Abstract
Lipids play a central role in lung physiology and pathology; however, a comprehensive lipidomic characterization of human pulmonary cells relevant to disease has not been performed. The cells involved in lung host defense, including alveolar macrophages (AMs), bronchial epithelial cells (BECs), and alveolar type II cells (ATIIs), were isolated from human subjects and lipidomic analysis by LC-MS and LC-MS/MS was performed. Additionally, pieces of lung tissue from the same donors were analyzed by MALDI imaging MS in order to determine lipid localization in the tissue. The unique distribution of phospholipids in ATIIs, BECs, and AMs from human subjects was accomplished by subjecting the large number of identified phospholipid molecular species to univariant statistical analysis. Specific MALDI images were generated based on the univariant statistical analysis data to reveal the location of specific cell types within the human lung slice. While the complex composition and function of the lipidome in various disease states is currently poorly understood, this method could be useful for the characterization of lipid alterations in pulmonary disease and may aid in a better understanding of disease pathogenesis.
Keywords: bronchial epithelial cells, alveolar type II cells, alveolar macrophages
Lipids play an important role in lung physiology and pathology. A well-known example is that of leukotrienes and prostaglandins, which are lipid mediators derived from enzymatic metabolism of arachidonic acid (AA) released from phospholipids in cell membranes. These lipid mediators are involved in lung inflammation and responsible for impaired mucociliary clearance, bronchoconstriction, enhanced mucous production, neutrophil recruitment to the airways, and pulmonary edema (1, 2). In fact, therapeutic intervention based on either interference with eicosanoid production or receptor antagonists are frequently used as approaches to manage asthma, pulmonary hypertension, COPD, and cystic fibrosis (3–7). Additionally, other phospholipids also play a role in lung biology. Dipalmitoylphosphatidylcholine is the major surface-active component of pulmonary surfactant and is essential for reducing surface tension in the alveoli and for efficient gas exchange (8). Lastly, exogenously added palmitoyl-oleoyl-phosphatidylglycerol vesicles have been shown to reduce inflammation in the lung and act as a potent antiviral agent (9).
The human lung is composed of up to 40 different types of cells, as well as pulmonary surfactant, and each type of cell contains a unique distribution of phospholipid molecular species. The diversity of phospholipids observed in biological samples is due to variations in the polar headgroup (choline, ethanolamine, serine, inositol, and glycerol) at the sn-3 position of the glycerol backbone. Additional complexity is introduced by the numerous combinations of fatty acyl constituents at the sn-1 and sn-2 position of the glycerol backbone. Lipidomic analyses by electrospray MS/MS of pulmonary surfactant in subjects with asthma (10), cystic fibrosis (11), and acute respiratory distress syndrome (12) have demonstrated that an altered distribution of phospholipid molecular species in the surfactant is related to the pathology of the disease.
Pulmonary cells of interest in inflammatory disease pathogenesis, such as asthma, COPD, and cystic fibrosis, are those involved in host defense and therefore responsible for protection against inhaled particulate matter, toxic gases, and infectious agents (13–16). The first cells to encounter inhaled substances are located in the bronchial epithelium. The bronchial epithelium is composed of three different cell types, including basal, ciliated, and secretory cells, and acts as a defensive barrier for the maintenance of normal airway function and plays a central role in the initiation of inflammatory responses (17). Additionally alveolar type II cells (ATIIs) and alveolar macrophages (AMs) are also involved in host defense in the lung, but are present in the alveolar space where gas exchange occurs. Alveolar type II epithelial cells contribute to lung defense due to their participation in epithelial repair and ion transport, secretion of antimicrobial products, and production of surfactant (18). AMs also participate in the defense of the respiratory tract, as these cells have a high phagocytic capacity and play an important role in the initiation and resolution of inflammatory responses in the alveolar region (19).
While it is clear that lipids play a central role in pulmonary disease and biology, the lipidome of human pulmonary cells responsible for host defense has not been well-characterized. Previously, an examination of the lipidome of cultured fetal ATIIs was performed and the molecular species of phospholipids present were identified to the fatty acid level (20). Additionally, the mole percentage of various lipids identified to the number of acyl carbon atoms and double bonds in human airway epithelial cells has been reported (21). In the current study, we present an alternative lipidomics approach to investigate the localization of phospholipid molecular species in the human lung. This new approach combines LC-MS and LC-MS/MS lipidomic analysis of freshly isolated human ATIIs, bronchial epithelial cells (BECs), and AMs combined with MALDI imaging MS (IMS) of human lung tissue from the same donor. Application of univariant statistical analysis to the large number of phospholipid molecular species identified to the fatty acid level by lipidomic analysis in combination with MALDI IMS has been used to reveal the unique distribution of phospholipids in pulmonary cells from human subjects that are important to lung defense. While the complex composition and function of the lipidome in various disease states is currently poorly understood, this method could be useful for the characterization of lipid alterations in pulmonary disease and may aid in a better understanding of disease pathogenesis.
MATERIALS AND METHODS
Reagents
Polyvinyl alcohol 6-98, 2,5-dihydroxybenzoic acid, and 2,6-dihydroxyacetophenone (DHAP) were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO). HBSS (1×) was purchased from Invitrogen (Carlsbad, CA). Polypropylene glycol, average MW 2,000 g/mol (PPG 2000), and HPLC grade organic solvents were purchased from Fisher Scientific (Fair Lawn, NJ). OCT medium was obtained from Ted Pella, Inc. (Redding, CA) and was used to attach lung tissue to the cryostat chuck; modified OCT (mOCT) (described below) was used to inflate isolated human lung tissue after dissection.
Human lung tissue and cells
Human lung was obtained from de-identified organ donors whose lungs could not be transplanted and were donated for medical research. This research has been deemed nonhuman subject research and IRB exempt because the donors are deceased and de-identified. The lungs were procured through the International Institute for the Advancement of Medicine, as described elsewhere (22).
Isolation of individual cell types
AMs were isolated from lavage of the lung before the instillation of elastase, as described previously (23). ATIIs were isolated by elastase digestion of the lung, and purification by centrifugation on a discontinuous density gradient made of Optiprep (Accurate Chemical Scientific Corp., Westbury, NY) with densities of 1.085 and 1.040 and by positive selection with MACS MicroBeads human CD326 (EpCAM) (Miltenyi Biotech, Bergisch Gladbach, Germany) (24). BECs were obtained by direct brushing of the bronchi of the donor lung. The purity of the isolated cell types was based on immunostaining of cytocentrifuge preparations. ATIIs were identified based on proSP-C (Seven Hills Bioreagents, Cincinnati, OH) immunostaining, BECs based on staining for cytokeratin (CAM 5.2; DakoCytomation, Carpineria, CA) and keratin 5 (Thermo Fisher Scientific Inc., Waltham, MA), and AMs based on CD68 (DakoCytomation) staining. Immunostaining was done as described previously (23). Cells were mounted with Vectorshield medium containing DAPI (Vector Laboratories, Burlingame, CA) and analyzed by fluorescent microscopy (Zeiss Axioskop 2; Thornwood, NY). The purity of the AMs was 92.6 ± 2.8%, the purity of the BECs was 90.2 ± 5.0% and contained a mixture of mostly ciliated and secretory cells with very few keratin V basal cells, and the purity of ATIIs was 84.2 ± 2.8%. Immunostaining of representative fields is presented in supplemental Fig. S1.
Lipid extraction and normal phase HPLC/MS and HPLC/MS/MS analysis of phospholipids in pulmonary cells
The phospholipids from the freshly isolated AMs, ATIIs, and BECs (2 × 106 cells) were extracted according to the method of Bligh and Dyer (25). For LC/MS analysis, the Bligh-Dyer extract was introduced onto the mass spectrometer and the lipid classes present were separated using normal phase (NP)-HPLC with an Ascentis 5μ Si (15 cm × 2.1 mm) column (Supelco, Bellefonte, PA). A NP solvent system of 30:40 hexane/2-propanol (solvent A) and 30:40:7 hexane/2-propanol/water with a final concentration of 1 mM ammonium acetate (solvent B) was used. The initial mobile phase (46% solvent B) was held for 3 min, and a linear gradient was started from 46% solvent B to 70% solvent B in 22 min, followed by another linear gradient from 70% solvent B to 100% solvent B in 5 min. The final step was isocratic elution at 100% solvent B for 20 min. Using this gradient, the phosphatidylglycerol (PG) lipids eluted from 4 to 5 min, the phosphatidylethanolamine (PE) and phosphatidylinositol (PI) lipids eluted from 8 to 10 min, the phosphatidylserine (PS) lipids eluted from 12 to 14 min, and the phosphatidylcholine (PC) lipids eluted from 35 to 40 min. The phospholipid classes were well-separated, except for PI and PE, which coeluted; however, this was not problematic in determining the mole percentages because the mass ranges of the PE and PI lipids did not overlap.
The HPLC system was directly interfaced to the electrospray source of a triple quadrupole mass spectrometer (MDS Sciex API 3200; Applied Biosystems, Foster City, CA). Detection of both positive (PC) and negative (PI, PG, PE, and PS) ions during the entire chromatographic run was achieved by polarity switching every 3 s and full scans were obtained in both the positive and negative ion mode from m/z 500 to 1,100. The positive ion mode mass spectrometric experimental parameters in the positive ion mode were an electrospray voltage of 4,000 V and a declustering potential of 50 V and in negative ion mode were an electrospray voltage of −4,500 V and a declustering potential of −45 V. The mass spectrum across each NP chromatographic peak was integrated and the molecular ion abundance was determined. The mole percentages reported are based on this molecular ion abundance after correction for naturally occurring isotopes (26). For PG, PS, PE, and PI molecular species the [M-H]− ion was used and for PC the [M+H]+ ion was used in determining the mole percentages of each lipid in AMs, BECs, and ATIIs. The identity of the phospholipids was determined by collision-induced dissociation (CID) of the [M-H]− for PE, PG, PI, and PS lipids, which provides information about the fatty acids esterified to the glycerol backbone (27), and allowed for the identification of phospholipids to the fatty acyl level. Additionally, the [M-CH3]− of PC lipids in the negative ion mode were collisionally activated to determine fatty acyl constituents. These demethylated PC anions were generated by setting a high declustering potential (−150 V) so that both acetate and chloride adduct ions were not observed.
Statistical analysis
Lipidomics data consisting of identified phospholipid molecular species and their mole percentages were analyzed using R-based statistical methods (available at the site www.metabolomicsworkbench.org that is a resource supported by the National Institutes of Health Common Fund Metabolomics Program). These R-based methods calculate standard deviations of the observed abundance of each molecular species of lipid, as measured in a single sample (same human subject) of the three lung cell types. The standard error of the mean of the three samples (supplemental Tables S1–S5) was determined by calculating the sample standard deviation (using n − 1 as the number of samples).
Preparation of human lungs for MALDI IMS
Three chunks of lung tissue (approximately 15 × 15 mm) were obtained from the same three human lungs used for the isolation of the pulmonary cells. The human lung sections taken for imaging were approximately 1.5 cm3 and were inflated by injecting 2 ml mOCT mixture with an 18 gauge needle in several areas until the mOCT could be seen emerging on the surface of the tissue section. The degree of inflation was preserved by keeping the tissue below −20°C at all times during cryosectioning. The preparation of mOCT [10% polyvinyl alcohol 6-98 and 8% PPG 2000 in HBSS (1×)] has been described previously (28). The pieces of inflated human lung were placed in standard cryomolds, covered with mOCT, placed at 20°C for 2 h, and stored until slicing at 70°C. The embedded lungs were sliced at a thickness of 14 μm with a cryostat (Reichert-Jung Cryocut 1800) at −16°C. Greater than six individual slices of lung tissue from each of the three human subjects were analyzed by MALDI IMS.
Matrix application
The glass coverslips containing the tissue sections were attached to stainless steel MALDI plate inserts (OptiToF; Applied Biosystems) using copper tape and the 2,5-dihydroxybenzoic acid or DHAP matrix was deposited onto the tissues by sublimation (29). After MALDI IMS, the glass coverslip with the lung section was removed from the MALDI plate, dipped in methanol to remove the matrix and fix the tissue prior, and subject, to modified Giemsa staining using standard protocols.
MALDI IMS
A quadrupole-TOF tandem mass spectrometer with an orthogonal MALDI source (QSTAR XL; Applied Biosystems/MDS Sciex, Thornhill, Ontario, Canada) was used to acquire images. MALDI mass spectra were obtained using a solid state laser (355 nm) at a power of 100 μJ and a pulse rate of 20 Hz with an accumulation time of 243 ms per image spot. The MALDI plate was moved at a rate of 12.75 mm/min and after each horizontal line was completed, the plate was moved vertically 50 μm. The mass spectrometric data were processed using a specialized script for Analyst software (Applied Biosystems/MDS Sciex) at a mass resolution of 0.1 amu and images were visualized using TissueView software (Applied Biosystems/MDS Sciex). The lateral resolution of this MALDI IMS technique is approximately 50 μm. From the mass spectra obtained from each pixel across the human lung tissue, an ion of interest can be extracted and an image of that particular ion distribution in the tissue can be visualized. Molecular species were identified in MALDI images by collisional activation in the negative ion mode directly from the tissue in separate experiments. The [M-H]− of PI, PE, PS, and PG and the [M+DHAP-H]− of the DHAP adducts of PC yielded fatty acid ions that allowed for the identification of the molecular ions observed during MALDI IMS. Collisional activation of selected ions was carried out using a relative collision energy of 40 V, with argon as collision gas.
RESULTS
Lipidomic analysis of pulmonary cells
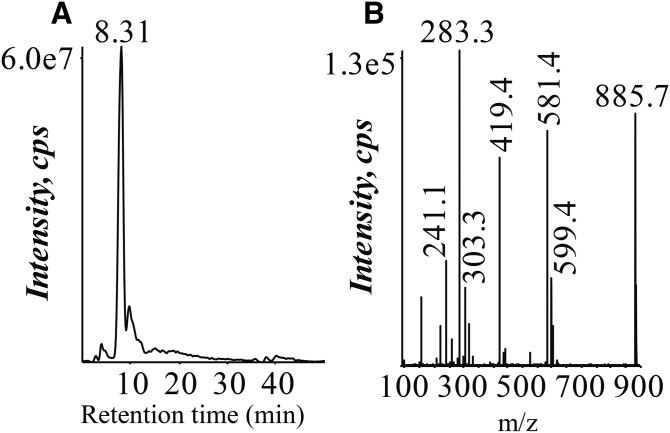
BECs, AMs, and ATIIs were analyzed by LC/MS and LC/MS/MS protocols that were developed to specifically determine phospholipid molecular species, including determination of the fatty acyl substituents present in each lipid molecular species. An example is illustrated using the particularly abundant AA-containing PI lipid at m/z 885.7 (Fig. 1). This lipid was identified by criteria that included the correct NP chromatographic retention time for the class of PI phospholipids (8–10 min) (Fig. 1A) and subsequent collisional activation of the ion at m/z 885.7 (Fig. 1B). Carboxylate anions corresponding to AA (m/z 303) as well stearic acid (m/z 283) were observed in the CID spectrum of m/z 885.7. The abundant product ions at m/z 581 and 599, which corresponded to the neutral loss of AA [M-H-R2COOH]− and the loss of AA as a neutral ketene [M-H-R2C=O]− , indicate AA at the sn-2 position. Additionally, several specific ions corresponding to the inositol polar head group were observed at m/z 315, 259, 241, and 223 (30). All of this information in the CID spectrum as well as the NP retention time lead to identification of the ion at m/z 885.7 as PI(18:0/20:4). No attempt was made to assign the positions of the fatty acyl groups to the sn-1 or sn-2 position of the glycerol backbone for most other phospholipid molecular species in this study.
Fig. 1.
Lipidomic LC/MS and LC/MS/MS analysis to identify phospholipid molecular species, including fatty acyl substituents in human pulmonary cells. A: Extraction of m/z 885.7, PI(18:0_20:4), from the negative ion NP-HPLC/MS analysis of the Bligh-Dyer extract of human BECs. B: CID of m/z 885.7 from the negative ion NP-HPLC/MS/MS analysis of the Bligh-Dyer extract of human BECs.
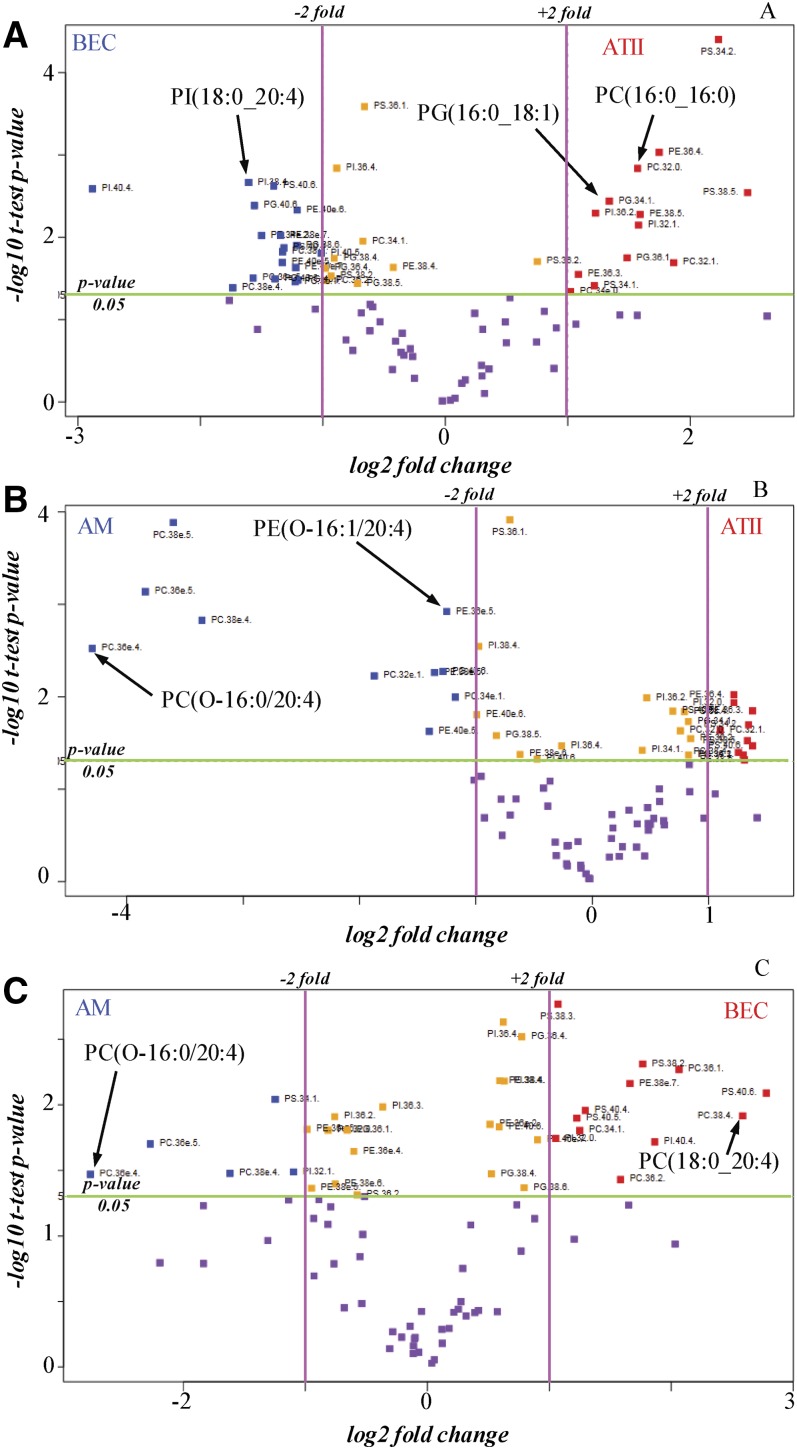
From the analysis of the phospholipids in BECs, AMs, and ATIIs, a list of molecular species of five different phospholipid classes was constructed, including PC, PE, PG, PI, and PS that were specifically identified to the molecular species level. The abundance of the molecular ions derived from each of the individual molecular species was normalized to each lipid class and presented as the mole fraction of lipid present in that class (supplemental Tables S1–S5). These lipidomics data were then processed by statistical methods using the mole fraction abundances to calculate fold differences for each of the phospholipid molecular species. The output of a univariant statistical analysis (volcano plot) derived from the lipidomic analysis of BECs, AMs, and ATIIs is presented in Fig. 2 in which the fold change is compared with the statistical significance level between the two types of cells under comparison. The phospholipids are labeled in Fig. 2 with the headgroup, total number of carbons, and double bonds in order to fit the labels onto the graph, but it should be noted that the molecular species identification had been determined for each phospholipid, as reported in supplemental Tables S1–S5. The extremities of the volcano plots indicate either highly negative or positive significant (t-test, P < 0.05) fold differences (log-based 2 > 1) and provide insight into those phospholipid molecular species that can be used to uniquely distinguish each of the cell types examined.
Fig. 2.
Volcano plots of differentially abundant lipids in human pulmonary cells identified by mass spectrometric lipid analysis. The −log10 t-test P values were plotted against the fold change in mole fraction abundances of the phospholipid molecular species. The horizontal green line represents a statistical significance of 0.05 and the pink vertical lines represent the phospholipid molecular species up or down 2-fold in the two compared cell types. The data points in purple have a P value >0.05 and, therefore, are not significantly different in the two types of cells compared, while those data points in orange are significantly different (P < 0.05), but have a small fold change (<2-fold) between the two types of cells. When comparing two types of cells, the blue data points indicate phospholipids that are significantly abundant by more than 2-fold in the first cell type and the red data points reveal phospholipid molecular species that are significantly abundant by more than 2-fold in the second cell type. BECs versus ATIIs (A), AMs versus ATIIs (B), and AMs versus BECs (C).
Three volcano plots were generated from the lipidomics data from human BECs, AMs, and ATIIs to cross compare the phospholipids present in these three different pulmonary cell types (Fig. 2). For example, Fig. 2A compares the mole fractions of the phospholipid molecular species in BECs to those observed in ATIIs. The data points in purple have a P value >0.05 and, therefore, are not significantly different in BECs compared with ATIIs, while many phospholipid molecular species were significantly different (P < 0.01) and furthermore were greater than 2-fold differentially present in the two types of cells. For example, PI(18:0/20:4), indicated by a blue data point in the upper left hand corner of Fig. 2A, was abundant by greater than 3-fold (P < 0.003) in BECs relative to ATIIs (supplemental Table S6). In contrast, the red data points in the upper right hand corner of Fig. 2A reveal that phospholipid molecular species, such as PC(16:0_16:0) and PG(16:0_18:1), were significantly abundant (P < 0.05) by more than 2-fold in ATIIs. In addition, many other phospholipids differed with a P value <0.05 and more than 2-fold change upon comparison of BECs and ATIIs (supplemental Table S6). The volcano plot in Fig. 2B compares the phospholipid distribution of the AMs to the ATIIs. This figure demonstrates that phospholipids containing an ether linkage to the glycerol backbone, such as PC(O-16:0_20:4) and PE(O-16:1_20:4), are significantly abundant (P < 0.05) by more than 2-fold in AMs compared with ATIIs. This volcano plot also reveals that phospholipid species that were abundant in surfactant, such as PI(16:0_16:0) and PC(16:0_16:1), were significantly abundant by greater than 2-fold in ATIIs compared with AMs. Upon comparison of phospholipid molecular species in AMs compared with ATIIs, many other phospholipids were significantly different (supplemental Table S7). The fold change of phospholipid molecular species in AMs compared with BECs is revealed in Fig. 2C. Phospholipids containing an ether linkage to the glycerol backbone were once again significantly abundant (P < 0.05) by more than 2-fold in AMs compared with BECs. In contrast, phospholipid molecular species, such as PC(18:0_20:4), were significantly abundant in BECs compared with AMs. Upon comparison of AMs and BECs, other phospholipids also differed (supplemental Table S8).
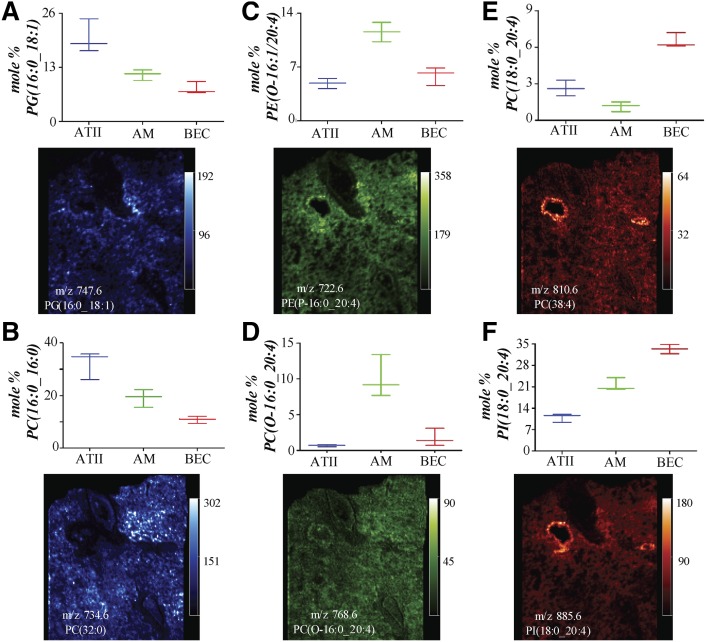
Phospholipid species of interest identified from each of the volcano plots were used to generate box plots to reveal the median distribution of the mole percent of representative molecular species within each of the three different cell types (Fig. 3). Examination of the box plots for PG(16:0_18:1) and PC(16:0_16:0) reveals that these phospholipid molecular species were most abundant in ATIIs, less abundant in AMs, and of lowest abundance in BECs (Fig. 3A, B). The box plots for PE(O-16:1/20:4) and PC(O-16:0_20:4) indicate that these ether lipids were of lowest abundance in ATIIs and the most abundant in AMs (Fig. 3C, D). Lastly, the box plots for PC(18:0_20:4) and PI(18:0_20:4) reveal that these diacyl AA-containing phospholipids were most abundant in the BECs (Fig. 3E, F).
Fig. 3.
Mole fraction abundances of specific phospholipid molecular species in ATIIs, AMs, and BECs of interest determined from the volcano plots in Fig. 2 and MALDI images of the same molecular species in human lung slices. The MALDI images for PG, PI, and PE molecular species are all from the same lung slice in negative ion mode and the MALDI images for the PC molecular species are in positive ion mode from an adjacent lung slice. PG(16:0_18:1) (A), PC(16:0_16:0) (B), PE(O-16:1_20:4) (C), PC(O-16:0_20:4) (D), PC(18:0_20:4) (E), and PI(18:0_20:4) (F).
Combined lipidomics and MALDI IMS data
Once the lipidomics analysis was complete, it was possible to directly interrogate MALDI IMS data of human lung tissue using the molecular species identified by the statistical analysis of the lipidomics data to generate specific images that would reveal the location of specific cell types within the MALDI image. This more focused examination of specific molecular species revealed from the lipidomics data, rather than merely mapping the tissue distribution of the most abundant ions observed in the MALDI IMS experiment, is a more direct method of searching for the unique distribution of specific molecular ions. The distribution of phospholipid molecular species uniquely abundant in human ATIIs, AMs, and BECs in MALDI images of human lung tissue were quite different (Fig. 3). ATIIs were found to be enriched in PG(16:0_18:1) and PC(16:0_16:0) from the lipidomics data and the MALDI images of these phospholipids (Fig. 3A, B) revealed regions of low signal intensity for these two lipid molecular ions along the luminal walls of the larger airways and blood vessels, and regions of higher signal intensity were interspersed throughout the alveolar region. Additionally, it was determined from lipidomics data that ether PC and PE lipids were most abundant in AMs and the MALDI images of PC(O-16:0_20:4) and PE(O-16:1_20:4) demonstrated that these phospholipid molecular species were rather uniformly present throughout the human lung tissue slice in alveolar regions, except for a somewhat higher abundance of the PE-phospholipid molecular species near the adventitia of the larger airways (Fig. 3C, D). Furthermore, the lipidomics data revealed that PC(18:0_20:4) and PI(18:0_20:4) were most abundant in BECs and the MALDI images indicated that these phospholipid molecular species were highly abundant at the luminal edge of the larger airways, with a much lower abundance in the parenchyma (Fig. 3E, F).
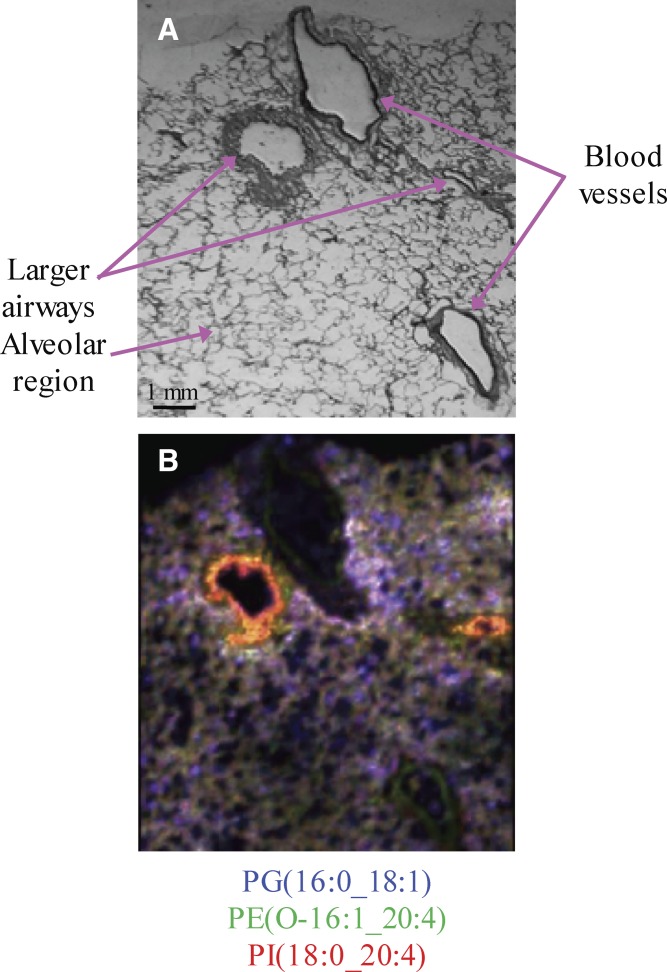
The MALDI images of PG(16:0_18:1) (blue), PE(O-16:1_20:4) (green), and PI(18:0_20:4) (red) were merged and the differences in the distribution of these molecular species across the lung tissue slice were distinctly highlighted (Fig. 4). It was clear from this merged image that, even though all three phospholipid molecular species, PG(16:0_18:1), PE(O-16:1_20:4), and PI(18:0_20:4), were present in the alveolar region, their distribution in the human lung slice was quite different (Fig. 4B). Most notably, PG(16:0_18:1) had regions of higher intensity throughout the entire lung slice, while PI(18:0_20:4) was of higher intensity near the larger airways. Additionally, the merged MALDI image suggested that the ether-containing AA lipid, PE(O-16:1_20:4), was colocalized in a few regions with the diacyl AA lipid, PI(18:0_20:4), as suggested by the yellow regions of the larger airway adventitia (Fig. 4B). However, the merged MALDI image showed a high intensity of PI(18:0_20:4) (red) at the luminal edges of larger airways.
Fig. 4.
Comparative localization of phospholipid molecular species in a human lung slice. A: Modified Giemsa stain of the human lung slice after MALDI IMS in negative ion mode. B: Merged negative ion MALDI image of PE(O-16:1_20:4) (green) at m/z 722.6, PI(18:0_20:4) (red) at m/z 885.6, and PG(16:0_18:1) (blue) at m/z 747.6.
DISCUSSION
The phospholipid composition of individual cell types present in the human lung is distinct due to the complex nature by which phospholipid molecular species are synthesized within each cell. Membrane phospholipids are the product of de novo synthesis by the Kennedy pathway, which results in saturated or monounsaturated fatty acyl groups esterified to the phospholipid glycerol backbone (31). Another mechanism for the generation of phospholipids, independent of de novo synthesis, is through the Lands pathway, which incorporates polyunsaturated fatty acids into individual molecular species of lysophospholipids through the action of lysophospholipid acyltransferases (32). There are numerous genes that regulate the production of the individual proteins engaged in this complex phospholipid biosynthetic pathway within cells and the corresponding expression of individual gene products can differ between cell types. The specificity of each of the enzyme types is also correlated to the number of enzymatic isoforms. Humans express four different lysophospholipid acyltransferases, each of which has very different substrate preferences in terms of utilization of fatty acyl CoAs to generate the individual phospholipid molecular species (33). For example, LPCAT3 (MBOAT5) is rather specific for the incorporation of AA into lysophosphatidylcholine and lysophosphatidylserine intermediates (34). Thus, the relatively high expression of these enzymes within inflammatory cells, such as the AMs and BECs, results in a larger relative proportion of AA-containing phospholipids within these cell types. The abundance of LPCAT1 in the type II cell results in a very active pathway for the production of PC(16:0/16:0) as a primary component of pulmonary surfactant (35). The current lipidomic study of ATIIs, BECs, and AMs reflects a unique combination of enzymatic activity within each cell type, including phospholipases, acyl-CoA synthetases, and lysophospholipid acyltransferases, that plays a major role in determination of phospholipid composition.
A comprehensive profile of phospholipid molecular species present in freshly isolated human BECs has been demonstrated for the first time. The lipid content of human BECs was previously examined; however, the phospholipids were only identified by the number of acyl carbon atoms and acyl double bonds, not the fatty acids esterified to the sn-1 and sn-2 position (21). One of the most interesting findings of the current study is that the human bronchial epithelium is highly enriched in diacyl AA-containing phospholipids according to both the lipidomics and MALDI IMS data (Figs. 2, 3). These results suggest that the bronchial epithelium could be a source of eicosanoids, potent lipid mediators derived from AA, in the lung. Eicosanoid production is initiated by the release of AA from membrane phospholipids by a calcium-dependent cytosolic phospholipase A2 (cPLA2α) (36). The 15-lipoxygenase (LO), which has been implicated in the pathogenesis of bronchial asthma, is an enzyme that uses AA as a substrate and is highly expressed in the bronchial epithelium (37, 38). The major AA metabolite formed via 15-LO is 15-HETE and increased amounts of this eicosanoid have been found in the BALF of antigen-challenged asthmatic patients, as well as in the sputum samples of asthmatic subjects (37, 39). Additionally, 15-HETE can also be esterified to PE in the bronchial epithelium, which results in increased mucus production (40). Cyclooxygenase (COX)-1 and COX-2 are enzymes also present in the bronchial epithelium that utilize AA released from phospholipids by cPLA2α as a substrate for the production of prostaglandins (41). Prostaglandins have been found to mediate both bronchoconstriction (PGD2 and PGF2α) and bronchoprotection (PGE2 and PGI2) (42). The level of prostaglandins in BALF is increased in asthma (43) and enhanced expression of COX-2 in the airways of asthmatics has been observed (44). The 5-LO is another enzyme that metabolizes AA and results in leukotrienes. Leukotrienes play a central role in asthma and mediate mucus secretion, bronchoconstriction, neutrophil recruitment to the airways, and pulmonary edema. Additionally, elevated levels of leukotrienes have been detected in BALF and sputum from subjects with asthma (45). One study has reported that primary BECs isolated from human airways express an inducible form of 5-LO (46), but it is generally considered that the major source of leukotrienes in the lung is from cells of myeloid origin, like the AMs or recruited inflammatory cells (47). While the current study does not identify the phospholipid source of eicosanoids, it is possible that the bronchial epithelium or AMs, which were found to contain a high mole percentage of AA-containing phospholipids, could be a source of lipid mediators and are at a location where these mediators could exert profound effects.
AMs are found in the alveolus and extend from the alveolar walls into the airway lumen, which results in the continuous exposure of these cells to inhaled matter (48). The current study is also the first to provide a detailed profile of the phospholipids present in freshly isolated human AMs. In particular, this lipidomic study indicated that PE and PC ether phospholipid molecular species were highly abundant in AMs compared with BECs and ATIIs (Figs. 2, 3). These ether phospholipids could be plasmalogen lipids that previously have been reported to be present in high abundance in inflammatory cells (49). One of the suggested roles of plasmalogen lipids is to act as antioxidant molecules within cellular membranes (50). This antioxidant activity is a result of the low dissociation energy of the hydrogen atoms adjacent to the vinyl ether bond, which results in preferential oxidation of plasmalogen lipids over diacyl phospholipids when exposed to reactive oxygen and nitrogen species. Due to the location of the AMs, the plasmalogen phospholipids that are present in this particular cell type are likely one of the initial targets of inhaled reactive gases (ozone, cigarette smoke, and nitric oxides) or endogenous reactive oxygen species made by inflammatory cells, and have the ability to impede these reactive oxygen species from causing further oxidative damage to cellular proteins or lipids.
The alveolar region of lungs is comprised of type I and type II alveolar epithelial cells. The ATIIs are responsible for epithelial reparation upon injury and for the production of pulmonary surfactant and contribute to lung host defense by the secretion of antimicrobial factors (18). Pulmonary surfactant, which regulates alveolar surface tension for efficient ventilation and alveolar stability, is stored in lamellar bodies in ATIIs upon synthesis, where it is stored until exocytosis. The composition of pulmonary surfactant is unique in that it contains 90% lipid and 10% protein. The most abundant class of phospholipids found in pulmonary surfactant is PC (∼80%) (51). The major PC molecular species present in human surfactant contain either saturated or monounsaturated fatty acids, with PC(16:0_16:0) as the most abundant (52). Additionally, PG lipids are present in relatively high amounts (∼10%) compared with their distribution in other tissues and fluids, with PG(16:0_18:1) as the most abundant. Because ATIIs are the source of pulmonary surfactant that is stored until exocytosis, it is expected that the lipidomic analysis of ATIIs somewhat reflects that of pulmonary surfactant. In fact, both PC(16:0_16:0) and PG(16:0_18:1) were found to have the highest relative mole percentage in ATIIs, as compared with the human AMs and BECs in this study (Figs. 2, 3). Until the current study, there was little information as to the lipid distribution in freshly isolated human ATIIs. Postle et al. (20) have reported the lipids present in fetal human ATIIs cultured in vitro under basal and hormone stimulated conditions. AMs contain an intermediate amount of surfactant lipids, such as PC(16:0_16:0) and PG(16:0_18:1), most likely because the phagocytic AMs participate in the turnover of surfactant components from the airspace.
The examination of phospholipid molecular species and their distribution within tissues and cell types remains a challenging task. The new lipidomics approach proposed in the current study to examine the localization of phospholipid molecular species in the human lung is based on LC-MS and LC-MS/MS analysis of freshly isolated human ATIIs, BECs, and AMs combined with MALDI IMS of human lung tissue from the same donor. The integrated data obtained from these complementary techniques provides unique information as to the lipid content and localization in lung tissue. This study provides the first in depth and comprehensive lipidomic analysis of freshly isolated ATIIs, BECs, and AMs from the same individuals. The method used herein could be useful for the characterization of lipid changes in inflammatory pulmonary disease states, such as asthma, COPD, and cystic fibrosis in order to gain a better understanding of disease pathogenesis.
Supplementary Material
Acknowledgments
The authors thank the families of the de-identified organ donors and the Institute for the Advancement of Medicine, who made this research possible.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- AM
- alveolar macrophage
- ATII
- alveolar type II cell
- BEC
- bronchial epithelial cell
- CID
- collision-induced dissociation
- COX
- cyclooxygenase
- DHAP
- 2,6-dihydroxyacetophenone
- IMS
- imaging MS
- LO
- lipoxygenase
- mOCT
- modified OCT
- NP
- normal phase
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PPG
- polypropylene glycol
- PS
- phosphatidylserine
This work was supported by a grant from the Exxon Mobil Foundation (R.J.M.), Flight Attendant Medical Research Institute Grant CIA130046 (B.K.), and National Institutes of Health Grants 7R01HL118171 (B.K.) and 4U54HL117798 (R.C.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Samuelsson B., Dahlén S. E., Lindgren J. A., Rouzer C. A., and Serhan C. N.. 1987. Leukotrienes and lipoxins: structures, biosynthesis, and biological effects. Science. 237: 1171–1176. [DOI] [PubMed] [Google Scholar]
- 2.Lewis R. A., Austen K. F., and Soberman R. J.. 1990. Leukotrienes and other products of the 5-lipoxygenase pathway. Biochemistry and relation to pathobiology in human diseases. N. Engl. J. Med. 323: 645–655. [DOI] [PubMed] [Google Scholar]
- 3.Riccioni G., Bucciarelli T., Mancini B., Di Ilio C., and D’Orazio N.. 2007. Antileukotriene drugs: clinical application, effectiveness and safety. Curr. Med. Chem. 14: 1966–1977. [DOI] [PubMed] [Google Scholar]
- 4.Bozyk P. D., and Moore B. B.. 2011. Prostaglandin E2 and the pathogenesis of pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 45: 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drakatos P., Lykouras D., Sampsonas F., Karkoulias K., and Spiropoulos K.. 2009. Targeting leukotrienes for the treatment of COPD? Inflamm. Allergy Drug Targets. 8: 297–306. [DOI] [PubMed] [Google Scholar]
- 6.Zaslona Z., and Peters-Golden M.. 2015. Prostanoids in asthma and COPD: actions, dysregulation, and therapeutic opportunities. Chest. 148: 1300–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konstan M. W., Döring G., Heltshe S. L., Lands L. C., Hilliard K. A., Koker P., Bhattacharya S., Staab A., and Hamilton A.; Investigators and Coordinators of BI Trial 543.45. 2014. A randomized double blind, placebo controlled phase 2 trial of BIIL 284 BS (an LTB4 receptor antagonist) for the treatment of lung disease in children and adults with cystic fibrosis. J. Cyst. Fibros. 13: 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veldhuizen R., Nag K., Orgeig S., and Possmayer F.. 1998. The role of lipids in pulmonary surfactant. Biochim. Biophys. Acta. 1408: 90–108. [DOI] [PubMed] [Google Scholar]
- 9.Numata M., Chu H. W., Dakhama A., and Voelker D. R.. 2010. Pulmonary surfactant phosphatidylglycerol inhibits respiratory syncytial virus-induced inflammation and infection. Proc. Natl. Acad. Sci. USA. 107: 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright S. M., Hockey P. M., Enhorning G., Strong P., Reid K. B., Holgate S. T., Djukanovic R., and Postle A. D.. 2000. Altered airway surfactant phospholipid composition and reduced lung function in asthma. J. Appl. Physiol. 89: 1283–1292. [DOI] [PubMed] [Google Scholar]
- 11.Mander A., Langton-Hewer S., Bernhard W., Warner J. O., and Postle A. D.. 2002. Altered phospholipid composition and aggregate structure of lung surfactant is associated with impaired lung function in young children with respiratory infections. Am. J. Respir. Cell Mol. Biol. 27: 714–721. [DOI] [PubMed] [Google Scholar]
- 12.Tibby S. M., Hatherill M., Wright S. M., Wilson P., Postle A. D., and Murdoch I. A.. 2000. Exogenous surfactant supplementation in infants with respiratory syncytial virus bronchiolitis. Am. J. Respir. Crit. Care Med. 162: 1251–1256. [DOI] [PubMed] [Google Scholar]
- 13.Balhara J., and Gounni A. S.. 2012. The alveolar macrophages in asthma: a double-edged sword. Mucosal Immunol. 5: 605–609. [DOI] [PubMed] [Google Scholar]
- 14.Zhao C. Z., Fang X. C., Wang D., Tang F. D., and Wang X. D.. 2010. Involvement of type II pneumocytes in the pathogenesis of chronic obstructive pulmonary disease. Respir. Med. 104: 1391–1395. [DOI] [PubMed] [Google Scholar]
- 15.Volsko T. A. 2013. Airway clearance therapy: finding the evidence. Respir. Care. 58: 1669–1678. [DOI] [PubMed] [Google Scholar]
- 16.Grainge C. L., and Davies D. E.. 2013. Epithelial injury and repair in airways diseases. Chest. 144: 1906–1912. [DOI] [PubMed] [Google Scholar]
- 17.Hiemstra P. S., McCray P. B., and Bals R.. 2015. The innate immune function of airway epithelial cells in inflammatory lung disease. Eur. Respir. J. 45: 1150–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fehrenbach H. 2001. Alveolar epithelial type II cell: defender of the alveolus revisited. Respir. Res. 2: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuquimia O. D., Petursdottir D. H., Periolo N., and Fernández C.. 2013. Alveolar epithelial cells are critical in protection of the respiratory tract by secretion of factors able to modulate the activity of pulmonary macrophages and directly control bacterial growth. Infect. Immun. 81: 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Postle A. D., Gonzales L. W., Bernhard W., Clark G. T., Godinez M. H., Godinez R. I., and Ballard P. L.. 2006. Lipidomics of cellular and secreted phospholipids from differentiated human fetal type II alveolar epithelial cells. J. Lipid Res. 47: 1322–1331. [DOI] [PubMed] [Google Scholar]
- 21.Zehethofer N., Bermbach S., Hagner S., Garn H., Müller J., Goldmann T., Lindner B., Schwudke D., and König P.. 2015. Lipid analysis of airway epithelial cells for studying respiratory diseases. Chromatographia. 78: 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J., Edeen K., Manzer R., Chang Y., Wang S., Chen X., Funk C. J., Cosgrove G. P., Fang X., and Mason R. J.. 2007. Differentiated human alveolar epithelial cells and reversibility of their phenotype in vitro. Am. J. Respir. Cell Mol. Biol. 36: 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., Nikrad M. P., Travanty E. A., Zhou B., Phang T., Gao B., Alford T., Ito Y., Nahreini P., Hartshorn K., et al. 2012. Innate immune response of human alveolar macrophages during influenza A infection. PLoS One. 7: e29879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito Y., Correll K., Schiel J. A., Finigan J. H., Prekeris R., and Mason R. J.. 2014. Lung fibroblasts accelerate wound closure in human alveolar epithelial cells through hepatocyte growth factor/c-Met signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 307: L94–L105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 26.Han X., and Gross R. W.. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24: 367–412. [DOI] [PubMed] [Google Scholar]
- 27.Pulfer M., and Murphy R. C.. 2003. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 22: 332–364. [DOI] [PubMed] [Google Scholar]
- 28.Berry K. A., Li B., Reynolds S. D., Barkley R. M., Gijón M. A., Hankin J. A., Henson P. M., and Murphy R. C.. 2011. MALDI imaging MS of phospholipids in the mouse lung. J. Lipid Res. 52: 1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hankin J. A., Barkley R. M., and Murphy R. C.. 2007. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18: 1646–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu F. F., and Turk J.. 2000. Characterization of phosphatidylinositol, phosphatidylinositol-4-phosphate, and phosphatidylinositol-4,5-bisphosphate by electrospray ionization tandem mass spectrometry: a mechanistic study. J. Am. Soc. Mass Spectrom. 11: 986–999. [DOI] [PubMed] [Google Scholar]
- 31.Kennedy E. P., and Weiss S. B.. 1956. The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 222: 193–214. [PubMed] [Google Scholar]
- 32.Lands W. E. 1960. Metabolism of glycerolipids. 2. The enzymatic acylation of lysolecithin. J. Biol. Chem. 235: 2233–2237. [PubMed] [Google Scholar]
- 33.Chang C. C. Y., Sun J., and Chang T-Y.. 2011. Membrane-bound O-acyltransferases (MBOATs). Front. Biol. 6: 177–182. [Google Scholar]
- 34.Gijón M. A., Riekhof W. R., Zarini S., Murphy R. C., and Voelker D. R.. 2008. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J. Biol. Chem. 283: 30235–30245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X., Hyatt B. A., Mucenski M. L., Mason R. J., and Shannon J. M.. 2006. Identification and characterization of a lysophosphatidylcholine acyltransferase in alveolar type II cells. Proc. Natl. Acad. Sci. USA. 103: 11724–11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh M., Tucker D. E., Burchett S. A., and Leslie C. C.. 2006. Properties of the group IV phospholipase A2 family. Prog. Lipid Res. 45: 487–510. [DOI] [PubMed] [Google Scholar]
- 37.Chu H. W., Balzar S., Westcott J. Y., Trudeau J. B., Sun Y., Conrad D. J., and Wenzel S. E.. 2002. Expression and activation of 15-lipoxygenase pathway in severe asthma: relationship to eosinophilic phenotype and collagen deposition. Clin. Exp. Allergy. 32: 1558–1565. [DOI] [PubMed] [Google Scholar]
- 38.Shannon V. R., Chanez P., Bousquet J., and Holtzman M. J.. 1993. Histochemical evidence for induction of arachidonate 15-lipoxygenase in airway disease. Am. Rev. Respir. Dis. 147: 1024–1028. [DOI] [PubMed] [Google Scholar]
- 39.Profita M., Sala A., Riccobono L., Paternò A., Mirabella A., Bonanno A., Guerrera D., Pace E., Bonsignore G., Bousquet J., et al. 2000. 15-Lipoxygenase expression and 15(S)-hydroxyeicoisatetraenoic acid release and reincorporation in induced sputum of asthmatic subjects. J. Allergy Clin. Immunol. 105: 711–716. [DOI] [PubMed] [Google Scholar]
- 40.Zhao J., O’Donnell V. B., Balzar S., St Croix C. M., Trudeau J. B., and Wenzel S. E.. 2011. 15-Lipoxygenase 1 interacts with phosphatidylethanolamine-binding protein to regulate MAPK signaling in human airway epithelial cells. Proc. Natl. Acad. Sci. USA. 108: 14246–14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demoly P., Jaffuel D., Lequeux N., Weksler B., Créminon C., Michel F. B., Godard P., and Bousquet J.. 1997. Prostaglandin H synthase 1 and 2 immunoreactivities in the bronchial mucosa of asthmatics. Am. J. Respir. Crit. Care Med. 155: 670–675. [DOI] [PubMed] [Google Scholar]
- 42.Claar D., Hartert T. V., and Peebles R. S.. 2015. The role of prostaglandins in allergic lung inflammation and asthma. Expert Rev. Respir. Med. 9: 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu M. C., Bleecker E. R., Lichtenstein L. M., Kagey-Sobotka A., Niv Y., McLemore T. L., Permutt S., Proud D., and Hubbard W. C.. 1990. Evidence for elevated levels of histamine, prostaglandin D2, and other bronchoconstricting prostaglandins in the airways of subjects with mild asthma. Am. Rev. Respir. Dis. 142: 126–132. [DOI] [PubMed] [Google Scholar]
- 44.Sousa A. r., Pfister R., Christie P. E., Lane S. J., Nasser S. M., Schmitz-Schumann M., and Lee T. H.. 1997. Enhanced expression of cyclo-oxygenase isoenzyme 2 (COX-2) in asthmatic airways and its cellular distribution in aspirin-sensitive asthma. Thorax. 52: 940–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montuschi P. 2010. Role of leukotrienes and leukotriene modifiers in asthma. Pharmaceuticals (Basel). 3: 1792–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jame A. J., Lackie P. M., Cazaly A. M., Sayers I., Penrose J. F., Holgate S. T., and Sampson A. P.. 2007. Human bronchial epithelial cells express an active and inducible biosynthetic pathway for leukotrienes B4 and C4. Clin. Exp. Allergy. 37: 880–892. [DOI] [PubMed] [Google Scholar]
- 47.Peters-Golden M. 1998. Cell biology of the 5-lipoxygenase pathway. Am. J. Respir. Crit. Care Med. 157: S227–S231. [PubMed] [Google Scholar]
- 48.Lambrecht B. N. 2006. Alveolar macrophage in the driver’s seat. Immunity. 24: 366–368. [DOI] [PubMed] [Google Scholar]
- 49.Zemski Berry K. A., and Murphy R. C.. 2004. Electrospray ionization tandem mass spectrometry of glycerophosphoethanolamine plasmalogen phospholipids. J. Am. Soc. Mass Spectrom. 15: 1499–1508. [DOI] [PubMed] [Google Scholar]
- 50.Engelmann B. 2004. Plasmalogens: targets for oxidants and major lipophilic antioxidants. Biochem. Soc. Trans. 32: 147–150. [DOI] [PubMed] [Google Scholar]
- 51.Bernhard W., Wang J. Y., Tschernig T., Tümmler B., Hedrich H. J., and von der Hardt H.. 1997. Lung surfactant in a cystic fibrosis animal model: increased alveolar phospholipid pool size without altered composition and surface tension function in cftrm1HGU/m1HGU mice. Thorax. 52: 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Postle A. D., Heeley E. L., and Wilton D. C.. 2001. A comparison of the molecular species compositions of mammalian lung surfactant phospholipids. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 129: 65–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.