). Results are representative of two samples per group.
DISCUSSION
Mass spectrometric detection and identification of lipids is an increasingly prevalent approach to lipidomics that provides a wealth of highly detailed information per experimental sample. However, when compared with other biochemical techniques, MS is expensive and time-consuming, which can unnecessarily limit the scope of lipidomic studies. Our combined HPLC-CAD/LC-MS workflow leverages the strengths of both MS and CAD lipidomics by enhancing the depth of information that may be obtained from HPLC-CAD experiments with a lower cost per sample.
The first published metabolic assays using BODIPY FL-C12 showed that in mammalian cell culture, this fluorescent FA is incorporated into diglycerides, TGs, and three classes of phospholipids identified by HPLC-fluorescence. However, this analysis was limited in that BODIPY-labeled phospholipids clustered by class and polar and nonpolar fluorescent lipids had to be analyzed separately (25). Our method provides both greater resolution of individual fluorescent complex lipids and a way to analyze all classes of complex lipids from a single sample. Furthermore, we have built upon earlier work (15) to expand the metabolic labeling toolbox for the larval zebrafish and other model systems by using this method to characterize the complex lipid product profiles of a wider range of fluorescent lipids delivered in a variety of diets (Table 7).
TABLE 7.
Summary of BODIPY lipids, diets, and product classes synthesized by larval zebrafish
| BODIPY-Lipid | Color | Diet | PL Products | TG Products | CE Products |
| BODIPY FL-C5 | Green | HF/HC | 5 | 26 | 1 |
| BODIPY FL-C12 | Green | HF/HC | 10 | 24 | 1 |
| BODIPY FL-C12 | Green | LF/LC | 15 | 18 | 1 |
| BODIPY FL-C12 | Green | LF/HC | 15 | 18 | 1 |
| BODIPY(558/568)-C12 | Red | HF/HC | 7 | 11 | 1 |
| BODIPY(558/568)-C12 | Red | LF/LC | 0 | 3 | 1 |
| BODIPY(558/568)-C12 | Red | LF/HC | 0 | 3 | 1 |
| TopFluor-C11 | Green | HF/HC | 11 | 22 | 0 |
| BODIPY FL-C16 | Green | HF/HC | 5 | 11 | 1 |
| Cholesterol BODIPY FL-C12 | Green | LF/LC | 3 | 11 | 1 |
| Cholesterol BODIPY FL-C12 | Green | LF/HC | 3 | 11 | 1 |
| Cholesterol BODIPY(576/589)-C11 | Red | LF/LC | 0 | 0 | 0 |
| Cholesterol BODIPY(576/589)-C11 | Red | LF/HC | 0 | 3 | 1 |
Each fluorescent HPLC peak is counted as a single product of its corresponding BODIPY-lipid.
Our results demonstrate that, although all BODIPY-lipids tested were metabolized into multiple fluorescent complex lipid products when fed to larval zebrafish, the resulting product profiles varied with the chain length of the labeled FA, BODIPY-lipid class, BODIPY fluorophore variant, and nutritional context (Table 7). Awareness of this variation in distribution of fluorescent signal among lipid classes is not only essential when selecting BODIPY-lipids for metabolic labeling experiments, but is also crucial to understanding and interpreting live imaging experiments using BODIPY-lipids. For example, emission wavelength is a reasonable primary criterion when choosing a BODIPY tag for imaging experiments because it is often necessary to choose a fluorescent lipid color different from that of the fluorescent proteins expressed in transgenic animal models or cell lines. However, when interpreting results obtained with different BODIPY variants or fluorescent FAs of different chain lengths, it is important to consider that differences in the distribution of fluorescent signal into lipid classes may have an effect on the distribution of fluorescent signal among subcellular membranes and structures. Additionally, feeding larval zebrafish lipids labeled with the red BODIPY variants generally results in a larger percentage of unincorporated substrate and a smaller range of complex lipid products when compared with green BODIPY-lipids. We hypothesize that the larger red BODIPY fluorophore slows uptake of fluorescent FAs into enterocytes and/or subsequent enzymatic interactions required for complex lipid synthesis; however, it is also possible that red BODIPY-lipids are more susceptible to lipolysis and that the larger amount of substrate observed is due to faster turnover. Even in experiments where labeling of cellular structures does not vary with the type of BODIPY fluorophore under one set of conditions (e.g., labeling of the same subcellular structures by red and green BODIPY-C12 in Fig. 4A despite their different metabolic product profiles in Fig. 4B), it cannot be assumed that this will hold true when conditions are varied or that any changes in labeling that occur under different experimental conditions will involve the same mechanisms for different BODIPY-lipids.
Of the BODIPY-lipids examined in this study, the partitioning of BODIPY FL-C12 into complex lipid classes most closely resembles the metabolism of palmitic acid, the most abundant FA in larval zebrafish. Radioactive palmitic acid fed to larval zebrafish was incorporated into complex lipids more quickly than BODIPY FL-FAs, but the distribution of signal among polar and nonpolar complex lipids was more similar to that observed with BODIPY FL-C12 than any other fluorescent FA examined (Table 2, Table 3). Furthermore, a similar array of individual complex lipid products were labeled by radioactive palmitate and BODIPY FL-C12, though HPLC retention times were consistently shifted earlier by the BODIPY tag (supplemental Fig. S2). Our results are consistent with a model in which the BODIPY fluorophore delays uptake of labeled FAs by cells, but does not alter the distribution of FAs among complex lipid products when compared with unlabeled FAs of similar overall length.
In contrast to the product profile of BODIPY FL-C12, incorporation of fluorescent lipids into phospholipid occurs at a lower rate with BODIPY FL-FAs much larger or smaller than the saturated FAs most commonly found in zebrafish phospholipids, 16:0, 18:1, and 18:0 (supplemental Table S1) (17). BODIPY FL-C5 and -C16, which correspond approximately in total length to 9- and 20-carbon unlabeled FAs, are incorporated into less phospholipid than BODIPY FL-C12 or 3H-palmitate. Incorporation of fluorescent FAs into phospholipid also appears to be more sensitive to the effects of the larger red fluorophore than incorporation into nonpolar lipids. Multiple mechanisms could account for these metabolic differences, including effects on digestion, uptake by enterocytes, esterification, and turnover of labeled lipids. One possibility is that BODIPY-FAs impede diglycerides from interacting with the phospholipid synthesis pathway enzymes, but do not hinder their ability to bind or be esterified by DGAT. As the FA composition of phospholipids is regulated to maintain membrane composition, we expect substrate FA specificity to be more stringent for enzymes involved in phospholipid synthesis and remodeling than for those involved in synthesis of the nonpolar storage lipid classes, TG and CE. Additionally, the relative contributions of de novo synthesis and remodeling of existing phospholipids to the incorporation of fluorescent FAs into phospholipid in the larval zebrafish intestine remain to be determined. Future work addressing these issues will clarify the physiological mechanisms underlying differential processing of the various BODIPY-lipids.
Despite early evidence for the synthesis of complex lipids from BODIPY FL-FAs in cell culture (25), there have been multiple studies in which metabolism of BODIPY-lipids by cultured mammalian cells was assayed by TLC and complex lipid products were not detected (23, 49, 50). In general, when metabolic products of BODIPY-lipids are observed, they are produced by a whole-animal model given BODIPY-lipids in the context of a mixed-lipid diet, primary cultured cells that would already contain lipid droplets (e.g., hepatocytes), or cell culture supplemented with one or more FAs (15, 26–29). Our results provide additional evidence supporting the hypothesis that metabolism of BODIPY-lipids is observed in some studies and not others due to differences in the nutritional context in which BODIPY-lipids are delivered. Larval zebrafish preferentially synthesize mixed-acyl TGs and phospholipids (supplemental Table S1), and the delivery of BODIPY FL-FAs to larval zebrafish in a mixed-lipid meal has been shown to promote their metabolism when compared with delivery in embryo media alone (15). To build on this previous work, we have shown that BODIPY-FAs are also metabolized to complex lipid products when delivered to larval zebrafish in a standard LF/LC diet, expanding this model for use in experiments where the previously validated HF/HC diet consisting of chicken egg yolk may be impractical or may confound results by delivering too much unlabeled lipid.
The BODIPY-lipid, dietary context, and feeding procedure best suited to an experiment depends on the application. For example, it is apparent from the results of this study that with current HPLC methods, the best BODIPY-lipids for monitoring CE synthesis in the larval zebrafish are BODIPY FL-C12 and -C16, as their fluorescent CE product peaks are well-isolated from neighboring fluorescent TGs, which ensures accurate quantitation. The actual effects of both ezetimibe and the ACAT inhibitor on CE synthesis from BODIPY FL-C5 and BODIPY(558/568)-C12 may be larger than what was observable by the HPLC-fluorescence method used, as unlike their counterparts derived from BODIPY FL-C12 and -C16, the cholesterol BODIPY FL-C5 and cholesterol BODIPY(558/568)-C12 peaks did not fully resolve from neighboring TG peaks (Figs. 3, 4). This would produce a background signal that would increase CE peak areas in both control and experimental groups, which would lead to an artificially high treated:control peak area ratio (Table 6). Further HPLC method development will address this issue.
Nutritional context was especially important in experiments investigating the metabolism of BODIPY-CEs. Addition of a large amount of cholesterol to a low-fat diet appeared to promote metabolism of the fluorescent FA group of cholesterol BODIPY(576/589)-C11 into TG and CE products, and there was no evidence of intestinal lipolysis of cholesterol BODIPY(576/589)-C11 when it was delivered in a LF/LC diet. (It is not practical to deliver BODIPY-CEs in a high-fat meal, as they are not digested well in this context, likely due to competition with TGs and other unlabeled CEs for intestinal lipases.) We hypothesize that a higher level of cholesterol than that provided by the LF/LC diet is needed to promote release of the enzymes necessary for digestion of BODIPY-CEs, and that cholesterol BODIPY FL-C12, with its smaller fluorescent tag, is more readily hydrolyzed by intestinal lipases than cholesterol BODIPY(576/589)-C11.
Historically, studies of dietary cholesterol metabolism have tracked esterification using radiolabeled cholesterol (30, 31). By using fluorescent FAs as metabolic tracers to monitor cholesterol esterification by dietary FAs, we were able to build upon the current model for partitioning of dietary cholesterol in enterocytes. Though a number of genes involved in cholesterol channeling have been identified in multiple animal models, including the larval zebrafish, the stepwise details of how these processes occur and are regulated in enterocytes are not fully characterized. Sorting of dietary sterols begins upon absorption: after entering the enterocyte through the NPC1L1 transport pathway, dietary cholesterol is largely retained in the cell, while the bulk of absorbed phytosterols are trafficked back to the intestinal lumen through the ABCG5/ABCG8 sterol efflux transporter (51). Once dietary cholesterol is transported to the endoplasmic reticulum (ER), it may be incorporated into enterocyte membranes or packaged into lipoproteins or lipid droplets with or without being esterified first at the ER membrane by ACAT2 (52).
Though the mechanisms regulating whether or not a newly absorbed dietary cholesterol molecule is esterified are still largely unknown, our results suggest that the amount of CE synthesized from newly absorbed dietary FA is primarily responsive to the amount of newly absorbed and metabolically available dietary cholesterol. Cholesterol was abundant in the digestive tissues of the 6 dpf larval zebrafish (determined by dissection and HPLC-CAD analysis; data not shown), but the cholesterol present before feeding appeared to be inaccessible to esterification by ACAT, likely due to being located in the plasma membrane (53). There was no statistically significant difference in the whole-body cholesterol content of larval zebrafish that had been fed HF/HC, LF/LC, or LF/HC meals when experimental samples were taken long enough after the withdrawal of food for the contents of the intestinal lumen to clear (Table 1, supplemental Fig. S4A). Additionally, neither ezetimibe nor an ACAT inhibitor produced a detectable change in total cholesterol within the time frame and treatment parameters of the experiments described herein (supplemental Fig. S4B). This illustrates the importance of metabolic labeling in the study of metabolism of a single lipid species, as without the fluorescent marker we employed, the signal-to-noise ratio would have been too low to detect changes in CE synthesis from newly absorbed dietary lipids in enterocytes. Physical segregation of ACAT2 from non-ER-located cholesterol pools and/or a lack of transport of cholesterol from other cellular structures to the ER may account for this effect; more work must be done to elucidate how ACAT2 activity is regulated. The method of tracking CE synthesis through metabolic labeling with fluorescent FAs will be an important complement to radioactive labeling methods in further studies of dietary cholesterol processing.
In summary, the HPLC-CAD/fluorescence lipidomics methods described allow for high-throughput analysis of both total lipids and the products of fluorescent metabolic tracers in a single sample, using equipment that requires a much lower initial investment and smaller support infrastructure when compared with MS methods. The ability to pair this metabolic labeling method with live imaging using the same fluorescent lipid reagents is a major advantage of this approach. Using this platform, we provide the most comprehensive analysis of fluorescent FA metabolism to date, new insights into zebrafish models for dyslipidemia, and evidence for coupling between dietary cholesterol uptake and esterification.
Supplementary Material
Acknowledgments
The authors acknowledge Marc Plante (formerly of Thermo Fisher Scientific) for providing excellent advice on setting up the HPLC method, Isaac Kim for providing HPLC assistance, and Carmen Tull, Andrew Rock, and Jennifer Anderson for maintaining fish stocks.
Footnotes
Abbreviations:
- BODIPY(558/568)
- 4,4-difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-s-indacene
- BODIPY FL
- 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene
- -C5, -C11, -C12, -C16
- saturated FAs (5, 11, 12, and 16 carbons, respectively) conjugated to BODIPY fluorophores
- CAD
- charged aerosol detection
- CE
- cholesterol ester
- dpf
- days post-fertilization
- ER
- endoplasmic reticulum
- HF/HC
- high-fat/high-cholesterol
- LF/HC
- low-fat/high-cholesterol
- LF/LC
- low-fat/low-cholesterol
- PL
- polar lipid
- TG
- triglyceride
This work was supported in part by National Institute on Alcohol Abuse and Alcoholism Grant F31AA023142 (to V.H.Q.), National Institute of Diabetes and Digestive and Kidney Diseases Grants R01DK093399 (to S.A.F.) and F32DK109592 (to M.H.W.), and National Institute of General Medicine Grant R01GM63904 to the Zebrafish Functional Genomics Consortium (Stephen Ekker and S.A.F.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support for this work was provided by the G. Harold and Leila Y. Mathers Charitable Foundation to the laboratory of S.A.F.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Hui D. Y., Labonte E. D., and Howles P. N.. 2008. Development and physiological regulation of intestinal lipid absorption. III. Intestinal transporters and cholesterol absorption. Am. J. Physiol. Gastrointest. Liver Physiol. 294: G839–G843. [DOI] [PubMed] [Google Scholar]
- 2.Drover V. A., Nguyen D. V., Bastie C. C., Darlington Y. F., Abumrad N. A., Pessin J. E., London E., Sahoo D., and Phillips M. C.. 2008. CD36 mediates both cellular uptake of very long chain fatty acids and their intestinal absorption in mice. J. Biol. Chem. 283: 13108–13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Aquila T., Hung Y. H., Carreiro A., and Buhman K. K.. 2016. Recent discoveries on absorption of dietary fat: presence, synthesis, and metabolism of cytoplasmic lipid droplets within enterocytes. Biochim. Biophys. Acta. 1861: 730–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson J. L., Carten J. D., and Farber S. A.. 2011. Zebrafish lipid metabolism: from mediating early patterning to the metabolism of dietary fat and cholesterol. Methods Cell Biol. 101: 111–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asaoka Y., Terai S., Sakaida I., and Nishina H.. 2013. The expanding role of fish models in understanding non-alcoholic fatty liver disease. Dis. Model. Mech. 6: 905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hölttä-Vuori M., Salo V. T., Nyberg L., Brackmann C., Enejder A., Panula P., and Ikonen E.. 2010. Zebrafish: gaining popularity in lipid research. Biochem. J. 429: 235–242. [DOI] [PubMed] [Google Scholar]
- 7.Fang L., Liu C., and Miller Y. I.. 2014. Zebrafish models of dyslipidemia: relevance to atherosclerosis and angiogenesis. Transl. Res. 163: 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlegel A., and Stainier D. Y.. 2007. Lessons from “lower” organisms: what worms, flies, and zebrafish can teach us about human energy metabolism. PLoS Genet. 3: e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong S., Stephens W. Z., Burns A. R., Stagaman K., David L. A., Bohannan B. J., Guillemin K., and Rawls J. F.. 2015. Ontogenetic differences in dietary fat influence microbiota assembly in the zebrafish gut. MBio. 6: e00687-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadler K. C., Rawls J. F., and Farber S. A.. 2013. Getting the inside tract: new frontiers in zebrafish digestive system biology. Zebrafish. 10: 129–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semova I., Carten J. D., Stombaugh J., Mackey L. C., Knight R., Farber S. A., and Rawls J. F.. 2012. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 12: 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farber S. A., Pack M., Ho S. Y., Johnson I. D., Wagner D. S., Dosch R., Mullins M. C., Hendrickson H. S., Hendrickson E. K., and Halpern M. E.. 2001. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 292: 1385–1388. [DOI] [PubMed] [Google Scholar]
- 13.Hama K., Provost E., Baranowski T. C., Rubinstein A. L., Anderson J. L., Leach S. D., and Farber S. A.. 2009. In vivo imaging of zebrafish digestive organ function using multiple quenched fluorescent reporters. Am. J. Physiol. Gastrointest. Liver Physiol. 296: G445–G453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clifton J. D., Lucumi E., Myers M. C., Napper A., Hama K., Farber S. A., Smith A. B. 3rd, Huryn D. M., Diamond S. L., and Pack M.. 2010. Identification of novel inhibitors of dietary lipid absorption using zebrafish. PLoS One. 5: e12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carten J. D., Bradford M. K., and Farber S. A.. 2011. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev. Biol. 360: 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteves A., Knoll-Gellida A., Canclini L., Silvarrey M. C., Andre M., and Babin P. J.. 2016. Fatty acid binding proteins have the potential to channel dietary fatty acids into enterocyte nuclei. J. Lipid Res. 57: 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyares R. L., de Rezende V. B., and Farber S. A.. 2014. Zebrafish yolk lipid processing: a tractable tool for the study of vertebrate lipid transport and metabolism. Dis. Model. Mech. 7: 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falcinelli S., Rodiles A., Unniappan S., Picchietti S., Gioacchini G., Merrifield D. L., and Carnevali O.. 2016. Probiotic treatment reduces appetite and glucose level in the zebrafish model. Sci. Rep. 6: 18061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otis J. P., and Farber S. A.. 2013. Imaging vertebrate digestive function and lipid metabolism in vivo. Drug Discov. Today Dis. Models. 10: e11–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraher D., Sanigorski A., Mellett N. A., Meikle P. J., Sinclair A. J., and Gibert Y.. 2016. Zebrafish embryonic lipidomic analysis reveals that the yolk cell is metabolically active in processing lipid. Cell Reports. 14: 1317–1329. [DOI] [PubMed] [Google Scholar]
- 21.Stoletov K., Fang L., Choi S. H., Hartvigsen K., Hansen L. F., Hall C., Pattison J., Juliano J., Miller E. R., Almazan F., et al. 2009. Vascular lipid accumulation, lipoprotein oxidation, and macrophage lipid uptake in hypercholesterolemic zebrafish. Circ. Res. 104: 952–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nealon J. R., Blanksby S. J., Donaldson P. J., Truscott R. J., and Mitchell T. W.. 2011. Fatty acid uptake and incorporation into phospholipids in the rat lens. Invest. Ophthalmol. Vis. Sci. 52: 804–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hostetler H. A., Balanarasimha M., Huang H., Kelzer M. S., Kaliappan A., Kier A. B., and Schroeder F.. 2010. Glucose regulates fatty acid binding protein interaction with lipids and peroxisome proliferator-activated receptor alpha. J. Lipid Res. 51: 3103–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang H., Starodub O., McIntosh A., Atshaves B. P., Woldegiorgis G., Kier A. B., and Schroeder F.. 2004. Liver fatty acid-binding protein colocalizes with peroxisome proliferator activated receptor alpha and enhances ligand distribution to nuclei of living cells. Biochemistry. 43: 2484–2500. [DOI] [PubMed] [Google Scholar]
- 25.Kasurinen J. 1992. A novel fluorescent fatty acid, 5-methyl-BDY-3-dodecanoic acid, is a potential probe in lipid transport studies by incorporating selectively to lipid classes of BHK cells. Biochem. Biophys. Res. Commun. 187: 1594–1601. [DOI] [PubMed] [Google Scholar]
- 26.Furlong S. T., Thibault K. S., Morbelli L. M., Quinn J. J., and Rogers R. A.. 1995. Uptake and compartmentalization of fluorescent lipid analogs in larval Schistosoma mansoni. J. Lipid Res. 36: 1–12. [PubMed] [Google Scholar]
- 27.Viktorova E. G., Ford-Siltz L. A., Nchoutmboube J., and Belov G. A.. 2014. Fluorescent fatty acid analogs as a tool to study development of the picornavirus replication organelles. J. Virol. Methods. 200: 15–21. [DOI] [PubMed] [Google Scholar]
- 28.Cheon H. G., and Cho Y. S.. 2014. Protection of palmitic acid-mediated lipotoxicity by arachidonic acid via channeling of palmitic acid into triglycerides in C2C12. J. Biomed. Sci. 21: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Wei E., Quiroga A. D., Sun X., Touret N., and Lehner R.. 2010. Altered lipid droplet dynamics in hepatocytes lacking triacylglycerol hydrolase expression. Mol. Biol. Cell. 21: 1991–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sylvén C., and Borgstrom B.. 1969. Intestinal absorption and lymphatic transport of cholesterol in the rat: influence of the fatty acid chain length of the carrier triglyceride. J. Lipid Res. 10: 351–355. [PubMed] [Google Scholar]
- 31.Huggins K. W., Camarota L. M., Howles P. N., and Hui D. Y.. 2003. Pancreatic triglyceride lipase deficiency minimally affects dietary fat absorption but dramatically decreases dietary cholesterol absorption in mice. J. Biol. Chem. 278: 42899–42905. [DOI] [PubMed] [Google Scholar]
- 32.Brady J. 1965. A simple technique for making very fine, durable dissecting needles by sharpening tungsten wire electrolytically. Bull. World Health Organ. 32: 143–144. [PMC free article] [PubMed] [Google Scholar]
- 33.Kates M. 1972. Techniques of Lipidology: Isolation, Analysis, and Identification of Lipids. American Elsevier Publishing Company, Inc., New York, NY. [Google Scholar]
- 34.Baek J. S., Fang L., Li A. C., and Miller Y. I.. 2012. Ezetimibe and simvastatin reduce cholesterol levels in zebrafish larvae fed a high-cholesterol diet. Cholesterol. 2012: 564705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeituni E. M., Wilson M. H., Zheng X., Iglesias P. A., Sepanski M., Siddiqi M. A., Anderson J. L., Zheng Y., and Farber S. A.. 2016. Endoplasmic reticulum lipid flux influences enterocyte nuclear morphology and lipid-dependent transcriptional responses. J. Biol. Chem. 291: 23804–23816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeb A. 2015. Chemistry and liquid chromatography methods for the analyses of primary oxidation products of triacylglycerols. Free Radic. Res. 49: 549–564. [DOI] [PubMed] [Google Scholar]
- 37.McCluer R. H., Ullman M. D., and Jungalwala F. B.. 1989. High-performance liquid chromatography of membrane lipids: glycosphingolipids and phospholipids. Methods Enzymol. 172: 538–575. [DOI] [PubMed] [Google Scholar]
- 38.Samet J. M., Friedman M., and Henke D. C.. 1989. High-performance liquid chromatography separation of phospholipid classes and arachidonic acid on cyanopropyl columns. Anal. Biochem. 182: 32–36. [DOI] [PubMed] [Google Scholar]
- 39.Hamilton J. G., and Comai K.. 1988. Separation of neutral lipid, free fatty acid and phospholipid classes by normal phase HPLC. Lipids. 23: 1150–1153. [DOI] [PubMed] [Google Scholar]
- 40.Hamilton J. G., and Karol R. J.. 1982. High performance liquid chromatography (HPLC) of arachidonic acid metabolites. Prog. Lipid Res. 21: 155–170. [DOI] [PubMed] [Google Scholar]
- 41.Khan G. R., and Scheinmann F.. 1978. Some recent advances in physical methods for analysis and characterization of polyunsaturated fatty acids. Prog. Chem. Fats Other Lipids. 15: 343–367. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz-Rodriguez A., Reglero G., and Ibanez E.. 2010. Recent trends in the advanced analysis of bioactive fatty acids. J. Pharm. Biomed. Anal. 51: 305–326. [DOI] [PubMed] [Google Scholar]
- 43.Ho S. Y., Lorent K., Pack M., and Farber S. A.. 2006. Zebrafish fat-free is required for intestinal lipid absorption and Golgi apparatus structure. Cell Metab. 3: 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S. O., Trimble R., Guo F., and Mak H. Y.. 2010. Lipid droplets as ubiquitous fat storage organelles in C. elegans. BMC Cell Biol. 11: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altmann S. W., Davis H. R. Jr., Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., et al. 2004. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 303: 1201–1204. [DOI] [PubMed] [Google Scholar]
- 46.Van Heek M., France C. F., Compton D. S., McLeod R. L., Yumibe N. P., Alton K. B., Sybertz E. J., and Davis H. R. Jr. 1997. In vivo metabolism-based discovery of a potent cholesterol absorption inhibitor, SCH58235, in the rat and rhesus monkey through the identification of the active metabolites of SCH48461. J. Pharmacol. Exp. Ther. 283: 157–163. [PubMed] [Google Scholar]
- 47.Bays H. E., Moore P. B., Drehobl M. A., Rosenblatt S., Toth P. D., Dujovne C. A., Knopp R. H., Lipka L. J., Lebeaut A. P., Yang B., et al. 2001. Effectiveness and tolerability of ezetimibe in patients with primary hypercholesterolemia: pooled analysis of two phase II studies. Clin. Ther. 23: 1209–1230. [DOI] [PubMed] [Google Scholar]
- 48.Lee S., Han J. M., Kim H., Kim E., Jeong T. S., Lee W. S., and Cho K. H.. 2004. Synthesis of cinnamic acid derivatives and their inhibitory effects on LDL-oxidation, acyl-CoA:cholesterol acyltransferase-1 and -2 activity, and decrease of HDL-particle size. Bioorg. Med. Chem. Lett. 14: 4677–4681. [DOI] [PubMed] [Google Scholar]
- 49.Huang H., Starodub O., McIntosh A., Kier A. B., and Schroeder F.. 2002. Liver fatty acid-binding protein targets fatty acids to the nucleus. Real time confocal and multiphoton fluorescence imaging in living cells. J. Biol. Chem. 277: 29139–29151. [DOI] [PubMed] [Google Scholar]
- 50.Atshaves B. P., Storey S. M., Huang H., and Schroeder F.. 2004. Liver fatty acid binding protein expression enhances branched-chain fatty acid metabolism. Mol. Cell. Biochem. 259: 115–129. [DOI] [PubMed] [Google Scholar]
- 51.Wang J., Mitsche M. A., Lutjohann D., Cohen J. C., Xie X. S., and Hobbs H. H.. 2015. Relative roles of ABCG5/ABCG8 in liver and intestine. J. Lipid Res. 56: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abumrad N. A., and Davidson N. O.. 2012. Role of the gut in lipid homeostasis. Physiol. Rev. 92: 1061–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Meer G., Voelker D. R., and Feigenson G. W.. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.