Abstract
Members of the gastrointestinal microbiota are known to convert glucocorticoids to androstanes, which are subsequently converted to potent androgens by other members of the gut microbiota or host tissues. Butyricicoccus desmolans and Clostridium cadaveris have previously been reported for steroid-17,20-desmolase and 20β-hydroxysteroid dehydrogenase (HSDH) activities that are responsible for androstane formation from cortisol; however, the genes encoding these enzymes have yet to be reported. In this work, we identified and located a gene encoding 20β-HSDH in both B. desmolans and C. cadaveris. The 20β-HSDH of B. desmolans was heterologously overexpressed and purified from Escherichia coli. The enzyme was determined to be a homotetramer with subunit molecular mass of 33.8 ± 3.7 kDa. The r20β-HSDH displayed pH optimum in the reductive direction at pH 9.0 and in the oxidative direction at pH 7.0–7.5 with (20β-dihydro)cortisol and NAD(H) as substrates. Cortisol is the preferred substrate with Km, 0.80 ± 0.06 μM; Vmax, 30.36 ± 1.97 μmol·min−1; Kcat, 607 ± 39 μmol·μM−1·min−1; Kcat/Km, 760 ± 7.67. Phylogenetic analysis of the 20β-HSDH from B. desmolans suggested that the 20β-HSDH is found in several Bifidobacterium spp., one of which was shown to express 20β-HSDH activity. Notably, we also identified a novel steroid-17,20-desmolase-elaborating bacterium, Propionimicrobium lymphophilum, a normal inhabitant of the urinary tract.
Keywords: American Type Culture Collection, 20β-hydroxysteroid dehydrogenase, glucocorticoids, gut microbiota
Studies by Nabarro et al. (1) and Wade et al. (2) reported that rectally infused cortisol in humans resulted in a 100-fold increase in urinary 17-ketosteroids, which was ablated by oral neomycin treatment. Winter and Bokkenheuser (3) and Cerone-McLernon et al. (4) reported that incubation of cortisol with mixed human and rat fecal microbiota resulted in the accumulation of C-19 17β-hydroxy metabolites. These studies first pointed to a bacterial pathway, known as steroid-17,20-desmolase, which converts cortisol into 11β-hydroxyandrostenedione (11β-OHAD). Quantification of steroid-17,20-desmolase activity in decimal dilutions of human and rat fecal samples suggests approximately 106 organisms per gram wet-weight feces (5).
Bokkenheuser et al. (5) isolated the first fecal bacterium in pure culture that expresses steroid-17,20-desmolase and 20α-hydroxysteroid dehydrogenase (HSDH). The gut bacterium was later named Clostridium scindens (type strain ATCC 35704) (6). Krafft and colleagues characterized the purified 20α-HSDH and partially-pure steroid-17,20-desmolase from cell extracts of C. scindens ATCC 35704 and reported substrate-specificity and cofactor requirements (7, 8). However, the genes encoding these enzymes and the mechanism by which anaerobic bacterial steroid-17,20-desmolase proceeds were unclear. Recent work by Ridlon et al. (9) identified a cortisol-inducible multi-gene operon (desABCD) hypothesized to encode bacterial steroid-17,20-desmolase and 20α-HSDH. The desAB genes are annotated as “transketolase” N-terminal and C-terminal subunits (9). The two-carbon fragment ketol group transferred via thiamine-pyrophosphate from a sugar phosphate donor to aldose acceptors during transketolation is identical to the side-chain of cortisol, suggesting that steroid-17,20-desmolase perhaps proceeds by a transketolation reaction (9). A cortisol-inducible “Na+-dependent melibiose transporter” encoded by the desD gene was also identified and hypothesized to encode a cortisol transport protein. Purified recombinant DesC was shown to have substrate-specificity and kinetics consistent with previously purified native 20α-HSDH (7, 8). In these studies, steroids with 20β-hydroxy groups were not substrates for DesC (9).
Steroid-17,20-desmolase activity has also been reported in Clostridium cadaveris and Butyricicoccus desmolans (formerly Eubacterium desmolans) (10, 11). Unlike C. scindens ATCC 35704, which elaborates 20α-HSDH, both C. cadaveris and B. desmolans have been reported to express 20β-HSDH activity (10). Following the report of the desABCD operon from C. scindens ATCC 35704, genome sequences for B. desmolans ATCC 43058 (Bio Project PRJNA223055) and C. cadaveris (Bio Projects PRJNA185698, PRJEB17278) became available, prompting us to determine whether these anaerobic bacteria encode the desAB genes in addition to an NAD(P)H-dependent dehydrogenase encoding 20β-HSDH.
MATERIALS AND METHODS
Bacterial strains and materials
B. desmolans ATCC 43058 and Bifidobacterium scardovii ATCC BAA-773 were purchased from ATCC. C. scindens ATCC 35704 was maintained as −80°C glycerol stocks in our laboratory. Propionimicrobium lymphophilum ACS-093-V-SCH5 was obtained from the Culture Collection, University of Götesborg, Sweden. Escherichia coli DH5α (turbo) competent cells were from New England Biolabs (Ipswich, MA) and E. coli BL21-CodonPlus(DE3) RIPL was purchased from Stratagene (La Jolla, CA). The pET-51b(+) vector was obtained from Novagen (San Diego, CA). Restriction enzymes were purchased from New England Biolabs; QIAprep Spin Miniprep kit was obtained from Qiagen (Valencia, CA). Isopropyl β-D-1-thiogalactopyranoside was purchased from Gold Biotechnology (St. Louis, MO). Strep-Tactin resin was purchased from IBA GmbH (Gottingen, Germany). Steroids were purchased from Steraloids (Newport, RI). Amicon Ultra-15 centrifugal filter units with 30 and 50 kDa molecular-mass cutoffs were obtained from Millipore (Billerica, MA). All other reagents were of the highest possible purity and were purchased from Fisher Scientific (Pittsburgh, PA).
Steroid conversion experiments
B. desmolans ATCC 43058, B. scardovii ATCC BAA-773, and C. scindens ATCC 35704 were cultivated in supplemented brain heart infusion (BHI) broth, containing 5 g l−1 of yeast extract. P. lymphophilum ACS-093-V-SCH5 was grown in Gifu anaerobic medium. Steroids were dissolved in methanol and added to the sterilized medium at a concentration of 50 μM, unless otherwise indicated; the concentration of methanol (v/v) per culture was kept at 0.5%. The medium was then seeded with 0.1 ml of a pure bacterial culture and incubated at 37°C for 2 days. After incubation, the products were extracted by vortexing the culture media with 2 vol of ethyl acetate for 1–2 min and then the organic phase was recovered. The organic phase was evaporated under nitrogen gas. The residue was dissolved in 50 μl methanol and analyzed using HPLC (Shimadzu, Japan) equipped with a C-18 analytical column (Capcell Pak c18; Shiseido, Japan). The mobile phase consisted of acetonitrile/water with 0.01% formic acid and the flow rate was maintained at 0.2 ml·min−1. A DAD detector was used at a wavelength of 254 nm. Peak retention times and peak areas of samples were compared with standard steroid molecules.
RNA isolation and transcriptional analysis.
B. desmolans ATCC 43058 was cultivated in BHI broth in the presence or absence of 50 μM cortisol until mid-log phase. Cultures were centrifuged at 16,000 g for 5 min and the pellet suspended in RNALater™ (Ambion) overnight at 4°C followed by centrifugation and storage at −70°C until further processing. Total RNA was isolated using the Ribopure bacteria kit (Ambion) according to the manufacturer’s instructions, including the DNase step. One microgram of total RNA was converted to cDNA using the Advantage RT-for-PCR kit (Clontech) with random hexamer primers. Intergenic PCR was performed using TITANIUM Taq PCR kit (Clontech) with oligonucleotides synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). All primers used in this study are listed in supplemental Table S1. The mRNA start-site was determined by 5′-RACE PCR using the SMARTer RACE PCR kit (Clontech) according to the manufacturer’s instructions with some modifications. RACE PCR products were purified from 1.5% agarose gels by GENECLEAN spin kit according to the manufacturer’s instructions (Bio 101) and cloned into a TA vector. The plasmid DNA was sequenced to identify the mRNA start sequence (W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign).
Gene cloning.
B. desmolans ATCC 43058 genomic DNA was extracted using the Fast DNA isolation kit from Mo-Bio (Carlsbad, CA) according to the manufacturer’s protocol. Oligonucleotide primers used for amplifying the 20β-HSDH gene are reported in supplemental Table S1. The coding sequence for 20β-HSDH was amplified using the Phusion high-fidelity polymerase (Stratagene) and cloned into a pET-51b vector using BamHI and HindIII restriction sites. Recombinant plasmid was transformed into chemically competent E. coli DH5α cells via the heat stock method, plated, and grown for 16 h at 37°C on lysogeny broth (LB) agar plates supplemented with ampicillin (100 μg/ml). A single colony from each transformation was inoculated into LB medium (5 ml) containing ampicillin (100 μg/ml) and grown to saturation. The cells were subsequently centrifuged (3,220 g, 15 min, 4°C) and plasmids were extracted from the resulting cell pellet using the QIAprep Spin Miniprep kit (Qiagen). The foreign DNA inserts in the recombinant plasmids were sequenced to confirm the correctness of the gene sequence (W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign).
Gene expression and protein purification
For protein expression, the correct recombinant plasmids extracted from the E. coli DH5α cells were transformed into E. coli BL-21 CodonPlus (DE3) RIPL chemically competent cells by the heat shock method and grown overnight at 37°C on LB agar plates supplemented with ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml). After 16 h, five isolated colonies were used to inoculate 10 ml of fresh LB medium supplemented with antibiotics and grown at 37°C for 6 h with vigorous aeration. The precultures were then added to fresh LB medium (1 liter), supplemented with the same antibiotics at the same concentrations, and grown with vigorous aeration at 37°C. At an OD600 of 0.3, isopropyl β-D-1-thiogalactopyranoside was added to each culture at a final concentration of 0.1 mM and the temperature was decreased to 16°C. Following 16 h of culturing, cells were pelleted by centrifugation (4,000 g, 30 min, 4°C) and resuspended in 30 ml of binding buffer [20 mM Tris-HCl, 150 mM NaCl, 20% glycerol, 10 mM 2-mercaptoethanol (pH 7.9)]. The cell suspension was subjected to four passages through an EmulsiFlex C-3 cell homogenizer (Avestin, Ottawa, Canada) and the cell lysate was clarified by centrifugation at 20,000 g for 30 min at 4°C.
The recombinant 20β-HSDH (r20β-HSDH) was then purified using Strep-Tactin® resin (IBA GmbH) as per the manufacturer’s protocol. The recombinant protein was eluted using an elution buffer composed of 20 mM Tris-HCl, 150 mM NaCl, 20% glycerol, 10 mM 2-mercaptoethanol (pH 7.9), and 2.5 mM desthiobiotin. The protein purity was assessed by SDS-PAGE and protein bands were visualized by staining with Coomassie brilliant blue G-250. The protein concentrations were calculated based on the molecular mass (41.3 kDa) and computed extinction coefficient (31.6 × 103 M−1·cm−1) (12).
Gel filtration chromatography
Gel filtration chromatography was carried out on Superdex 200 analytical column (GE Healthcare, Piscataway, NJ) attached to Akta FPLC (GE Healthcare) at 4°C. The eluted protein from the Strep-tag column was concentrated and loaded on the column with a buffer composed of 50 mM Tris-Cl, 300 mM NaCl, 10% glycerol, 10 mM 2- mercaptoethanol, and 0.1% Triton X-100 (pH 7.5). The native molecular mass of the protein was calculated by peak retention volume of protein to retention volume of standard proteins. Eluted fractions were visualized by staining with Coomassie brilliant blue G-250. The activity of the enzyme in the eluted fraction was calculated as described below in the standard enzyme assay section.
The 20β-HSDH enzyme assay
The 20β-HSDH activity was measured aerobically by monitoring the conversion of cortisol into 20β-dihydrocortisol using a continuous spectrophotometric method, monitored by measuring the oxidation of NADH at 340 nM (ε = 6,220 M−1·cm−1). Linearity with respect to time and enzyme concentration was determined. The standard reaction mixture contained 50 mM phosphate buffer (pH 7.5), 50 μM of cortisol, 150 μM NADH, 0.05 μM enzyme, and buffer to a final volume of 0.5 ml. The reaction was started by the addition of the enzyme. The initial velocity of enzyme-catalyzed reactions was plotted against the substrate concentrations, and the kinetic parameters were estimated by fitting the data to the Michaelis-Menten equation by the nonlinear regression method using the enzyme kinetics module in GraphPad Prism (GraphPad Software, La Jolla, CA).
Determination of optimal pH
The buffers used to study the pH profiling of 20β-HSDH were as follows: 50 mM sodium citrate, 150 mM NaCl, 20% glycerol and 10 mM 2-mercaptoethanol (pH 4.0–6.0), 50 mM sodium phosphate (pH 6.5–7.5), 50 mM Tris-Cl (pH 8–9), and 50 mM glycine-NaOH (pH 10–11). In order to determine the optimal pH in the reductive direction, purified recombinant 20β-HSDH enzyme at a final concentration of 0.05 μM was added to 50 μM cortisol and 150 μM NADH in 500 μl buffer. The oxidation of NADH was measured continuously at 340 nM for 5 min by spectrophotometry and the initial activities were calculated. For the oxidative reaction, 0.05 μM of enzyme was added to 50 μM 20β-dihydrocortisol and 150 μM NAD+ in 500 μl buffer. The formation of NADH was measured continuously at 340 nm for 5 min and initial velocity was calculated.
Phylogenetic analysis and bioinformatics
Candidate protein sequences for phylogeny reconstruction were retrieved from NCBI after BLAST using as query the short-chain dehydrogenase/reductase (SDR) family oxidoreductase (WP_051643274.1) from B. desmolans ATCC 43058. Seventy sequences with an identity cutoff of 30% with the 20β-HSDH were selected and aligned using MUSCLE v3.8.31 (13). The resulting alignment was manually examined and subsequent phylogeny reconstruction was performed with FastTree v2.1.7 (14) using the implemented Shimodaira-Hasegawa test to estimate the reliability of each split in the tree. iTOL (15) was used for visualization of the obtained phylogeny. Deduced protein molecular weight was determined using ExPASy toolkit (16).
RESULTS
B. desmolans expresses 20β-HSDH activity
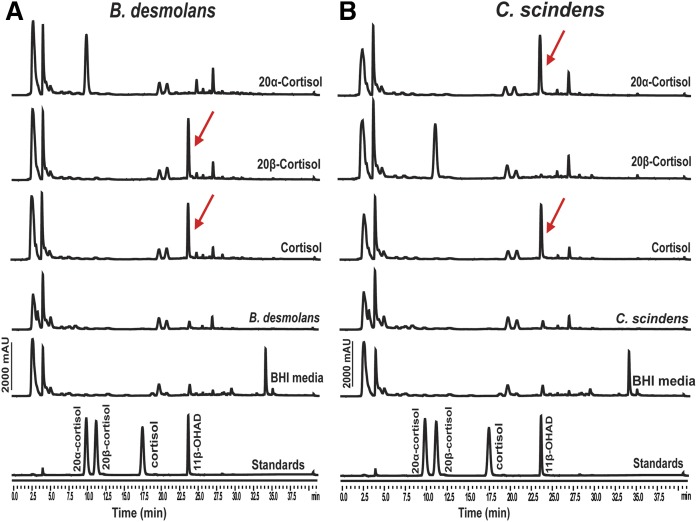
Net conversion of cortisol to 11β-OHAD was observed in the presence of cortisol by both C. scindens ATCC 35704 and B. desmolans ATCC 43058 (Fig. 1). Net conversion of 20α-dihydrocortisol to 11β-OHAD was observed after 24 h growth by C. scindens ATCC 35704. By contrast, 20α-dihydrocortisol was not metabolized after 48 h by B. desmolans ATCC 43058. As expected, net conversion of 20β-dihydrocortisol to 11β-OHAD was observed in cultures of B. desmolans ATCC 43058. These results confirm that B. desmolans ATCC 43058 elaborates a 20β-HSDH, but lacks 20α-HSDH activity.
Fig. 1.
Confirmation of 20β- and 20α-HSDH activity from B. desmolans ATCC 43058 (A) and C. scindens ATCC 35704 (B). Each steroid substrate (50 μM) was incubated with both B. desmolans ATCC 43058 and C. scindens ATCC 35704 in BHI media for 48 h. The products were extracted and analyzed using HPLC equipped with a C-18 analytical column (Capcell Pak c18). Peak retention times and peak areas of samples were compared with standard steroid molecules. Red arrows denote the formation of 11β-OHAD from cortisol substrates.
Identification of the steroid-17,20-desmolase operon and prediction of a novel 20β-HSDH gene in B. desmolans ATCC 43058
The desA-deduced amino acid sequence (CLOSCI_00899) from C. scindens ATCC 35704 was used to query the nonredundant protein database (BLASTP). DesA is annotated in the thiamine-pyrophosphate-dependent enzymes superfamily domain, particularly N-terminal subunit transketolase (EC 2.2.1.1). The highest hits (E value 0.0, 2e−178) were against amino acid sequences encoded by C. cadaveris (WP_027640052) and B. desmolans (WP_031476417) with 82% identity/89% similarity and 81% identity/88% similarity, respectively (supplemental Fig. S1). A deduced amino acid sequence (WP_016455356.1) from the taxon, P. lymphophilum ACS-093-V-SCH5, shared 75% identity/86% similarity with DesA from C. scindens ATCC 35704.
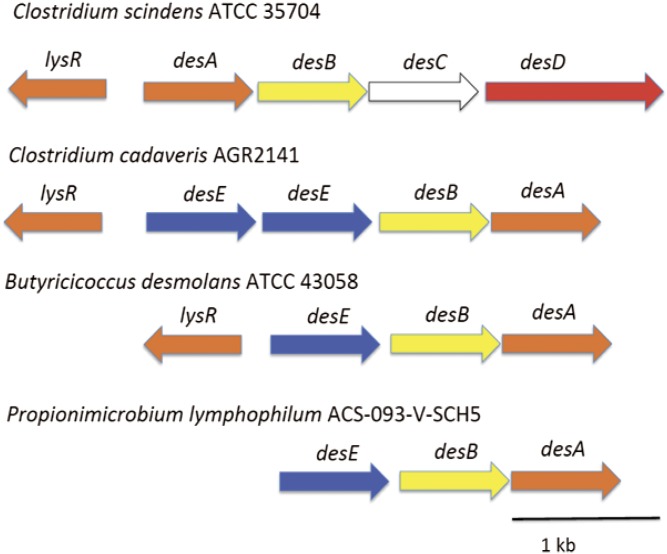
Further examination of the genes flanking desA revealed what appeared to be conserved desB gene in addition to an open reading frame named WP_051643274.1, whose gene product consisted of 282 amino acids containing the canonical Gly-X-X-X-Gly-X-Gly N-terminal pyridine nucleotide-binding motif typical of SDR family enzymes (supplemental Fig. S2) (17, 18). The deduced subunit molecular mass for the protein encoded by WP_051643274.1 is 30.29 kDa. Because desC from C. scindens ATCC 35704 encodes a 20α-HSDH with annotation in the L-threonine dehydrogenase family (EC 1.1.1.103) and B. desmolans and C. cadaveris are reported to express 20β-HSDH (10), we hypothesized that WP_051643274.1 encodes a 20β-HSDH. We are here referring to WP_051643274.1 as the desE gene. Comparison of the gene organization between C. scindens, C. cadaveris, B. desmolans, and P. lymphophylum is depicted in Fig. 2.
Fig. 2.
Identification and comparison of the desmolase operon of B. desmolans ATCC 43058 and C. cadaveris AGR2141 with C. scindens ATCC 35704. lysR, gene encoding putative LysR transcription factor; desA and desB, putative steroid-17,20-desmolase; desC, 20α-HSDH; desE, 20β-HSDH; desD, putative steroid transporter.
Characterization of mRNA encoding desEAB operon in B. desmolans
To determine whether the desEAB genes are coexpressed as a polycistronic operon, we isolated total RNA from mid-log phase B. desmolans and designed oligonucleotides (supplemental Table S1) spanning the intergenic regions between desE-desA and desA-desB. Amplification from the cDNA demonstrates that the 20β-HSDH (desE) is coexpressed with the putative steroid-17,20-desmolase (desAB) (supplemental Fig. S3). The transcription initiation site was determined by SMARTER-RACE PCR to be an adenine residue at position −144. The putative ribosome-binding site (AGGAGGA) was identified starting at position −9. Conserved upstream elements (−149 to −182 and −183 to −196) were identified in alignments between the region between lysR and desA in C. scindens ATCC 35704 and lysR and desE from C. cadavaris AGR2141 and B. desmolans ATCC 43058 (supplemental Fig. S4).
Cloning, expression, and purification of 20β-HSDH from B. desmolans
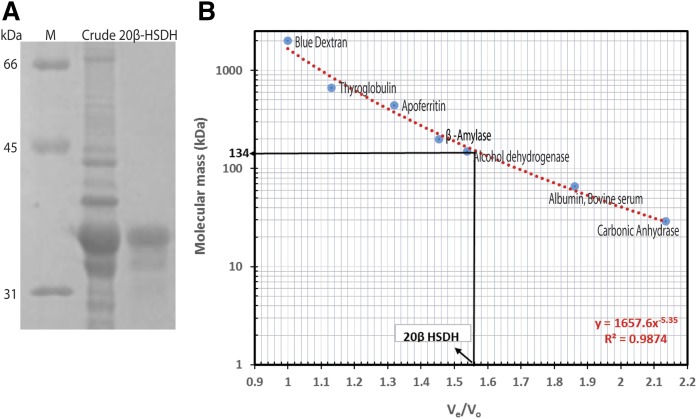
To determine whether WP_051643274.1 encodes 20β-HSDH, the gene was cloned into pET51b(+) expression vector. The N-terminal streptavidin-tagged recombinant protein was purified by Strep-Tactin® affinity chromatography. The recombinant 20β-HSDH protein was resolved as a single band on SDS-PAGE with subunit molecular mass of 33.8 ± 3.7 kDa (Fig. 3A).
Fig. 3.
Purified 20β-HSDH from B. desmolans ATCC 43058. A: SDS-PAGE of crude and purified 20β-HSDH. Lane M, prestained molecular mass marker (Bio-Rad); lane 1, crude cell lysate; lane 2, 20β-HSDH purified by Strep-tag affinity chromatography. B: Native molecular size analysis of purified 20β-HSDH by size-exclusion chromatography.
To determine the native molecular mass, the purified recombinant 20β-HSDH was separated by gel filtration chromatography and eluted at 1.56 Ve/Vo corresponding to a relative molecular mass of 134 ± 4 kDa, eluting after yeast alcohol dehydrogenase (150 kDa) (Fig. 3B). The 20β-HSDH activity of protein fractions corresponding to eluted volume was confirmed with cortisol and NADH by standard spectrophotometry assay (data not shown). The gel filtration elution profile, coupled with the subunit molecular mass determined by SDS-PAGE, suggests a tetrameric structure (3.96) with subunits of identical molecular mass.
Biochemical characterization of recombinant 20β-HSDH
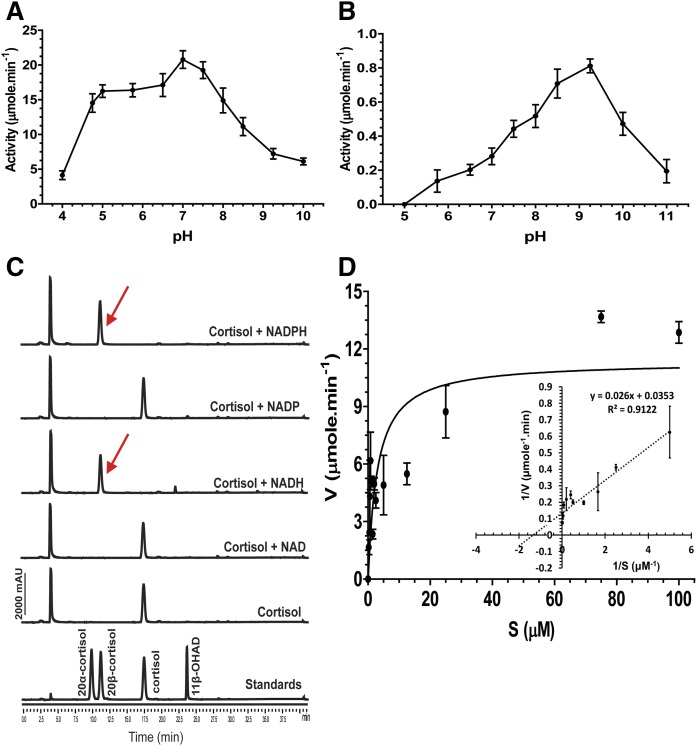
The pH optima of the 20β-HSDH enzyme was determined in the oxidative and reductive directions by spectrophotometry. Steroid reduction was observed within the pH range 4.0–10, retaining 19.9 ± 3.1% of maximal activity at pH 4.0 and 29.5 ± 3.7% of maximal activity at pH 10.0 (Fig. 4A). Between pH 6.5 and pH 7.0 (82.6 ± 10.8% to 100%), we found a statistically significant difference and maximum activity was reached at around pH 7.0–7.5 (100% activity at pH 7.0 vs. 93.2 ± 7.6% at pH 7.5; P < 0.05).
Fig. 4.
Biochemical characterization of recombinant 20β-HSDH from B. desmolans ATCC 43058. A: Effect of pH on reductive activity of 20β-HSDH. The 20β-HSDH (0.05 μM) was incubated with cortisol (50 μM) and NADH (150 μM) in sodium citrate buffer (pH 4–6), sodium phosphate buffer (pH 6.5–7.5), Tris-Cl buffer (pH 8–9), and glycine-NaOH buffer (pH 10) for 5 min. B: Effect of pH on oxidative activity of 20β-HSDH. The 20β-HSDH (0.05 μM) was incubated with cortisol (50 μM) and NAD+ (150 μM) in sodium citrate buffer (pH 5–6), sodium phosphate buffer (pH 6.5–7.5), Tris-Cl buffer (pH 8–9), and glycine-NaOH buffer (pH 10–11) for 5 min. One-way ANOVA followed by Tukey’s multiple comparison tests between the pH values using GraphPad Prism 5.04 (GraphPad Software Inc.). C: Cofactor requirement of 20β-HSDH activity from B. desmolans: 20β-HSDH (0.05 μM) was incubated with cortisol (50 μM) and respective pyridine nucleotides (150 μM) in sodium phosphate buffer (pH 7) overnight. D: Michaelis-Menten and Lineweaver-Burk plot of purified 20β-HSDH using cortisol as substrate.
Similarly steroid oxidation, as measured by the conversion of NAD+ to NADH in the presence of 20β-dihydrocortisol, was observed over a broad pH range, from pH 5.2 to 11.0 with maximal enzyme activity between pH 8.5 and 9.25 (Fig. 4B). Strikingly, even at maximal activity (0.81 ± 0.04 μmol·min−1), the oxidative direction represented only 3.9% of the maximal activity in the reductive direction (20.8 ± 1.2 μmol·min−1). These results indicate that under in vitro conditions, the enzyme catalyzes reductive reactions far more readily than oxidative reactions.
Next, we tested pyridine-nucleotide cofactor specificity in both the reductive and oxidative directions. When NADPH was used as a cosubstrate under optimum conditions for reduction of cortisol, the initial reaction rate observed was <5% of that with NADH, indicating that NAD(H) is the preferred pyridine nucleotide. Nevertheless, the purified enzyme (50 nM) catalyzed the net reduction of cortisol with either NADH or NADPH as the cosubstrate after 12 h incubation; however, formation of 20β-dihydrocortisol was not detected if NAD+ or NADP+ was added as cofactor, as expected (Fig. 4C). Km and Vmax values for cortisol in the presence of saturating NADH were measured from the linear portion of the Lineweaver-Burk plot (Fig. 4D). The calculated kinetic parameters are reported in Table 1.
TABLE 1.
Kinetic parameters for cortisol and NADH with purified 20β-HSDH from B. desmolans ATCC 43058
| Kinetic Parameters | Value |
| Km (μM) | 0.80 ± 0.06 |
| Vmax (μmol·min−1) | 30.36 ± 1.97 |
| Kcat (μmol·μM−1·min−1) | 607 ± 39 |
| Kcat/Km | 760 ± 7.69 |
The substrate-specificity of recombinant 20β-HSDH is shown in Table 2. The equilibrium for recombinant 20β-HSDH clearly favors the reductive direction. Cortisol is the preferred substrate, as it was more rapidly converted to 20β-dihydrocortisol relative to other structurally similar steroids tested (Table 2). Substrates lacking 11β-hydroxy or 17α-hydroxy resulted, respectively, in 49.8% and 39.8% of the activity observed with cortisol. Interestingly, recombinant 20β-HSDH showed only 49.2% of the activity observed with cortisol when 5β-tetrahydrocortisol was substrate, suggesting recognition of the 3-hydroxy/keto and/or AB-ring structure. In the oxidative direction, 20β-hydroxy substrates ranged from no activity to 4.1% activity relative to cortisol, and recombinant 20β-HSDH was not active against 20α-hydroxy substrates.
TABLE 2.
Substrate specificity of purified 20β-HSDH from B. desmolans
| Steroids | Co-substrate | Activity (μmol/min) | Relative Activity (%) |
| 4-pregnen-11β,17,21-triol-20,3-dione | NADH | 21 ± 1.4 | 100 |
| 4-pregnen-17,21-diol-3,20-dione | NADH | 10.5 ± 1.3 | 49.8 |
| 5β-pregnan-3α,11β,17,21-tetrol-20-one | NADH | 10.4 ± 1.7 | 49.2 |
| 4-pregnen-11β,21-diol-3,20-dione | NADH | 8.4 ± 1.3 | 39.8 |
| 4-pregnen-11β,17,20β,21-tetrol-3-one | NAD+ | 0.80 ± 0.02 | 4.1 |
| 4-pregnen-17,20β-diol-3-one | NAD+ | 0.43 ± 0.03 | 2.1 |
| 4-pregnen-11β,20β,21-triol-3-one | NAD+ | NA | NA |
| 4-pregnen-11β,17,20α,21-tetrol-3-one | NAD+ | NA | NA |
| 4-pregnen-17,20α-diol-3-one | NAD+ | NA | NA |
The reactions were initiated by addition of 50 nM purified enzyme to 0.5 ml of 0.05 M sodium phosphate buffer (pH 7) containing 0.15 M NaCl, 20% glycerol, 0.01 M 2-mercaptoethanol, 50 μM steroid, and 150 μM NAD(H) at 37°C. All values are the means of four replicates. NA, no activity.
Phylogenetic analysis of 20β-HSDH
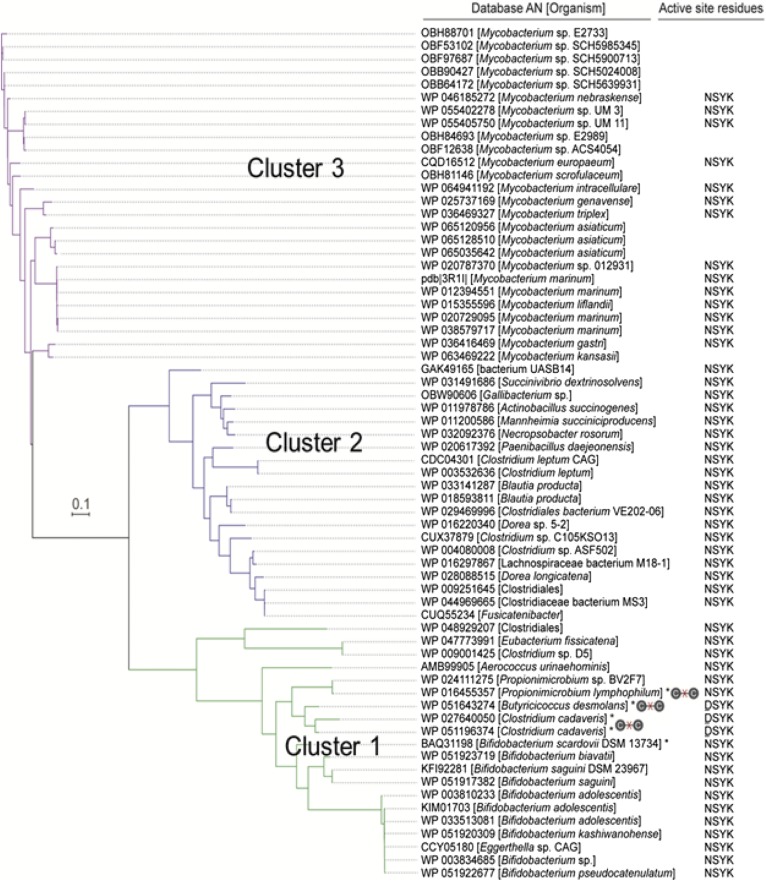
Of the 70 sequences that made the 30% amino acid sequence identity cutoff with desE, the sequences formed three major clusters (Fig. 5), the most distantly related of which was composed exclusively of Mycobacterium spp. (cluster 3), followed by a cluster inhabited by anaerobic gut bacteria inhabiting the gastrointestinal tracts of ruminants and monogastric fermenters (cluster 2). Several of these genes are found in succinate-producing gram-negative ruminants (Succinivibrio dextrinosolvens, Gallibacterium spp., Actinobacillus succinogenes, Mannheimia succiniciproducens). Several others belong to members of Clostridium cluster XVIa, including species of Dorea and Blautia producta, and Clostridium cluster IV, including Clostridium leptum. Notably, a conserved active-site tetrad, NSYK, was identified in the majority of sequences within each cluster, with the exception of B. desmolans and C. cadaveris, whose active-site residues, according to the multiple-sequence alignment, are DSYK (Fig. 5; supplemental Fig. S2).
Fig. 5.
Phylogenetic analysis of identified 20β-HSDH. The amino acid sequences of candidate proteins with an identity cutoff of 30% retrieved from NCBI. The phylogenetic tree was reconstructed using FastTree v2.1.7. The active sites of 20β-HSDH of different organisms are mentioned on right side. Steroid-17,20-desmolase-expressing strains are denoted by an “x” through a C-C bond. Predicted active-site residues are depicted adjacent to strain designations.
Proteins from C. cadaveris are represented twice in cluster 1 (encoded by WP_027640050, WP_051196374), and most closely related to the 20β-HSDH from B. desmolans. Examination of their genomic location revealed that the two putative 20β-HSDH encoding genes share 91% amino acid sequence similarity, suggesting the likelihood of gene duplication (Fig. 2). The two desE genes in C. cadaveris are located adjacent to desAB genes. Closely related to the desE amino acid sequence from B. desmolans and C. cadaveris are SDR-family proteins from two strains of P. lymphophilum, which are detected in urine and have been isolated (Fig. 5). Like B. desmolans and C. cadaveris, the genes encoding putative 20β-HSDH from both strains of P. lymphophilum are adjacent to desAB genes, suggesting that these bacterial strains are capable of converting cortisol to 11β-OHAD.
Cluster 1 also contains several species of bifidobacteria. The desAB genes were not found in the available genome sequences of the bifidobacteria species represented in the phylogeny, suggesting that these strains may harbor 20β-HSDH, but not steroid-17,20-desmolase activity. Indeed, desAB genes were not found in any other members of clusters 1, 2, and 3, except for B. desmolans, C. cadaveris, and P. lymphophilum.
Verification of functional steroid metabolic activity by selected members of cluster III of the 20β-HSDH phylogeny
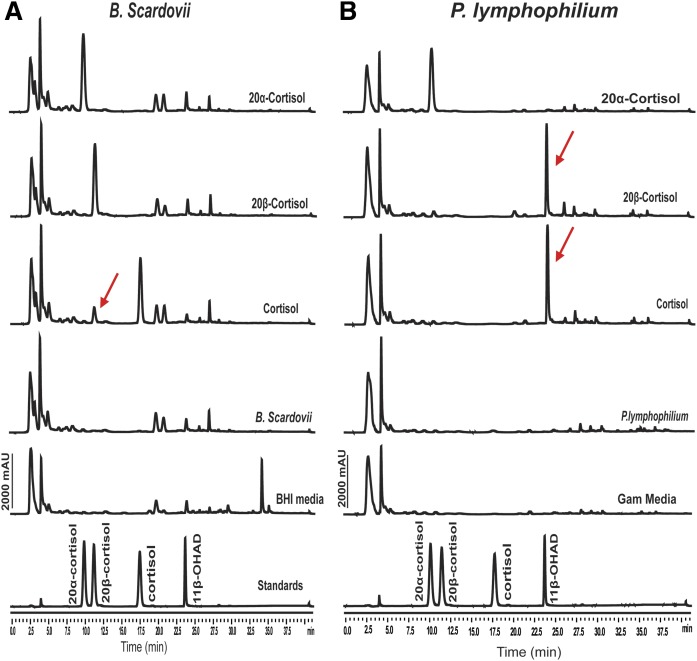
We next sought to determine whether two organisms, whose gene products cluster closely with the 20β-HSDH from B. desmolans ATCC 43058, exhibited 20β-HSDH activity. We tested B. scardovii ATCC BAA-773 and P. lymphophilum ACS-093-V-SCH5 for 20β-HSDH and steroid-17,20-desmolase activity, the latter strain hypothesized to express both activities because it encodes the desABE operon (Fig. 2). Neither organism was capable of metabolizing 20α-dihydrocortisol. We detected the formation of 20β-dihydrocortisol when cortisol was the substrate; however, we did not detect the oxidation of 20β-dihydrocortisol by whole-cells of B. scardovii, which is consistent with the observation that, at equilibrium, the recombinant 20β-HSDH from B. desmolans converted ∼98% of cortisol to 20β-dihydrocortisol in vitro over a 24 h period. We did not detect the formation of 11β-OHAD by cultures of B. scardovii ATCC BAA-773. The 20β-HSDH and steroid-17,20-desmolase activity were confirmed in P. lymphophilum ACS-093-V-SCH5 by the fact that only 20β-hydroxy substrate was side-chain cleaved, forming 11β-OHAD (Fig. 6).
Fig. 6.
Confirmation of 20β-HSDH activity from B. scardovii ATCC BAA-773 (A) and steroid-17,20-desmolase activity in P. lymphophilum ACS-093-V-SCH5 (B). Each steroid substrate (50 μM) was incubated with both B. scardovii in BHI medium and P. lymphophilum in Gifu anaerobic medium for 48 h. The products were extracted and analyzed using HPLC equipped with a C-18 analytical column (Capcell Pak c18). Peak retention times and peak areas of samples were compared with retention times of standards.
DISCUSSION
HSDH enzymes play an important role in steroid and bile acid metabolism for gut bacteria. C. scindens ATCC 35704 expresses 20α-HSDH and steroid-17,20-desmolase, but also 3α-HSDH and 7α-HSDH as part of the complex bile acid 7α-dehydroxylation pathway (19). The 7α-HSDH may serve two functions: 1) convert 7-oxo-bile acids generated by other gut bacteria (20) to 7α-hydroxy bile acids, which can enter the bile acid 7α-dehydroxylation pathway; and 2) regulate the flux of bile acids into the 7α-dehydroxylation pathway by reversible oxidation-reduction of the 7α-hydroxy group (19). Gut bacteria, in human and other mammalian hosts, are also capable of forming 7β-hydroxy bile acids, including, but not limited to, Clostridium baratii (21), B. producta (22), and Ruminococcus gnavus (23). While lacking a 7β-HSDH, necessary to epimerize the 7-hydroxy, C. scindens and other bile acid 7-dehydroxylating bacteria circumvent this need by expressing a 7β-dehydratase (20). Interestingly, epimerization of the 3α-hydroxy group to 3β-hydroxy “iso-bile acids” (24) prevents 7α-dehydroxylation (J. M. Ridlon and P. B. Hylemon, unpublished observations). C. scindens lacks 3β-HSDH, and the first enzymatic step following bile acid CoA-ligation is oxidation of the 3α-hydroxy group (20).
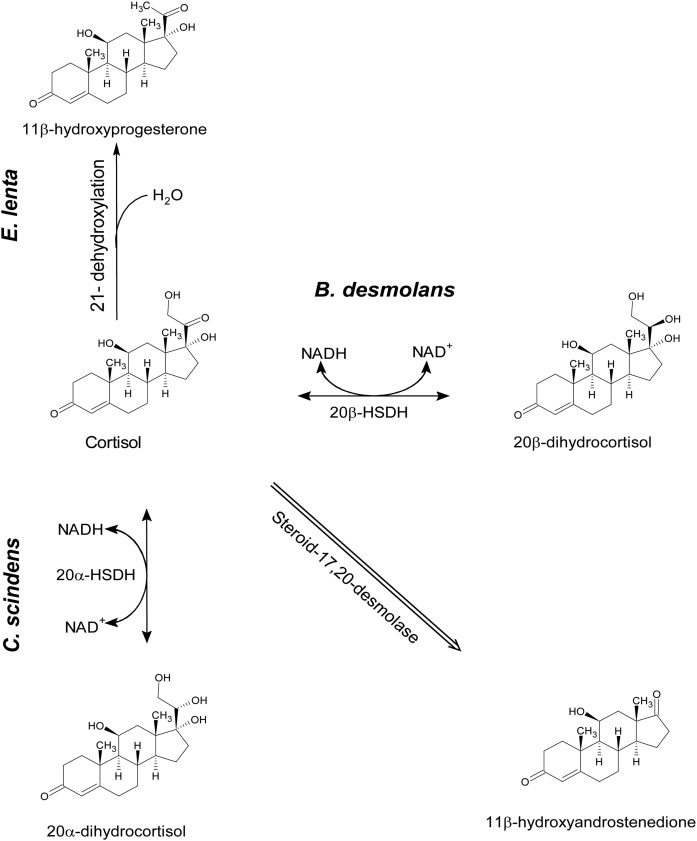
Steroid-17,20-desmolase and 21-dehydroxylation of glucocorticoids are similar to 7α-dehydroxylation of bile acids in that certain bacterial modifications of steroid substrates preclude these pathways. Namely, steroid-17,20-desmolase requires a 20-keto group on 17α,21-dihydroxy corticosteroids (6). C. scindens ATCC 35704 is capable of converting 20α-dihydrocortisol, but not 20β-dihydrocortisol, to 11β-OHAD due to expression of 20α-HSDH, but lacking a gene encoding 20β-HSDH, as we and others (6) have demonstrated. The reverse is true of B. desmolans, C. cadavaris (10), and P. lymphophilum. The rate of 21-dehydroxylation of cortisol, corticosterone, or deoxycorticosterone to progestin-derivatives by Eggerthella lenta may be altered by gut bacterial conversion of 20-keto to 20-hydroxy because E. lenta lacks 20α-HSDH and 20β-HSDH activity, and 21-dehydroxylase requires 20-keto substrates (25, 26). By working out the biochemical pathways in steroid and other microbial metabolic pathways, it may be possible to alter the composition of steroids and bile acids through probiotics and/or develop specific inhibitors of particular bacterial steroid modifying enzymes, resulting in a measurable alteration in the metabolomic profile and phenotypic effects on the host.
Winter et al. (27) isolated a strain of Bifidobacterium adolescentis from human feces that displayed 20β-HSDH activity. Phylogenetic analysis of the desE gene in B. desmolans resulted in identification of several Bifidobacterium spp. strains, including B. adolescentis, encoding desE but lacking desAB. This is consistent with the observation that B. scardovii ATCC BAA-773 produces 20β-dihydrocortisol from cortisol (Fig. 6). The observation that some bifidobacteria strains encode and express 20β-HSDH, but not steroid-17,20-desmolase, is interesting in this regard. It is possible that 20β-HSDH activity in bifidobacteria may reduce the conversion of cortisol to 11β-OHAD in GI tracts dominated by 20α-HSDH-elaborating bacteria, such as C. scindens ATCC 35704.
Metabolites of gut bacterial steroid-17,20-desmolase and steroid 21-dehydroxylase are potent inhibitors of host 11β-HSDH isoform 2 (11β-HSD2) (28–31). The absorption of 11β-hydroxyandrogens and 11β-hydroxyprogestins from the GI tract into the host may have varied effects depending on the target tissue. Inhibition of 11β-HSD2 in colonic epithelium may be protective against colorectal cancer (32). However, inhibition of 11β-HSD2 in vascular tissue (also by inhibition of 11β-HSD1) and kidney is a possible contributing factor in essential hypertension (33). A current method of determining the functionality of 11β-HSDH isoforms in patients is urinary measurement of the cortisol:cortisone ratio and potassium levels (33).
Of particular note is the observation that some strains of P. lymphophilum, which may be members of the normal urinary microbiota (34, 35), previously unrecognized to encode steroid-17,20-desmolase and 20β-HSDH, have been shown in the current study to convert cortisol to 11β-OHAD. This could potentially be of significance given that renal activity of 11β-HSDH-2 is currently measured by the ratio of cortisol:cortisone. However, if significant metabolism of either metabolite or both in the bladder occurs in some individuals, precise measurement of this ratio may not reflect accurate renal activity of this enzyme, and consideration of the urinary microbiome may be necessary.
Finally, our phylogenic analysis of desE also included numerous genes from Mycobacterium spp. (Fig. 7), including M. tuberculosis (supplemental Fig. S5). Mycobacteria encode numerous genes involved in the catabolism of host fatty acids, cholesterol, and oxysterols for the generation of energy and mycolic acid (36, 37), but also endocrine disruption by altering the structure of host immune signaling steroids (38). A 3α,20β-HSDH (Rv2002) has been reported in M. tuberculosis, which is suggested to be involved in steroid metabolism (39). Rv2002 did not show up in our phylogeny (Fig. 5) or in our extended phylogeny (supplemental Fig. S5), which included several M. tuberculosis SDR-family proteins. It is possible that the proteins identified in our phylogeny are utilized by mycobacteria to metabolize host sterols or perhaps environmental sterols from plants and animal material; however, this remains the subject of future investigation.
Fig. 7.
Reactions in the steroid-17,20-desmolase and 21-dehydroxylase pathways by gut bacteria.
Supplementary Material
Footnotes
Abbreviations:
- BHI
- brain heart infusion
- HSDH
- hydroxysteroid dehydrogenase
- 11β-HSD2
- 11β-HSDH isoform 2
- LB
- lysogeny broth
- 11β-OHAD
- 11β-hydroxyandrostenedione
- SDR
- short-chain dehydrogenase/reductase
This work was supported by University of Illinois at Urbana-Champaign (Hatch ILLU-538-916) for new faculty startup (to J.M.R.).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Nabarro J. D. N., Moxham A., Walker G., and Slater J. D. H.. 1957. Rectal hydrocortisone. BMJ. 2: 272–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wade A. P., Slater J. D. M., Kellie A. E., and Holliday M. E.. 1959. Urinary excretion of 17-ketosteroids following rectal infusion of cortisol. J. Clin. Endocrinol. Metab. 19: 444–453. [DOI] [PubMed] [Google Scholar]
- 3.Winter J., and Bokkenheuser V. D.. 1978. 21-dehydroxylation of corticoids by anaerobic bacteria isolated from human fecal flora. J. Steroid Biochem. 9: 379–384. [DOI] [PubMed] [Google Scholar]
- 4.Cerone-McLernon A. M., Winter E. H., Mosbach J., and Bokkenheuser V. D.. 1981. Side-chain cleavage of cortisol by fecal flora. Biochim. Biophys. Acta. 666: 341–347. [DOI] [PubMed] [Google Scholar]
- 5.Bokkenheuser V. D., Morris G. N., Ritchie A. E., Holdeman L. V., and Winter J.. 1984. Biosynthesis of androgen from cortisol by a species of Clostridium recovered from human fecal flora. J. Infect. Dis. 149: 489–494. [DOI] [PubMed] [Google Scholar]
- 6.Winter J., Morris G. N., O’Rourke-Locascio S., Bokkenheuser V. D., Mosbach E. H., Cohen B. I., and Hylemon P. B.. 1984. Mode of action of steroid desmolase and reductases synthesized by Clostridium “scindens” (formerly Clostridium strain 19). J. Lipid Res. 25: 1124–1131. [PubMed] [Google Scholar]
- 7.Krafft A. E., Winter J., Bokkenheuser V. D., and Hylemon P. B.. 1987. Cofactor requirements of steroid-17-20-desmolase and 20 alpha-hydroxysteroid dehydrogenase activities in cell extracts of Clostridium scindens. J. Steroid Biochem. 28: 49–54. [DOI] [PubMed] [Google Scholar]
- 8.Krafft A. E., Winter J., and Hylemon P. B.. 1989. Purification and characterization of 20α-hydroxysteroid dehydrogenase from Clostridium scindens. J. Bacteriol. 171: 2925–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ridlon J. M., Ikegawa S., Alves J. M. P., Zhou B., Kobayashi A., Iida T., Mitamura K., Tanabe G., Serrano M., De Guzman A., et al. 2013. Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J. Lipid Res. 54: 2437–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bokkenheuser V. D., Winter J., Morris G. N., and Locascio S.. 1986. Steroid desmolase synthesis by Eubacterium desmolans and Clostridium cadavaris. Appl. Environ. Microbiol. 52: 1153–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takada T., Watanabe K., Makino H., and Kushiro A.. 2016. Reclassification of Eubacterium desmolans as Butyricicoccus desmolans comb. nov., and Butyricicoccus faecihominis sp. nov., a novel butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 66: 4125–4131. [DOI] [PubMed] [Google Scholar]
- 12.Gill S. C., and von Hippel P. H.. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182: 319–326. [DOI] [PubMed] [Google Scholar]
- 13.Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price M. N., Dehal P. S., and Arkin A. P.. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Letunic I., and Bork P.. 2007. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23: 127–128. [DOI] [PubMed] [Google Scholar]
- 16.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., and Bairoch A.. 2005. Protein identification and analysis tools on the ExPASy server. In Walker J. M., editor. Humana Press Inc., Totowa, NJ: 571–607. [Google Scholar]
- 17.Kallberg Y., Oppermann U., Jörnvall H., and Persson B.. 2002. Short-chain dehydrogenases/reductases (SDRs). Eur. J. Biochem. 269: 4409–4417. [DOI] [PubMed] [Google Scholar]
- 18.Kavanagh K. L., Jörnvall H., Persson B., and Oppermann U.. 2008. The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65: 3895–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ridlon J. M., Harris S. C., Bhowmik S., Kang D. J., and Hylemon P. B.. 2016. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 7: 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ridlon J. M., Kang D. J., and Hylemon P. B.. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47: 241–259. [DOI] [PubMed] [Google Scholar]
- 21.Lepercq P., Gérard P., Béguet F., Raibaud P., Grill J. P., Relano P., Cayuela C., and Juste C.. 2004. Epimerization of chenodeoxycholic acid to ursodeoxycholic acid by Clostridium baratii isolated from human feces. FEMS Microbiol. Lett. 235: 65–72. [DOI] [PubMed] [Google Scholar]
- 22.Hirano S., and Masuda N.. 1982. Characterization of NADP-dependent 7β-hydroxysteroid dehydrogenases from Peptostreptococcus productus and Eubacterium aerofaciens. Appl. Environ. Microbiol. 43: 1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J. Y., Arai H., Nakamura Y., Fukiya S., Wada M., and Yokota A.. 2013. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J. Lipid Res. 54: 3062–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devlin A. S., and Fischbach M. A.. 2015. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 11: 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feighner S. D., and Hylemon P. B.. 1980. Characterization of a corticosteroid 21-dehydroxylase from the intestinal anaerobic bacterium, Eubacterium lentum. J. Lipid Res. 21: 585–593. [PubMed] [Google Scholar]
- 26.Feighner S. D., Bokkenheuser V. D., Winter J., and Hylemon P. B.. 1979. Characterization of a C21 neutral steroid hormone transforming enzyme, 21-dehydroxylase, in crude cell extracts of Eubacterium lentum. Biochim. Biophys. Acta. 574: 154–163. [DOI] [PubMed] [Google Scholar]
- 27.Winter J., Cerone-McLernon A., O’Rourke S., Ponticorvo L., and Bokkenheuser V. D.. 1982. Formation of 20β-dihydrosteroids by anaerobic bacteria. J. Steroid Biochem. 17: 661–667. [DOI] [PubMed] [Google Scholar]
- 28.Morris D. J., and Souness G. W.. 1996. Endogenous 11β-hydroxysteroid dehydrogenase inhibitors and their role in glucocorticoid Na+ retention and hypertension. Endocr. Res. 22: 793–801. [DOI] [PubMed] [Google Scholar]
- 29.Morris D. J., Latif S. A., Hardy M. P., and Brem A. S.. 2007. Endogenous inhibitors (GALFs) of 11β-hydroxysteroid dehydrogenase isoforms 1 and 2: derivatives of adrenally produced corticosterone and cortisol. J. Steroid Biochem. Mol. Biol. 104: 161–168. [DOI] [PubMed] [Google Scholar]
- 30.Lo Y. H., Sheff M. F., Latif S. A., Ribeiro C., Silver H., Brem A. S., and Morris D. J.. 1997. Kidney 11β-HSD2 is inhibited by glycyrrhetinic acid-like factors in human urine. Hypertension. 29: 500–505. [DOI] [PubMed] [Google Scholar]
- 31.Latif S. A., Pardo H. A., Hardy M. P., and Morris D. J.. 2005. Endogenous selective inhibitors of 11β-hydroxysteroid dehydrogenase isoforms 1 and 2 of adrenal origin. Mol. Cell. Endocrinol. 243: 43–50. [DOI] [PubMed] [Google Scholar]
- 32.Stewart P. M., and Prescott S. M.. 2009. Can licorice lick colon cancer? J. Clin. Invest. 119: 760–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrari P. 2010. The role of 11β-hydroxysteroid dehydrogenase type 2 in human hypertension. Biochim. Biophys. Acta. 1802: 1178–1187. [DOI] [PubMed] [Google Scholar]
- 34.Lewis D. A., Brown R., Williams J., White P., Jacobson S. K., Marchesi J. R., and Drake M. J.. 2013. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front. Cell. Infect. Microbiol. 3: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stackebrandt E., Schumann P., Schaal K. P., and Weiss N.. 2002. Propionimicrobium gen. nov., a new genus to accommodate Propionibacterium lymphophilum (Torrey 1916) Johnson and Cummins 1972, 1057AL as Propionimicrobium lymphophilum comb. nov. Int. J. Syst. Evol. Microbiol. 52: 1925–1927. [DOI] [PubMed] [Google Scholar]
- 36.Egorova O. V., Nikolayeva V. M., Suzina N. E., and Donova M. V.. 2005. Localization of 17β-hydroxysteroid dehydrogenase in Mycobacterium sp. VKM Ac-1815D mutant strain. J. Steroid Biochem. Mol. Biol. 94: 519–525. [DOI] [PubMed] [Google Scholar]
- 37.Yang X., Dubnau E., Smith I., and Sampson N. S.. 2007. Rv1106c from Mycobacterium tuberculosis is a 3β-hydroxysteroid dehydrogenase. Biochemistry. 46: 9058–9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bah S. Y., Dickinson P., Forster T., Kampmann B., and Ghazal P.. Immune oxysterols: role in mycobacterial infection and inflammation. J. Steroid Biochem. Mol. Biol. Epub ahead of print. May 4, 2016; doi:10.1016/j.jsbmb.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 39.Yang J. K., Park M. S., Waldo G. S., and Suh S. W.. 2003. Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 100: 455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.