Abstract
Alzheimer’s disease (AD) is one of the fastest-growing causes of death and disability in persons 65 years of age or older, affecting more than 5 million Americans alone. Clinical manifestations of AD include progressive decline in memory, executive function, language, and other cognitive domains. Research efforts within the last three decades have identified APOE as the most significant genetic risk factor for late-onset AD, which accounts for >99% of cases. The apoE protein is hypothesized to affect AD pathogenesis through a variety of mechanisms, from its effects on the blood-brain barrier, the innate immune system, and synaptic function to the accumulation of amyloid-β (Aβ). Here, we discuss the role of apoE on the biophysical properties and metabolism of the Aβ peptide, the principal component of amyloid plaques and cerebral amyloid angiopathy (CAA). CAA is characterized by the deposition of amyloid proteins (including Aβ) in the leptomeningeal medium and small arteries, which is found in most AD cases but sometimes occurs as an independent entity. Accumulation of these pathologies in the brain is one of the pathological hallmarks of AD. Beyond Aβ, we will extend the discussion to the potential role of apoE on other amyloidogenic proteins found in AD, and also a number of diverse neurodegenerative diseases.
Keywords: ATP binding cassete A1, apolipoproteins, brain lipids, high density lipoprotein
ApoE: PHYSIOLOGIC FUNCTIONS AND RISK FACTOR FOR ALZHEIMER’S DISEASE
In 1907, the German psychiatrist, Alois Alzheimer, published a case report, “On an unusual illness of the cerebral cortex,” in which he described what we now know as Alzheimer’s disease (AD) (1). More than a century after this landmark discovery, AD is now recognized as the most common cause of late-life dementia. AD pathology is characterized by the cerebral accumulation of amyloid plaques and neurofibrillary tangles, both of which consist predominantly of specific insoluble protein aggregates. The majority of AD cases are sporadic and have a relatively late onset, predominantly after the age of 65. This form of AD is known as late-onset AD (LOAD), in contrast to early-onset AD, which has a strong genetic component, is often autosomal dominant, and accounts for less than 1% of AD cases (2). While most genetic risk factors for LOAD identified in the past two decades have a relatively small impact on AD risk, extensive epidemiological, clinical, and pathological studies have established the APOE gene on chromosome 19 as the most important genetic risk factor for developing LOAD (3–5). This locus encodes a 299 amino acid glycoprotein (apoE) that is expressed in several cell types, with highest expression levels found in the liver and the brain, where it is expressed predominantly by astrocytes (6) and, to a lesser extent, microglia (7, 8). The human APOE gene contains several single nucleotide polymorphisms that result in changes to the coding region of the apoE protein. The three most common variants of apoE are apoE2 (cys112, cys158), apoE3 (cys112, arg158), and apoE4 (arg112, arg158). Although the three most common isoforms differ by only one or two amino acids, they have distinct structure and function, which may explain the differential effects they exert on AD risks. Relative to the prevalent ε3/ε3 genotype, carriers with one copy of the ε4 allele have an ∼3.7-fold increase, while those with two copies have a 12-fold increase in risks for developing AD. The ε2 allele appears to be protective [odds ratio (OR) = 0.4] relative to the ε3/ε3 genotype (9–11).
Like other apolipoproteins, apoE primarily functions to maintain the structure of specific lipoprotein particles, as well as to direct lipoproteins to specific cell surface receptors. apoE is found in a class of HDL-like lipoprotein in the cerebrospinal fluid (CSF) and interstitial fluid (ISF) of the brain (12, 13). In the periphery, apoE is associated with many different classes of lipoproteins, including VLDLs, intermediate density lipoproteins, chylomicron remnants, and a subclass of HDL (6). Mice (14) and humans (15) lacking apoE suffer from marked peripheral hypercholesterolemia. In the CNS, in vitro experiments have suggested that apoE supports synaptogenesis (16) and the maintenance of synaptic connections (17), by virtue of the cholesterol species it carries. However, there is not strong evidence that it has this function in vivo. Furthermore, both in vitro (18, 19) and in vivo (20, 21) evidence support a role of apoE in neuronal sprouting after injury. Perhaps unexpectedly, in the absence of injury, brain function appears to be grossly normal in the absence of apoE, as observed in both mice (22, 23) and humans (15). This raises the question of whether the increased risk of ε4 is due to a loss of protective function or a gain of toxic function. Though the answer to this question is not definitive, numerous studies have suggested that one major mechanism by which apoE affects AD pathology is through its influence on the accumulation of amyloid plaques in the brain and cerebrovasculature.
APOE AND THE AMYLOID-β PROTEIN
Amyloid plaques are one of the pathological hallmarks of AD and consist mostly of aggregated fibrils of amyloid-β (Aβ). Aβ peptides are 38–43 amino acids in length, although the most common species found in AD brains by far are Aβ1-40 (Aβ40) and Aβ1-42 (Aβ42). Aβ is generated by sequential cleavage of the amyloid precursor protein (APP) by β- and γ-secretases. The precise location and the number of cleavages determine the ultimate length of the peptide. Autosomal dominantly inherited missense mutations in APP or components of the human γ-secretase complex, such as presenilin (PS)1 or PS2 (24), appear to cause early-onset AD by increasing Aβ production or by increasing the ratio of Aβ42 relative to Aβ40 (25). In vitro kinetic studies indicate that Aβ42 is more prone to aggregation and that Aβ peptides ending at amino acid 42 (e.g., Aβ1-42 or Aβ3-42), specifically, play a critical role in determining the rate of amyloid formation (26).
The link between apoE and AD, and apoE and Aβ particularly, was first suggested in the early 1990s, when apoE was found to colocalize with amyloid plaques (27, 28). Subsequently, the ε4 allele of apoE was identified as a strong genetic risk factor for AD (5, 29, 30). Histopathologic examination of postmortem brains from AD patients found a positive correlation between senile plaque density and dosage of the APOE-ε4 allele (29, 31). While some studies did not reproduce the aforementioned findings (32, 33), a more recent study that examined a large (n = 296) cohort of autopsied AD brains found a significant correlation between the APOE-ε4 allele and neuritic plaques (34). The development (35) and clinical utilization (36) of Pittsburgh compound-B (PiB) as a positron emission topography tracer for amyloid deposits revolutionized how AD is diagnosed and monitored. A thioflavin T derivative, radioisotope-labeled PiB readily crosses the blood-brain barrier (BBB) (37) and provides a quantitative in vivo detection of amyloid plaques. Early clinical studies utilizing PiB in cognitively normal middle-aged and older people found APOE-ε4 gene dosage to be associated with fibrillar Aβ burden (38, 39), along with low CSF Aβ42 (39, 40). Consistent with prior epidemiological observations, APOE-ε2 carriers rarely develop fibrillar Aβ that is detected by PiB (39). Altogether, data from human studies make a strong case for the apoE genotype as a strong susceptibility factor for cerebral amyloid plaque accumulation and eventual development of AD.
Aside from clinical investigations, a significant amount of basic research effort in the field has focused on understanding the effects of apoE on the accumulation of amyloid plaques and cerebral amyloid angiopathy (CAA) in the brain. Deposition of Aβ into insoluble plaques depends on several factors, including the rate of production, clearance from the brain ISF (where amyloid plaques are found), and the rate of fibrillization, all of which may be influenced by apoE. We discuss the current state of knowledge on each aspect of the process below.
APOE and Aβ production
Among the multitude of factors that have been hypothesized to affect Aβ deposition, studies on apoE’s role in Aβ production are probably the most controversial. In vitro studies from one group using rat neuroblastoma B103 cells that express human APP suggested that lipid-poor apoE4 enhanced Aβ production compared with lipid-poor apoE3, and that this effect was mediated through the LRP pathway (41). A very recent study shows that recombinant and HEK cell-derived apoE can increase Aβ synthesis in human embryonic stem cell-derived neurons in vitro in an isoform-dependent fashion (apoE4 > apoE3 > apoE2) (Fig. 1, dashed red arrow)(42). The observed phenomenon was driven by differences in the isoforms’ ability to bind and activate surface apoE receptors, which ultimately activate cFos-containing AP-1 transcription factors and increase APP transcription. Of note, APP transcription was maximally activated when human neurons were cocultured with glial cells (which secrete several glial-derived factors), and the isoform-specific effects were abolished under such conditions. However, other in vitro and in vivo studies found no apparent apoE isoform effect on APP processing and Aβ production (43–45). In agreement with the latter findings, in vivo data from APP transgenic (Tg) mice found no changes in amyloidogenic processing of APP or Aβ synthesis rates according to human apoE isoforms (46). These conflicting findings might be explained by their different experimental conditions, such as the lipidation state of apoE and the experimental model system used to generate Aβ. Harmonizing the disparate in vitro and in vivo findings on the role of APOE on Aβ production will require additional studies. Future work assessing Aβ synthesis in the presence of different apoE isoforms in vivo in animal models that have the endogenous APP promoter, as well as in humans, will provide insight into this important question.
Fig. 1.
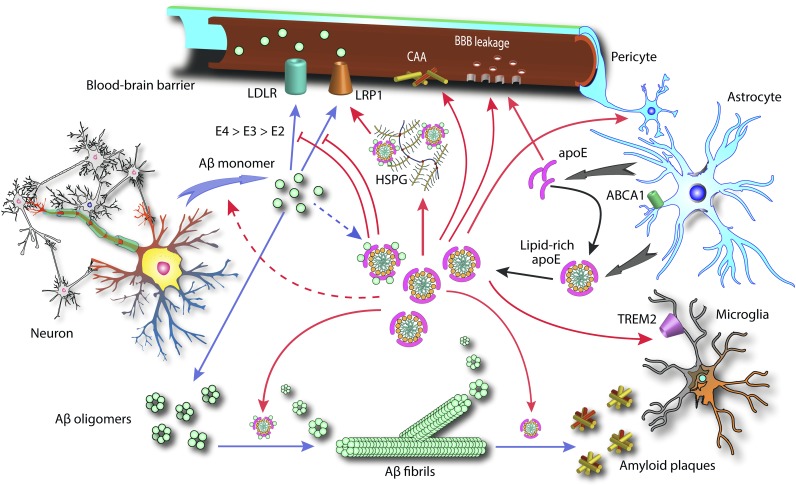
Pathways by which apoE and Aβ interact in the brain. Solid black arrows indicate normal secretion and lipidation of apoE by astrocytes. Solid blue arrows indicate the aggregation cascade and potential clearance route for Aβ. Solid red arrows indicate pathways or processes that apoE has been shown to influence. Dashed blue arrow indicates a small pool of Aβ-apoE complexes. Dashed red arrow indicates apoE’s proposed effect on Aβ production.
APOE-Aβ interaction
Early studies that focused on the direct interaction between apoE and Aβ yielded contradictory findings. apoE was first proposed to be an Aβ binding partner in 1992 (28). Various studies showed that synthetic Aβ avidly forms a complex with apoE purified from human CSF (5), plasma (47), or the conditioned media of apoE-expressing HEK cells (48). However, the affinity of different apoE isoforms for Aβ appears to be highly dependent on the preparation condition of apoE and the species of Aβ involved (soluble versus fibrillar), as well as the pH level. Allan Roses’ group found apoE4 to bind synthetic Aβ more rapidly than apoE3 (47, 49), while LaDu et al. (48) found lipidated apoE3 to be a much more efficient binding partner of Aβ (20-fold) than apoE4. These differences might be explained by the lipidation state of the apoE species being used, as several other groups confirmed that the efficiency of complex formation between lipidated apoE and Aβ follows the order of apoE2 > apoE3 >> apoE4 (50–52). However, one study noted that the observed apoE-Aβ complex represents only a small proportion of the incubated apoE, despite a large molar excess of Aβ (48). In support of this notion, a previous study from our group provided both in vitro and in vivo evidence that more than 95% of the soluble Aβ (sAβ) detected is not associated with apoE-containing lipoproteins (53). sAβ is defined as any Aβ species that is soluble in phosphate-buffered saline, including monomeric and oligomeric Aβ. Using various techniques, including density-gradient centrifugation, size-exclusion chromatography, and fluorescence correlation spectroscopy, the aforementioned study provided multiple lines of evidence that sAβ is a very poor binding partner of apoE-containing lipoproteins (Fig. 1, dashed blue arrow). The metastable nature of Aβ oligomers had made it difficult to study in the past, but it is important for future studies to further dissect out the potentially different role of monomeric versus oligomeric Aβ in the context of apoE interactions. Indeed, a study using an ELISA assay specific for Aβ oligomers found the latter species to be rapidly sequestered away from the ISF and CSF in J20 hAPP Tg mice (54). From these results, if oligomeric Aβ is present in the ISF, it could be the predominant species that interacts with apoE in the ISF prior to binding to other cell surface molecules, such as receptors, thus leaving monomeric Aβ to be the predominant species to be detected in the fluid phase. A recently developed method for isolating and characterizing the different oligomeric Aβ species may facilitate more rigorous studies on the relative contribution of monomeric and oligomeric Aβ to the APOE-mediated effects on amyloid pathology progression (55). This is an important question to address, considering the substantial body of literature supporting the role of Aβ oligomers in facilitating neurotoxicity (56).
Upon examining functional consequences of the apoE/Aβ interaction, Tamamizu-Kato et al. (57) found that Aβ binding to apoE compromises its lipid-binding function in vitro. Thus, it is possible that apoE interacts with Aβ directly through the carboxy terminal domain that also overlaps with the lipid-binding region. Furthermore, some studies suggest that Aβ peptides modulate the binding of apoE isoforms differently to apoE receptors (58, 59). These in vitro data suggest that Aβ peptides can modulate or interfere with the normal function of apoE.
APOE and Aβ aggregation
The aggregation of Aβ into higher-order species follows nucleation-dependent kinetics with a typical three-phase sigmoidal curve when assessed in vitro. The initial “lag phase” is slow and requires random collision of Aβ monomers (26, 60). Upon formation of a sufficient amount of nucleating higher-order species of Aβ (termed “nuclei” or “seeds”), perhaps consisting mostly of Aβ oligomers, the incorporation of additional monomers (growth phase) occurs at a much faster rate. The rate of fibril growth slows down (plateau phase) when it is outcompeted by newly formed nuclei or, possibly, depletion of the local pool of monomer. Thus, apoE could potentially exert pro-amyloidogenic effects by influencing either the formation of Aβ seeds or the subsequent elongation of fibrils (Fig. 1).
All three isoforms of apoE have been previously shown to promote the fibrillization of Aβ in vitro, and the potency follows the order of apoE4 > apoE3 > apoE2 (49, 61, 62). On the contrary, other studies suggest that apoE isoforms can also inhibit Aβ aggregation, with apoE4 being least effective (63–66). These seemingly contradicting findings may be explained by the variable experimental conditions, such as the species and source of Aβ used (67) (Aβ40 versus Aβ42, recombinant versus synthetic) and, especially, the preparation and the (resulting) lipidation state of apoE. Some studies utilized poorly lipidated apoE, while the most abundant and biologically active species of apoE found in vivo are lipidated. Indeed, in vivo studies have shown that alteration of the apoE lipidation state in the brain can exert a profound impact on Aβ fibrillization. Nascent apoE peptides in the brain are normally lipidated by ABCA1. Interestingly, APP Tg mice crossed onto an Abca1−/− background exhibit a marked increase in amyloid deposition, along with a decrease in apoE lipidation (68–72). Conversely, overexpression of ABCA1 in the brain leads to an increase in apoE lipidation and a corresponding decrease in amyloid deposition (68).
To initially assess the effects of human apoE isoforms on Aβ in mouse models, Tg mice expressing human apoE isoforms under the control of the astrocyte-specific GFAP promoter (in the absence of endogenous murine apoE) were crossed with APP Tg mice that develop Aβ plaques. These studies showed that, relative to mouse apoE, human apoE delayed Aβ deposition, but the order and amount of Aβ deposition was apoE4 > apoE3 > apoE2, as is seen in humans (73, 74). Early in vivo studies on the role of apoE on AD-related pathology utilized PDAPP mice, a model of Aβ amyloidosis that harbors the human APP transgene with the Indiana mutation (V717F) (75, 76). Introduction of the human apoE leads to delayed Aβ deposition in comparison to PDAPP mice expressing murine apoE or even those lacking apoE (74, 77), as was seen in previous studies. In 1997, Sullivan et al. (78) generated the first mouse strains carrying various isoforms of human apoE through targeted replacement. These lines allow for better characterization of the isoform-dependent effects on Aβ aggregation propensity in that the Apoe gene is regulated by the endogenous factors that control its expression. When these mice were crossed with different APP Tg mice, there was also a strong human apoE isoform-specific effect on Aβ deposition in the order of apoE4 > apoE3 > apoE2 (46, 79). These results are consistent with studies in humans and confirm the relevance of genetically engineered mouse models that mimic at least some aspects of the Aβ aggregation component of the disease. Using these human apoE knock-in mouse models, the allele-dependent effects of apoE on Aβ accumulation have been studied extensively.
In addition to the isoform effects of APOE, the expression level of APOE also appears to influence Aβ pathology. Deletion of endogenous murine apoE in PDAPP mice, as well as in the Tg2576 or APPswe models, leads to significantly less Aβ deposition and virtually abolishes fibrillar Aβ deposits in the brain parenchyma as well as CAA and CAA-associated microhemorrhage (80–83). These results suggest that apoE is a critical factor that facilitates Aβ aggregation in vivo into a fibrillar form, though the aforementioned studies did not offer insight into the mechanism for such process. To examine the effects of human APOE gene dosage, human APOE knock-in mouse lines were crossed to APP Tg (APP/PS1-21) mice. Interestingly, APP Tg mice expressing only one copy of human apoE have significantly less Aβ plaque deposition compared with mice expressing two copies of the same apoE isoform (either APOE-ε3 or APOE-ε4) (84). Very similar findings were also found by another group (85), and have potentially important therapeutic implications because there has been a long debate about whether increasing or decreasing apoE is beneficial for Aβ pathology. The caveat of these findings is that the animals carried the APOE gene and gene dosage differences since birth, and there could be developmental compensation in the genetic APOE haploinsufficiency model that could account for the protective effect against Aβ deposition. Whether reducing APOE levels in adult mice would similarly affect Aβ burden is unknown. Additionally, reduction of APOE expression could influence both the aggregation and clearance of Aβ, the latter being another major effect of apoE on Aβ metabolism that is well-described, as discussed in the following section.
APOE and Aβ clearance
Substantial evidence exists to support the role of apoE isoforms on monomeric Aβ clearance. Specifically, apoE has been proposed to influence Aβ clearance through several mechanisms: enzymatic degradation (86, 87), transport across the BBB (88), ISF-CSF bulk flow, and cellular uptake/subsequent degradation (89, 90).
The role of APOE in Aβ transport and clearance.
A large number of studies support the hypothesis that apoE influences Aβ transport and clearance from the ISF. The level of sAβ in the ISF can be measured continuously in live freely moving animals through utilization of an in vivo microdialysis technique (91). Clearance of Aβ from the ISF was found to be faster in apoE-KO mice (83), suggesting an important role for apoE in regulating Aβ levels in the brain. apoE isoforms differentially impact the level of ISF Aβ and the rate of Aβ clearance in vivo (46). ApoE4-expressing mice exhibited higher ISF levels of Aβ and a slower rate of Aβ clearance from the ISF than apoE3-expressing mice. Conversely, apoE2-expressing mice exhibited a lower level of ISF Aβ and a faster Aβ clearance rate relative to apoE3. The isoform-dependent effect of apoE was observed in aged mice, but also at young ages, well before plaque accumulation occurs. These findings suggest that the difference in Aβ accumulation between isoforms is likely due, in part, to their differential effects on Aβ clearance from the ISF, which can occur through a number of mechanisms.
One potential mechanism subject to apoE-dependent modulation is the transport of Aβ across the BBB to the systemic circulation (Fig. 1). The BBB is a highly selective permeability barrier that separates the blood from brain ISF. It is formed by continuous capillary endothelial cells containing tight junctions, which are surrounded by basal lamina, astrocytic perivascular endfeet, and pericytes. Through specific transporters, the BBB and pericytes work together to control entry of substances from the blood and promote clearance from brain of various potentially neurotoxic and vasculotoxic macromolecules (92). Similar to CAA, AD is associated with microvascular dysfunction and a locally defective BBB (93–95), both of which could be influenced by Aβ. Intriguingly, carriers of the APOE-ε4 allele are predisposed to CAA (96–99). In Tg2576 mice, apoE4 promotes a shift of Aβ deposition from the brain parenchyma to arterioles in the form of CAA, relative to apoE3 or murine apoE (100). Consistent with these findings, a recent study using 5×-FAD mice (APP Tg mice expressing these specific mutations in APP-APP KM670/671NL and V717I) harboring both human apoE4 and murine apoE found a higher degree of colocalization between murine apoE and parenchymal plaques, while plaques in the vasculature contained more apoE4 (101). Mechanistic studies linking apoE to Aβ transport from the ISF have identified a number of involved transporters, including P-glycoprotein (88), LDL receptor-related protein 1 (LRP1) (92), and LDL receptor (LDLR) (102).
LRP1 was found in amyloid plaques (103) and can bind Aβ directly (104) or indirectly via its ligands, including apoE (67, 105) and others (106, 107). Additionally, LRP1 participates in a lipid transport pathway that involves binding to heparan sulfate proteoglycans (HSPGs) (108, 109). Intriguingly, HSPG was found in senile plaques as well as CAA (110–112), and had been shown to directly interact with Aβ (113), accelerating its oligomerization and aggregation (Fig. 1) (114–116). A recent study found that ablation of HSPG in postnatal forebrain neurons of APP/PS1 mice led to a reduction in both Aβ oligomerization and the deposition of amyloid plaques (117). These reductions were driven by an accelerated rate of Aβ clearance from the ISF. Moreover, the authors also found a significantly increased level of various HSPG species in postmortem brain tissues from AD patients relative to controls. Deletion (118) or overexpression (119) of LRP1 causes increases or decreases of brain apoE levels, respectively. Whereas Aβ40 is cleared across the BBB at a much more rapid rate than via the ISF flow, binding to apoE-ε3 reduces the Aβ40 efflux rate from the ISF by 5- to 7-fold (120). Another study found that Aβ binding to apoE4 redirects its clearance from LRP1 to VLDL receptor, which internalizes Aβ-apoE4 complexes at the BBB more slowly than LRP1 (121). In contrast, Aβ-apoE2 and Aβ-apoE3 complexes are cleared through the BBB via both VLDL receptor and LRP1 at an accelerated rate compared with that of Aβ-apoE4 complexes.
In support of LDLR’s role in apoE transport across the BBB, deletion of the LDLR gene in the mouse brain leads to higher apoE levels in the brain and CSF (122), while overexpression of LDLR in the brain decreases brain apoE (123). Furthermore, deletion of LDLR caused a decrease in Aβ uptake, whereas increasing LDLR levels significantly enhanced both the uptake and clearance of Aβ by primary astrocytes in culture, even in the absence of apoE (124). Taken together, these data suggest that the influence of apoE on sAβ metabolism may not require direct binding of apoE with sAβ in solution, and that Aβ binding to apoE may lead to reduced clearance. This raises the possibility that APOE isoforms can significantly inhibit the uptake of sAβ by competing for the same pathway, facilitated through either LRP1 or LDLR. More rigorous in vivo studies are needed to provide direct evidence that the aforementioned mechanism is responsible for the increase in Aβ clearance in mice with a reduced apoE level.
Independent from its effects on Aβ clearance, there are also isoform-specific effects of apoE on the BBB, as expression of APOE-ε4 and lack of murine apoE, but not APOE-ε2 and APOE-ε3, was found to lead to BBB breakdown in young mice (125). The increase in vascular permeability in these mice was linked to a decrease in apoE-dependent pericyte-expressed LRP1 activation and a concomitant increase in CypA-MMP9 activity. In support of these findings, a human study using postmortem brain samples from control and AD subjects found accelerated pericyte degeneration in AD APOE-ε4 carriers compared with AD APOE-ε3 carriers and non-AD controls (126). Intriguingly, a significant increase in CypA and MMP-9 was also detected in pericytes and endothelial cells of APOE-ε4 subjects relative to APOE-ε3 subjects. However, in situ brain perfusion using [14C]sucrose as a vascular space marker found no differences in cerebral vascular volume among mice carrying any of the APOE isoforms, despite an observed reduction in cerebral vascularization and basement membrane thickness in 12-month-old APOE-ε4 mice compared with APOE-ε2 and APOE-ε3 mice (127). Additionally, a relatively recently study reported no apparent differences in the level of brain IgG and radiotracer uptake in apoE4 or apoE−/− mice when assessed at 2–3 months of age (128). Though the latter results might not be expected, they suggest that the global homeostatic capacity of the BBB might be intact in APOE-ε4 mice, despite highly localized disruption in BBB integrity.
APOE and cellular metabolism of Aβ.
Various cell types in the CNS have been shown to possess the ability to internalize Aβ, including astrocytes (129), neurons (130), and microglia (131–133), albeit with different capabilities. Primary hippocampal neurons are more efficient at internalizing Aβ in the presence of apoE3 compared with apoE4 (134). Nevertheless, neurons appear to lack the ability to effectively degrade Aβ, resulting in formation of high molecular weight Aβ species in endosomal vesicles (119, 135). The rest of this section will focus on astrocytes and microglia, which probably account for a significant amount of cellular Aβ metabolism.
Immunohistological studies on human AD brains using antibodies against various Aβ epitopes identified N-terminal-truncated fragments of Aβ40/42 that are found inside lipofuscin-like granules within astrocytes (89, 90). In agreement with human studies, in vitro studies using primary astrocytes showed that astrocytes can internalize and degrade both soluble (136, 137) and insoluble (138) Aβ. Interestingly, the ability of astrocytes to engulf and degrade Aβ is compromised in apoE KO astrocytes, or in WT astrocytes upon the addition of either an antibody against Aβ, apoE, or an LDLR family antagonist (129). These results suggest an essential role of apoE in a receptor-mediated Aβ uptake mechanism by astrocytes. However, more recent studies provided evidence that such processes can occur in the absence of apoE (124), and apoE can actually compete with Aβ for binding/uptake by LDLR receptors despite minimal interaction with sAβ (53).
Microglia are the resident macrophages and primary immune effectors in the brain, whose roles in AD are increasingly getting more attention, both in the context of disease pathogenesis as well as therapeutic opportunities. Microglia have the ability to clear sAβ as well as insoluble Aβ in vitro, followed by rapid degradation through the late endocytic/lysosomal pathway (133, 139). ApoE has been shown to be essential for efficient Aβ degradation via microglia by neprilysin or in the extracellular milieu by insulin degrading enzymes (87). More importantly, this process is dependent on the lipidation status of APOE, suggesting that apoE exerts its effects via manipulation of cellular cholesterol levels (87, 140). More recently, the newly discovered AD risk gene that encodes the triggering receptor expressed on myeloid cells 2 (TREM2) (141, 142) protein was found to have apoE as one of its ligands (143, 144). Accordingly, TREM2-deficient microglia or microglia with an AD risk variant (R47H) lose the ability to effectively bind lipoproteins (including apoE), which decreases their phagocytic capabilities (145). Furthermore, the lipid-sensing component of TREM2 allows it to sustain the microglial response that is necessary to cluster and potentially play a role in more efficient plaque phagocytosis, a phenomenon that is attenuated in TREM2-deficient microglia (146), as well as R47H mutants (147). Intriguingly, APOE-ε4-expressing microglia exhibited more pronounced downregulation of TREM2 than those that express APOE-ε3 in response to toll-like receptor (TLR) activation (148). In the future, generation of murine models with human TREM2 and mutant variants should allow for more rigorous in vivo studies that will fill in the gaps in knowledge about TREM2’s role in AD pathogenesis and its potential link with apoE.
APOE AND OTHER AMYLOIDOGENIC PROTEINS
The pro-amyloidogenic nature of APOE has prompted investigation of its role in other neurodegenerative diseases, many of which share the common feature of pathogenic protein aggregation. Specifically, APOE had been linked to Parkinson disease (PD) (149, 150), chronic traumatic encephalopathy (151), Huntington disease (152, 153), frontotemporal dementia (FTD) (154), and certain subsets of amyotrophic lateral sclerosis (155–157), though the latter findings conflicted with some other studies (158–160). Here, we discuss the current state of knowledge on the specific interaction between APOE and the aggregation-prone proteins associated with some of these diseases.
APOE and Tau
The strong correlation between APOE genotypes and AD, and the presence of immunoreactive apoE in neurons containing neurofibrillary tangles (27, 28) led to numerous clinical and epidemiological studies on the potential interaction between APOE and Tau, both in the context of AD as well as other tauopathies, including FTD. The majority of clinical studies found an over-representation of the APOE-ε4 allele in both AD and FTD (161–164), while histopathologic examinations revealed a significant positive correlation between APOE genotype and stage of neurofibrillary pathology (165) according to Braak staging (166). In support of these findings, the presence of the APOE-ε4 allele significantly correlates with brain atrophy in disease-specific brain regions in both AD and FTD (154). However, studies have not demonstrated a consistent effect of APOE genotype on cognition in tauopathies (154, 167). It should be mentioned that a few studies found either no association between APOE status and FTD (168), or that the association was only applicable to male cohorts (169). The disagreement of these findings might be explained by the population being studied and/or the difference in the statistical power of the studies. More sufficiently powered longitudinal studies are necessary to investigate whether APOE status has any effect on cognitive status, as well as other aspects of neuropathology, in patients with non-AD tauopathies.
Early studies by Allen Roses, Warren Strittmatter, and colleagues found apoE3, but not apoE4, to interact with Tau in an in vitro binding assay (170). This interaction is facilitated through the microtubule-binding domain of Tau and the LDLR binding domain on apoE3, which is distinct from its binding site for Aβ. Interestingly, expression of full-length human apoE4, specifically in neurons alone, is sufficient to induce NFT-like inclusions and hyperphosphorylation of Tau in WT mice (171, 172). Along with these findings, a C-terminal-truncated fragment of apoE (Δ272-299) (172, 173) was found to accumulate in the brains of AD patients and mice with neuronal expression of APOE (174). The same group also linked the increase in pTau to changes in the zinc-induced Erk activation pathway (175). Considering the availability of mice that develop Tau pathology (176) and APOE knock-in (78) mouse models, future in vivo studies on the APOE/Tau interaction are feasible and warrant new insights on the relationship between one of AD’s signature hallmarks and its biggest risk factor.
APOE and α-synuclein
α-Synuclein (α-syn) is a 140 amino acid protein that is normally localized to presynaptic terminals. Aggregates of α-syn form the main constituent of Lewy bodies and Lewy neurites (177–179), the pathological hallmarks of a class of neurodegenerative diseases termed “synucleinopathies,” which includes PD and the related disorders, PD dementia and dementia with Lewy bodies [reviewed extensively in (180, 181)]. Interestingly, up to 50% of patients with synucleinopathy and dementia also have Aβ plaques, while a smaller subset also has concomitant neurofibrillary tangles (182–188). Historically, this overlap presented a challenge with regard to disease nomenclature. The overlapping features between PD and AD have been demonstrated by clinical (189, 190), pathologic (191, 192), and genetic data (193, 194). Intriguingly, some of the findings in this area suggest a possible connection between apoE and α-syn.
Initial epidemiological studies found contradictory results as to whether APOE genotype influences the risk of PD. A meta-analysis of 22 clinical studies found the APOE-ε2 allele to be positively associated with risk for developing sporadic PD (OR = 1.20), whereas no association was found for APOE-ε3 or APOE-ε4 (195). On the contrary, an independent study by Li et al. (149) genotyped 658 PD-affected families for APOE functional polymorphisms and found that the APOE-ε4 allele increased the risk and decreased the age at onset for PD, while the APOE-ε3 allele had the opposite effects. Recent efforts to combine data across multiple genetic association studies have demonstrated that, compared with APOE-ε3, the APOE-ε2 allele modestly increases risk of PD [OR = 1.11 (95% CI 1.02, 1.21)], while APOE-ε4 has no effect on overall PD risk [OR = 1.00 (95% CI 0.91, 1.10)] (196). In addition to the genetic diversity in study populations and differences in statistical power, the disparity in results may also be attributed to the heterogeneity in both clinical and pathologic entities within the PD spectrum: that is, many patients with a diagnosis of PD also have cognitive impairment and a large, but not necessarily overlapping, proportion have Aβ plaque pathology. Numerous studies support the association of APOE-ε4 with increased Aβ and, to some extent, Tau pathology in individuals with PD, as well as other neurodegenerative disorders (197, 198). The effect of APOE-ε4 on cognitive impairment in PD may be mediated, at least in part, through Aβ, as illustrated by a prospective cohort study of 45 patients with PD with at least one yearly follow-up visit in which any observed correlation between APOE-ε4 status and cognitive impairment was abolished after adjusting for CSF levels of Aβ42. However, this phenomenon does not account for the effect of APOE genotype on dementia in PD patients who do not have concomitant Aβ plaque pathology (199). Several studies have found that the presence of one or more APOE-ε4 alleles not only increases the Aβ plaque load, but also the α-syn pathology burden in PD cases (183, 200, 201). Interestingly, when a relatively large group of PD cases was stratified pathologically and cases with concomitant Aβ pathology were excluded (leaving a cohort with pure α-syn pathology), APOE-ε4 was still strongly associated with dementia and even more strongly associated with early development of dementia (202). Taken together, these findings suggest that the impact of APOE genotype on synucleinopathy and dementia is likely multifactorial and that apoE may directly influence α-syn pathology.
Basic research on the role of APOE on α-syn has lagged behind clinical and epidemiological studies, but several important findings have emerged. A study in Tg mice expressing mutations in the SNCA gene, which codes for α-syn, found that aged symptomatic mice had elevated levels of endogenous murine Aβ40 and Aβ42, as well as apoE, relative to asymptomatic littermates (203). Interestingly, deletion of the murine Apoe gene prolonged survival and decreased levels of insoluble Aβ40, Aβ42, and α-syn, in addition to decreasing levels of ubiquitinated proteins. These data suggest that apoE may directly influence proteostatic mechanisms that regulate α-syn and Aβ. More recently, an in vitro study found that apoE, especially the apoE4 isoform, appeared to have a stimulatory effect on α-syn aggregation at low concentrations (low nanomoles); however, this effect was attenuated at higher concentrations (204). One caveat to this study is that the apoE particles used were lipid-poor, while most physiologically active apoE species found in vivo are lipid-rich. It is also not yet clear how apoE, a predominantly secreted molecule found in the extracellular space, interacts with α-syn, which localizes to the intracellular space in both monomeric and aggregated forms. Further research is clearly needed to elucidate molecular mechanisms of this interaction and to explore opportunities for therapeutic intervention.
FUTURE DIRECTIONS
More than 20 years have passed since the landmark discovery of APOE being linked with AD, and APOE still remains the strongest genetic risk factor for LOAD. Intensive research efforts have undoubtedly unveiled several important insights regarding apoE and its role in AD. Nevertheless, no APOE-based (or any) disease-modifying therapy has been approved for AD treatment within this period. That means many questions remain unanswered, including the most important question: How does APOE gene polymorphism confer AD risk? This review focuses mostly on the relationship between apoE and Aβ, and to a lesser extent, Tau and α-syn. Taken together, the evidence that APOE influences AD risk substantially via effects on Aβ is strong. It should be noted that a relatively large body of literature also exists to support APOE’s roles in AD pathogenesis that are Aβ-independent. This area needs much further exploration. It is possible that apoE participates in an unknown fundamental mechanistic event (such as a general signaling cascade) that influences brain homeostasis. If true, the single amino acid change among the different isoforms (particularly apoE4) somehow alters the behavior/function of apoE in this process that ultimately lowers the brain’s ability to tolerate neurotoxic insults (such as Aβ or Tau accumulations). This increased vulnerability, in turn, could affect AD risk. Similarly, apoE’s potential role in facilitating the immune response to AD pathology needs further investigation. Considering TREM2’s newly described role as a lipid-sensing receptor, it is necessary to definitively determine whether there is a true meaningful connection between APOE and TREM2.
Along with the affordability and availability of big data science, there needs to be more studies that look at apoE isoforms’ differential effects on specific cell populations in the brain in an unbiased fashion. Such studies might identify novel targets that will allow us to gain a deeper understanding as to why AD occurs when it does (old age) and where it does (susceptibility regions). Concurrently, researchers should be cautious that considerable species differences between rodents and humans exist and might challenge our ability to generate findings that are relevant and directly translatable to humans from studies in mice and rats. Apparent differences in physiological function and metabolism, such as lipid metabolism and immune response, between humans and rodents might preclude important discoveries that are relevant to disease mechanism. While the current generation of APOE knock-in mice harbors the human gene sequence, they retain the regulatory elements found in mice. It is critical to address this caveat in rodent studies, in lieu of recent studies including one showing evidence that apoE can act as a transcription factor and influence a wide variety of signaling pathways (205). To circumvent the limitation of existing animal models, new experimental paradigms, particularly those derived from humans (such as iPSCs and brain organoids) need to be developed and characterized. An alternative approach is to engineer human Aβ and/or Tau into species with more human-like physiology than rodents. These new experimental platforms will undoubtedly complement the current animal models and fuel a new wave of discovery that is necessary to fully understand AD pathology and its relationship with apoE, and to design new therapies.
In the next 25 years, the advancement of technology and the solid foundation built by many scientists in the last 25 years should aid researchers with new tools and freedom to push the field toward a meaningful understanding of APOE and AD pathogenesis. From that insight, new and (hopefully) effective therapies can be designed that can alter the course of the disease.
Footnotes
Abbreviations:
- Aβ
- amyloid-β Aβ40, amyloid-β1-40
- Aβ42
- amyloid-β1-42
- AD
- Alzheimer’s disease
- APP
- amyloid-β precursor protein
- α-syn
- α-synuclein
- BBB
- blood-brain barrier
- CAA
- cerebral amyloid angiopathy
- CSF
- cerebrospinal fluid
- FTD
- frontotemporal dementia
- HSPG
- heparan sulfate proteoglycan
- ISF
- interstitial fluid
- LDLR
- LDL receptor
- LOAD
- late-onset Alzheimer’s disease
- LRP1
- LDL receptor-related protein 1
- OR
- odds ratio
- PD
- Parkinson disease
- PiB
- Pittsburgh compound-B
- PS
- presenilin
- sAβ
- soluble amyloid-β
- Tg
- transgenic
- TREM2
- triggering receptor expressed on myeloid cells 2
This work was supported by National Institutes of Health Grants NS090934 and AG047644. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Alzheimer A. 1907. Über eine eigenartige Erkrankung der Hirnrinde. Allg. Z. Psychiatr. 64: 146–148. [Google Scholar]
- 2.Bateman R. J., Aisen P. S., De Strooper B., Fox N. C., Lemere C. A., Ringman J. M., Salloway S., Sperling R. A., Windisch M., and Xiong C.. 2011. Autosomal-dominant Alzheimer’s disease: a review and proposal for the prevention of Alzheimer’s disease. Alzheimers Res. Ther. 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corder E. H., Saunders A. M., Risch N. J., Strittmatter W. J., Schmechel D. E., Gaskell P. C. Jr., Rimmler J. B., Locke P. A., Conneally P. M., Schmader K. E., et al. 1994. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet. 7: 180–184. [DOI] [PubMed] [Google Scholar]
- 4.Corder E. H., Saunders A. M., Strittmatter W. J., Schmechel D. E., Gaskell P. C., Small G. W., Roses A. D., Haines J. L., and Pericak-Vance M. A.. 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 261: 921–923. [DOI] [PubMed] [Google Scholar]
- 5.Strittmatter W. J., Saunders A. M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G. S., and Roses A. D.. 1993. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA. 90: 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahley R. W. 1988. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 240: 622–630. [DOI] [PubMed] [Google Scholar]
- 7.Pitas R. E., Boyles J. K., Lee S. H., Foss D., and Mahley R. W.. 1987. Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins. Biochim. Biophys. Acta. 917: 148–161. [DOI] [PubMed] [Google Scholar]
- 8.Grehan S., Tse E., and Taylor J. M.. 2001. Two distal downstream enhancers direct expression of the human apolipoprotein E gene to astrocytes in the brain. J. Neurosci. 21: 812–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roses A. D. 1994. Apolipoprotein E affects the rate of Alzheimer disease expression: beta-amyloid burden is a secondary consequence dependent on APOE genotype and duration of disease. J. Neuropathol. Exp. Neurol. 53: 429–437. [DOI] [PubMed] [Google Scholar]
- 10.Farrer L. A., Cupples L. A., Haines J. L., Hyman B., Kukull W. A., Mayeux R., Myers R. H., Pericak-Vance M. A., Risch N., and van Duijn C. M.. 1997. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 278: 1349–1356. [PubMed] [Google Scholar]
- 11. Alzgene. 2010. AlzGene - Gene overview of all published AD-association studies for APOE_e2/3/4. Accessed January 31, 2017, at http://www.alzgene.org/geneoverview.asp?geneid=83. [Google Scholar]
- 12.LaDu M. J., Gilligan S. M., Lukens J. R., Cabana V. G., Reardon C. A., Van Eldik L. J., and Holtzman D. M.. 1998. Nascent astrocyte particles differ from lipoproteins in CSF. J. Neurochem. 70: 2070–2081. [DOI] [PubMed] [Google Scholar]
- 13.Ulrich J. D., Burchett J. M., Restivo J. L., Schuler D. R., Verghese P. B., Mahan T. E., Landreth G. E., Castellano J. M., Jiang H., Cirrito J. R., et al. 2013. In vivo measurement of apolipoprotein E from the brain interstitial fluid using microdialysis. Mol. Neurodegener. 8: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S. H., Reddick R. L., Piedrahita J. A., and Maeda N.. 1992. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 258: 468–471. [DOI] [PubMed] [Google Scholar]
- 15.Mak A. C., Pullinger C. R., Tang L. F., Wong J. S., Deo R. C., Schwarz J. M., Gugliucci A., Movsesyan I., Ishida B. Y., Chu C., et al. 2014. Effects of the absence of apolipoprotein e on lipoproteins, neurocognitive function, and retinal function. JAMA Neurol. 71: 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mauch D. H., Nagler K., Schumacher S., Goritz C., Muller E. C., Otto A., and Pfrieger F. W.. 2001. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 294: 1354–1357. [DOI] [PubMed] [Google Scholar]
- 17.Pfrieger F. W. 2003. Cholesterol homeostasis and function in neurons of the central nervous system. Cell. Mol. Life Sci. 60: 1158–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nathan B. P., Bellosta S., Sanan D. A., Weisgraber K. H., Mahley R. W., and Pitas R. E.. 1994. Differential effects of apolipoproteins E3 and E4 on neuronal growth in vitro. Science. 264: 850–852. [DOI] [PubMed] [Google Scholar]
- 19.Holtzman D. M., Pitas R. E., Kilbridge J., Nathan B., Mahley R. W., Bu G., and Schwartz A. L.. 1995. Low density lipoprotein receptor-related protein mediates apolipoprotein E-dependent neurite outgrowth in a central nervous system-derived neuronal cell line. Proc. Natl. Acad. Sci. USA. 92: 9480–9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masliah E., Mallory M., Ge N., Alford M., Veinbergs I., and Roses A. D.. 1995. Neurodegeneration in the central nervous system of apoE-deficient mice. Exp. Neurol. 136: 107–122. [DOI] [PubMed] [Google Scholar]
- 21.Poirier J. 2003. Apolipoprotein E and cholesterol metabolism in the pathogenesis and treatment of Alzheimer’s disease. Trends Mol. Med. 9: 94–101. [DOI] [PubMed] [Google Scholar]
- 22.Fagan A. M., Murphy B. A., Patel S. N., Kilbridge J. F., Mobley W. C., Bu G., and Holtzman D. M.. 1998. Evidence for normal aging of the septo-hippocampal cholinergic system in apoE (-/-) mice but impaired clearance of axonal degeneration products following injury. Exp. Neurol. 151: 314–325. [DOI] [PubMed] [Google Scholar]
- 23.Anderson R., Barnes J. C., Bliss T. V., Cain D. P., Cambon K., Davies H. A., Errington M. L., Fellows L. A., Gray R. A., Hoh T., et al. 1998. Behavioural, physiological and morphological analysis of a line of apolipoprotein E knockout mouse. Neuroscience. 85: 93–110. [DOI] [PubMed] [Google Scholar]
- 24.Hardy J., and Selkoe D. J.. 2002. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 297: 353–356. [DOI] [PubMed] [Google Scholar]
- 25.Szaruga M., Veugelen S., Benurwar M., Lismont S., Sepulveda-Falla D., Lleo A., Ryan N. S., Lashley T., Fox N. C., Murayama S., et al. 2015. Qualitative changes in human gamma-secretase underlie familial Alzheimer’s disease. J. Exp. Med. 212: 2003–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarrett J. T., and Lansbury P. T. Jr. 1993. Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 73: 1055–1058. [DOI] [PubMed] [Google Scholar]
- 27.Namba Y., Tomonaga M., Kawasaki H., Otomo E., and Ikeda K.. 1991. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 541: 163–166. [DOI] [PubMed] [Google Scholar]
- 28.Wisniewski T., and Frangione B.. 1992. Apolipoprotein E: a pathological chaperone protein in patients with cerebral and systemic amyloid. Neurosci. Lett. 135: 235–238. [DOI] [PubMed] [Google Scholar]
- 29.Schmechel D. E., Saunders A. M., Strittmatter W. J., Crain B. J., Hulette C. M., Joo S. H., Pericak-Vance M. A., Goldgaber D., and Roses A. D.. 1993. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc. Natl. Acad. Sci. USA. 90: 9649–9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertram L., McQueen M. B., Mullin K., Blacker D., and Tanzi R. E.. 2007. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 39: 17–23. [DOI] [PubMed] [Google Scholar]
- 31.Rebeck G. W., Reiter J. S., Strickland D. K., and Hyman B. T.. 1993. Apolipoprotein E in sporadic Alzheimer’s disease: allelic variation and receptor interactions. Neuron. 11: 575–580. [DOI] [PubMed] [Google Scholar]
- 32.Benjamin R., Leake A., Ince P. G., Perry R. H., McKeith I. G., Edwardson J. A., and Morris C. M.. 1995. Effects of apolipoprotein E genotype on cortical neuropathology in senile dementia of the Lewy body and Alzheimer’s disease. Neurodegeneration. 4: 443–448. [DOI] [PubMed] [Google Scholar]
- 33.Heinonen O., Lehtovirta M., Soininen H., Helisalmi S., Mannermaa A., Sorvari H., Kosunen O., Paljarvi L., Ryynanen M., and Riekkinen P. J. Sr. 1995. Alzheimer pathology of patients carrying apolipoprotein E epsilon 4 allele. Neurobiol. Aging. 16: 505–513. [DOI] [PubMed] [Google Scholar]
- 34.Tiraboschi P., Hansen L. A., Masliah E., Alford M., Thal L. J., and Corey-Bloom J.. 2004. Impact of APOE genotype on neuropathologic and neurochemical markers of Alzheimer disease. Neurology. 62: 1977–1983. [DOI] [PubMed] [Google Scholar]
- 35.Klunk W. E., Wang Y., Huang G. F., Debnath M. L., Holt D. P., and Mathis C. A.. 2001. Uncharged thioflavin-T derivatives bind to amyloid-beta protein with high affinity and readily enter the brain. Life Sci. 69: 1471–1484. [DOI] [PubMed] [Google Scholar]
- 36.Klunk W. E., Engler H., Nordberg A., Wang Y., Blomqvist G., Holt D. P., Bergstrom M., Savitcheva I., Huang G. F., Estrada S., et al. 2004. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh compound-B. Ann. Neurol. 55: 306–319. [DOI] [PubMed] [Google Scholar]
- 37.Mathis C. A., Mason N. S., Lopresti B. J., and Klunk W. E.. 2012. Development of positron emission tomography beta-amyloid plaque imaging agents. Semin. Nucl. Med. 42: 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reiman E. M., Chen K., Liu X., Bandy D., Yu M., Lee W., Ayutyanont N., Keppler J., Reeder S. A., Langbaum J. B., et al. 2009. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 106: 6820–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris J. C., Roe C. M., Xiong C., Fagan A. M., Goate A. M., Holtzman D. M., and Mintun M. A.. 2010. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann. Neurol. 67: 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sunderland T., Mirza N., Putnam K. T., Linker G., Bhupali D., Durham R., Soares H., Kimmel L., Friedman D., Bergeson J., et al. 2004. Cerebrospinal fluid beta-amyloid1–42 and tau in control subjects at risk for Alzheimer’s disease: the effect of APOE epsilon4 allele. Biol. Psychiatry. 56: 670–676. [DOI] [PubMed] [Google Scholar]
- 41.Ye S., Huang Y., Mullendorff K., Dong L., Giedt G., Meng E. C., Cohen F. E., Kuntz I. D., Weisgraber K. H., and Mahley R. W.. 2005. Apolipoprotein (apo) E4 enhances amyloid beta peptide production in cultured neuronal cells: apoE structure as a potential therapeutic target. Proc. Natl. Acad. Sci. USA. 102: 18700–18705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y. A., Zhou B., Wernig M., and Sudhof T. C.. 2017. ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription and Aβ secretion. Cell. 168: 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biere A. L., Ostaszewski B., Zhao H., Gillespie S., Younkin S. G., and Selkoe D. J.. 1995. Co-expression of beta-amyloid precursor protein (betaAPP) and apolipoprotein E in cell culture: analysis of betaAPP processing. Neurobiol. Dis. 2: 177–187. [DOI] [PubMed] [Google Scholar]
- 44.Cedazo-Mínguez A., Wiehager B., Winblad B., Huttinger M., and Cowburn R. F.. 2001. Effects of apolipoprotein E (apoE) isoforms, beta-amyloid (Abeta) and apoE/Abeta complexes on protein kinase C-alpha (PKC-alpha) translocation and amyloid precursor protein (APP) processing in human SH-SY5Y neuroblastoma cells and fibroblasts. Neurochem. Int. 38: 615–625. [DOI] [PubMed] [Google Scholar]
- 45.Irizarry M. C., Deng A., Lleo A., Berezovska O., Von Arnim C. A., Martin-Rehrmann M., Manelli A., LaDu M. J., Hyman B. T., and Rebeck G. W.. 2004. Apolipoprotein E modulates gamma-secretase cleavage of the amyloid precursor protein. J. Neurochem. 90: 1132–1143. [DOI] [PubMed] [Google Scholar]
- 46.Castellano J. M., Kim J., Stewart F. R., Jiang H., DeMattos R. B., Patterson B. W., Fagan A. M., Morris J. C., Mawuenyega K. G., Cruchaga C., et al. 2011. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci. Transl. Med. 3: 89ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strittmatter W. J., Weisgraber K. H., Huang D. Y., Dong L. M., Salvesen G. S., Pericak-Vance M., Schmechel D., Saunders A. M., Goldgaber D., and Roses A. D.. 1993. Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc. Natl. Acad. Sci. USA. 90: 8098–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaDu M. J., Falduto M. T., Manelli A. M., Reardon C. A., Getz G. S., and Frail D. E.. 1994. Isoform-specific binding of apolipoprotein E to beta-amyloid. J. Biol. Chem. 269: 23403–23406. [PubMed] [Google Scholar]
- 49.Sanan D. A., Weisgraber K. H., Russell S. J., Mahley R. W., Huang D., Saunders A., Schmechel D., Wisniewski T., Frangione B., Roses A. D., et al. 1994. Apolipoprotein E associates with beta amyloid peptide of Alzheimer’s disease to form novel monofibrils. Isoform apoE4 associates more efficiently than apoE3. J. Clin. Invest. 94: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aleshkov S., Abraham C. R., and Zannis V. I.. 1997. Interaction of nascent ApoE2, ApoE3, and ApoE4 isoforms expressed in mammalian cells with amyloid peptide beta (1-40). Relevance to Alzheimer’s disease. Biochemistry. 36: 10571–10580. [DOI] [PubMed] [Google Scholar]
- 51.Yang D. S., Smith J. D., Zhou Z., Gandy S. E., and Martins R. N.. 1997. Characterization of the binding of amyloid-beta peptide to cell culture-derived native apolipoprotein E2, E3, and E4 isoforms and to isoforms from human plasma. J. Neurochem. 68: 721–725. [DOI] [PubMed] [Google Scholar]
- 52.Tokuda T., Calero M., Matsubara E., Vidal R., Kumar A., Permanne B., Zlokovic B., Smith J. D., Ladu M. J., Rostagno A., et al. 2000. Lipidation of apolipoprotein E influences its isoform-specific interaction with Alzheimer’s amyloid beta peptides. Biochem. J. 348: 359–365. [PMC free article] [PubMed] [Google Scholar]
- 53.Verghese P. B., Castellano J. M., Garai K., Wang Y., Jiang H., Shah A., Bu G., Frieden C., and Holtzman D. M.. 2013. ApoE influences amyloid-β (Aβ) clearance despite minimal apoE/Aβ association in physiological conditions. Proc. Natl. Acad. Sci. USA. 110: E1807–E1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong S., Ostaszewski B. L., Yang T., O’Malley T. T., Jin M., Yanagisawa K., Li S., Bartels T., and Selkoe D. J.. 2014. Soluble Aβ oligomers are rapidly sequestered from brain ISF in vivo and bind GM1 ganglioside on cellular membranes. Neuron. 82: 308–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esparza T. J., Wildburger N. C., Jiang H., Gangolli M., Cairns N. J., Bateman R. J., and Brody D. L.. 2016. Soluble amyloid-beta aggregates from human Alzheimer’s disease brains. Sci. Rep. 6: 38187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benilova I., Karran E., and De Strooper B.. 2012. The toxic Aβ oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat. Neurosci. 15: 349–357. [DOI] [PubMed] [Google Scholar]
- 57.Tamamizu-Kato S., Cohen J. K., Drake C. B., Kosaraju M. G., Drury J., and Narayanaswami V.. 2008. Interaction with amyloid beta peptide compromises the lipid binding function of apolipoprotein E. Biochemistry. 47: 5225–5234. [DOI] [PubMed] [Google Scholar]
- 58.Beffert U., Aumont N., Dea D., Lussier-Cacan S., Davignon J., and Poirier J.. 1998. Beta-amyloid peptides increase the binding and internalization of apolipoprotein E to hippocampal neurons. J. Neurochem. 70: 1458–1466. [DOI] [PubMed] [Google Scholar]
- 59.Hone E., Martins I. J., Jeoung M., Ji T. H., Gandy S. E., and Martins R. N.. 2005. Alzheimer’s disease amyloid-beta peptide modulates apolipoprotein E isoform specific receptor binding. J. Alzheimers Dis. 7: 303–314. [DOI] [PubMed] [Google Scholar]
- 60.Roychaudhuri R., Yang M., Hoshi M. M., and Teplow D. B.. 2009. Amyloid beta-protein assembly and Alzheimer disease. J. Biol. Chem. 284: 4749–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma J., Yee A., Brewer H. B. Jr., Das S., and Potter H.. 1994. Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature. 372: 92–94. [DOI] [PubMed] [Google Scholar]
- 62.Wisniewski T., Castano E. M., Golabek A., Vogel T., and Frangione B.. 1994. Acceleration of Alzheimer’s fibril formation by apolipoprotein E in vitro. Am. J. Pathol. 145: 1030–1035. [PMC free article] [PubMed] [Google Scholar]
- 63.Evans K. C., Berger E. P., Cho C. G., Weisgraber K. H., and Lansbury P. T. Jr. 1995. Apolipoprotein E is a kinetic but not a thermodynamic inhibitor of amyloid formation: implications for the pathogenesis and treatment of Alzheimer disease. Proc. Natl. Acad. Sci. USA. 92: 763–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood S. J., Chan W., and Wetzel R.. 1996. Seeding of A beta fibril formation is inhibited by all three isotypes of apolipoprotein E. Biochemistry. 35: 12623–12628. [DOI] [PubMed] [Google Scholar]
- 65.Beffert U., and Poirier J.. 1998. ApoE associated with lipid has a reduced capacity to inhibit beta-amyloid fibril formation. Neuroreport. 9: 3321–3323. [DOI] [PubMed] [Google Scholar]
- 66.Garai K., Verghese P. B., Baban B., Holtzman D. M., and Frieden C.. 2014. The binding of apolipoprotein E to oligomers and fibrils of amyloid-beta alters the kinetics of amyloid aggregation. Biochemistry. 53: 6323–6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J., Basak J. M., and Holtzman D. M.. 2009. The role of apolipoprotein E in Alzheimer’s disease. Neuron. 63: 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wahrle S. E., Jiang H., Parsadanian M., Kim J., Li A., Knoten A., Jain S., Hirsch-Reinshagen V., Wellington C. L., Bales K. R., et al. 2008. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J. Clin. Invest. 118: 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koldamova R., Staufenbiel M., and Lefterov I.. 2005. Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. J. Biol. Chem. 280: 43224–43235. [DOI] [PubMed] [Google Scholar]
- 70.Wahrle S. E., Jiang H., Parsadanian M., Hartman R. E., Bales K. R., Paul S. M., and Holtzman D. M.. 2005. Deletion of Abca1 increases Abeta deposition in the PDAPP transgenic mouse model of Alzheimer disease. J. Biol. Chem. 280: 43236–43242. [DOI] [PubMed] [Google Scholar]
- 71.Hirsch-Reinshagen V., Zhou S., Burgess B. L., Bernier L., McIsaac S. A., Chan J. Y., Tansley G. H., Cohn J. S., Hayden M. R., and Wellington C. L.. 2004. Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain. J. Biol. Chem. 279: 41197–41207. [DOI] [PubMed] [Google Scholar]
- 72.Hirsch-Reinshagen V., Maia L. F., Burgess B. L., Blain J. F., Naus K. E., McIsaac S. A., Parkinson P. F., Chan J. Y., Tansley G. H., Hayden M. R., et al. 2005. The absence of ABCA1 decreases soluble ApoE levels but does not diminish amyloid deposition in two murine models of Alzheimer disease. J. Biol. Chem. 280: 43243–43256. [DOI] [PubMed] [Google Scholar]
- 73.Holtzman D. M., Bales K. R., Tenkova T., Fagan A. M., Parsadanian M., Sartorius L. J., Mackey B., Olney J., McKeel D., Wozniak D., et al. 2000. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 97: 2892–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fagan A. M., Watson M., Parsadanian M., Bales K. R., Paul S. M., and Holtzman D. M.. 2002. Human and murine ApoE markedly alters A beta metabolism before and after plaque formation in a mouse model of Alzheimer’s disease. Neurobiol. Dis. 9: 305–318. [DOI] [PubMed] [Google Scholar]
- 75.Games D., Adams D., Alessandrini R., Barbour R., Berthelette P., Blackwell C., Carr T., Clemens J., Donaldson T. , Gillespie F., et al. 1995. Alzheimer-type neuropathology in transgenic mice overexpressing V717F beta-amyloid precursor protein. Nature. 373: 523–527. [DOI] [PubMed] [Google Scholar]
- 76.Rockenstein E. M., McConlogue L., Tan H., Power M., Masliah E., and Mucke L.. 1995. Levels and alternative splicing of amyloid beta protein precursor (APP) transcripts in brains of APP transgenic mice and humans with Alzheimer’s disease. J. Biol. Chem. 270: 28257–28267. [DOI] [PubMed] [Google Scholar]
- 77.Holtzman D. M., Bales K. R., Wu S., Bhat P., Parsadanian M., Fagan A. M., Chang L. K., Sun Y., and Paul S. M.. 1999. Expression of human apolipoprotein E reduces amyloid-beta deposition in a mouse model of Alzheimer’s disease. J. Clin. Invest. 103: R15–R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sullivan P. M., Mezdour H., Aratani Y., Knouff C., Najib J., Reddick R. L., Quarfordt S. H., and Maeda N.. 1997. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J. Biol. Chem. 272: 17972–17980. [DOI] [PubMed] [Google Scholar]
- 79.Bales K. R., Liu F., Wu S., Lin S., Koger D., DeLong C., Hansen J. C., Sullivan P. M., and Paul S. M.. 2009. Human APOE isoform-dependent effects on brain beta-amyloid levels in PDAPP transgenic mice. J. Neurosci. 29: 6771–6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bales K. R., Verina T., Cummins D. J., Du Y., Dodel R. C., Saura J., Fishman C. E., DeLong C. A., Piccardo P., Petegnief V., et al. 1999. Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 96: 15233–15238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bales K. R., Verina T., Dodel R. C., Du Y., Altstiel L., Bender M., Hyslop P., Johnstone E. M., Little S. P., Cummins D. J., et al. 1997. Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat. Genet. 17: 263–264. [DOI] [PubMed] [Google Scholar]
- 82.Holtzman D. M., Fagan A. M., Mackey B., Tenkova T., Sartorius L., Paul S. M., Bales K., Ashe K. H., Irizarry M. C., and Hyman B. T.. 2000. Apolipoprotein E facilitates neuritic and cerebrovascular plaque formation in an Alzheimer’s disease model. Ann. Neurol. 47: 739–747. [PubMed] [Google Scholar]
- 83.DeMattos R. B., Cirrito J. R., Parsadanian M., May P. C., O’Dell M. A., Taylor J. W., Harmony J. A., Aronow B. J., Bales K. R., Paul S. M., et al. 2004. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 41: 193–202. [DOI] [PubMed] [Google Scholar]
- 84.Kim J., Jiang H., Park S., Eltorai A. E., Stewart F. R., Yoon H., Basak J. M., Finn M. B., and Holtzman D. M.. 2011. Haploinsufficiency of human APOE reduces amyloid deposition in a mouse model of amyloid-beta amyloidosis. J. Neurosci. 31: 18007–18012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bien-Ly N., Gillespie A. K., Walker D., Yoon S. Y., and Huang Y.. 2012. Reducing human apolipoprotein E levels attenuates age-dependent Abeta accumulation in mutant human amyloid precursor protein transgenic mice. J. Neurosci. 32: 4803–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nalivaeva N. N., Beckett C., Belyaev N. D., and Turner A. J.. 2012. Are amyloid-degrading enzymes viable therapeutic targets in Alzheimer’s disease? J. Neurochem. 120(Suppl 1): 167–185. [DOI] [PubMed] [Google Scholar]
- 87.Jiang Q., Lee C. Y., Mandrekar S., Wilkinson B., Cramer P., Zelcer N., Mann K., Lamb B., Willson T. M., Collins J. L., et al. 2008. ApoE promotes the proteolytic degradation of Abeta. Neuron. 58: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cirrito J. R., Deane R., Fagan A. M., Spinner M. L., Parsadanian M., Finn M. B., Jiang H., Prior J. L., Sagare A., Bales K. R., et al. 2005. P-glycoprotein deficiency at the blood-brain barrier increases amyloid-beta deposition in an Alzheimer disease mouse model. J. Clin. Invest. 115: 3285–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Funato H., Yoshimura M., Yamazaki T., Saido T. C., Ito Y., Yokofujita J., Okeda R., and Ihara Y.. 1998. Astrocytes containing amyloid beta-protein (Abeta)-positive granules are associated with Abeta40-positive diffuse plaques in the aged human brain. Am. J. Pathol. 152: 983–992. [PMC free article] [PubMed] [Google Scholar]
- 90.Thal D. R., Schultz C., Dehghani F., Yamaguchi H., Braak H., and Braak E.. 2000. Amyloid beta-protein (Abeta)-containing astrocytes are located preferentially near N-terminal-truncated Abeta deposits in the human entorhinal cortex. Acta Neuropathol. 100: 608–617. [DOI] [PubMed] [Google Scholar]
- 91.Cirrito J. R., May P. C., O’Dell M. A., Taylor J. W., Parsadanian M., Cramer J. W., Audia J. E., Nissen J. S., Bales K. R., Paul S. M., et al. 2003. In vivo assessment of brain interstitial fluid with microdialysis reveals plaque-associated changes in amyloid-beta metabolism and half-life. J. Neurosci. 23: 8844–8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shibata M., Yamada S., Kumar S. R., Calero M., Bading J., Frangione B., Holtzman D. M., Miller C. A., Strickland D. K., Ghiso J., et al. 2000. Clearance of Alzheimer’s amyloid-ss(1-40) peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier. J. Clin. Invest. 106: 1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zlokovic B. V. 2005. Neurovascular mechanisms of Alzheimer’s neurodegeneration. Trends Neurosci. 28: 202–208. [DOI] [PubMed] [Google Scholar]
- 94.de la Torre J. C. 2010. Vascular risk factor detection and control may prevent Alzheimer’s disease. Ageing Res. Rev. 9: 218–225. [DOI] [PubMed] [Google Scholar]
- 95.Marchesi V. T. 2011. Alzheimer’s dementia begins as a disease of small blood vessels, damaged by oxidative-induced inflammation and dysregulated amyloid metabolism: implications for early detection and therapy. FASEB J. 25: 5–13. [DOI] [PubMed] [Google Scholar]
- 96.Caselli R. J., Walker D., Sue L., Sabbagh M., and Beach T.. 2010. Amyloid load in nondemented brains correlates with APOE e4. Neurosci. Lett. 473: 168–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Greenberg S. M., Vonsattel J. P., Segal A. Z., Chiu R. I., Clatworthy A. E., Liao A., Hyman B. T., and Rebeck G. W.. 1998. Association of apolipoprotein E epsilon2 and vasculopathy in cerebral amyloid angiopathy. Neurology. 50: 961–965. [DOI] [PubMed] [Google Scholar]
- 98.Nelson P. T., Pious N. M., Jicha G. A., Wilcock D. M., Fardo D. W., Estus S., and Rebeck G. W.. 2013. APOE-epsilon2 and APOE-epsilon4 correlate with increased amyloid accumulation in cerebral vasculature. J. Neuropathol. Exp. Neurol. 72: 708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pfeifer L. A., White L. R., Ross G. W., Petrovitch H., and Launer L. J.. 2002. Cerebral amyloid angiopathy and cognitive function: the HAAS autopsy study. Neurology. 58: 1629–1634. [DOI] [PubMed] [Google Scholar]
- 100.Fryer J. D., Simmons K., Parsadanian M., Bales K. R., Paul S. M., Sullivan P. M., and Holtzman D. M.. 2005. Human apolipoprotein E4 alters the amyloid-beta 40:42 ratio and promotes the formation of cerebral amyloid angiopathy in an amyloid precursor protein transgenic model. J. Neurosci. 25: 2803–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liao F., Zhang T. J., Jiang H., Lefton K. B., Robinson G. O., Vassar R., Sullivan P. M., and Holtzman D. M.. 2015. Murine versus human apolipoprotein E4: differential facilitation of and co-localization in cerebral amyloid angiopathy and amyloid plaques in APP transgenic mouse models. Acta Neuropathol. Commun. 3: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Castellano J. M., Deane R., Gottesdiener A. J., Verghese P. B., Stewart F. R., West T., Paoletti A. C., Kasper T. R., DeMattos R. B., Zlokovic B. V., et al. 2012. Low-density lipoprotein receptor overexpression enhances the rate of brain-to-blood Abeta clearance in a mouse model of beta-amyloidosis. Proc. Natl. Acad. Sci. USA. 109: 15502–15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rebeck G. W., Harr S. D., Strickland D. K., and Hyman B. T.. 1995. Multiple, diverse senile plaque-associated proteins are ligands of an apolipoprotein E receptor, the alpha 2-macroglobulin receptor/low-density-lipoprotein receptor-related protein. Ann. Neurol. 37: 211–217. [DOI] [PubMed] [Google Scholar]
- 104.Deane R., Wu Z., Sagare A., Davis J., Du Yan S., Hamm K., Xu F., Parisi M., LaRue B., Hu H. W., et al. 2004. LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron. 43: 333–344. [DOI] [PubMed] [Google Scholar]
- 105.Bu G. 2009. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat. Rev. Neurosci. 10: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Narita M., Holtzman D. M., Schwartz A. L., and Bu G.. 1997. Alpha2-macroglobulin complexes with and mediates the endocytosis of beta-amyloid peptide via cell surface low-density lipoprotein receptor-related protein. J. Neurochem. 69: 1904–1911. [DOI] [PubMed] [Google Scholar]
- 107.Kanekiyo T., and Bu G.. 2009. Receptor-associated protein interacts with amyloid-beta peptide and promotes its cellular uptake. J. Biol. Chem. 284: 33352–33359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cooper A. D. 1997. Hepatic uptake of chylomicron remnants. J. Lipid Res. 38: 2173–2192. [PubMed] [Google Scholar]
- 109.Mahley R. W., and Ji Z. S.. 1999. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J. Lipid Res. 40: 1–16. [PubMed] [Google Scholar]
- 110.Cotman S. L., Halfter W., and Cole G. J.. 2000. Agrin binds to beta-amyloid (Abeta), accelerates abeta fibril formation, and is localized to Abeta deposits in Alzheimer’s disease brain. Mol. Cell. Neurosci. 15: 183–198. [DOI] [PubMed] [Google Scholar]
- 111.van Horssen J., Otte-Holler I., David G., Maat-Schieman M. L., van den Heuvel L. P., Wesseling P., de Waal R. M., and Verbeek M. M.. 2001. Heparan sulfate proteoglycan expression in cerebrovascular amyloid beta deposits in Alzheimer’s disease and hereditary cerebral hemorrhage with amyloidosis (Dutch) brains. Acta Neuropathol. 102: 604–614. [DOI] [PubMed] [Google Scholar]
- 112.Van Gool D., David G., Lammens M., Baro F., and Dom R.. 1993. Heparan sulfate expression patterns in the amyloid deposits of patients with Alzheimer’s and Lewy body type dementia. Dementia. 4: 308–314. [DOI] [PubMed] [Google Scholar]
- 113.Zhang G. L., Zhang X., Wang X. M., and Li J. P.. 2014. Towards understanding the roles of heparan sulfate proteoglycans in Alzheimer’s disease. BioMed Res. Int. 2014: 516028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Watanabe N., Araki W., Chui D. H., Makifuchi T., Ihara Y., and Tabira T.. 2004. Glypican-1 as an Abeta binding HSPG in the human brain: its localization in DIG domains and possible roles in the pathogenesis of Alzheimer’s disease. FASEB J. 18: 1013–1015. [DOI] [PubMed] [Google Scholar]
- 115.Cheng F., Ruscher K., Fransson L. A., and Mani K.. 2013. Non-toxic amyloid beta formed in the presence of glypican-1 or its deaminatively generated heparan sulfate degradation products. Glycobiology. 23: 1510–1519. [DOI] [PubMed] [Google Scholar]
- 116.Brunden K. R., Richter-Cook N. J., Chaturvedi N., and Frederickson R. C.. 1993. pH-dependent binding of synthetic beta-amyloid peptides to glycosaminoglycans. J. Neurochem. 61: 2147–2154. [DOI] [PubMed] [Google Scholar]
- 117.Liu C. C., Zhao N., Yamaguchi Y., Cirrito J. R., Kanekiyo T., Holtzman D. M., and Bu G.. 2016. Neuronal heparan sulfates promote amyloid pathology by modulating brain amyloid-beta clearance and aggregation in Alzheimer’s disease. Sci. Transl. Med. 8: 332ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu Q., Zerbinatti C. V., Zhang J., Hoe H. S., Wang B., Cole S. L., Herz J., Muglia L., and Bu G.. 2007. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron. 56: 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zerbinatti C. V., Wahrle S. E., Kim H., Cam J. A., Bales K., Paul S. M., Holtzman D. M., and Bu G.. 2006. Apolipoprotein E and low density lipoprotein receptor-related protein facilitate intraneuronal Abeta42 accumulation in amyloid model mice. J. Biol. Chem. 281: 36180–36186. [DOI] [PubMed] [Google Scholar]
- 120.Bell R. D., Sagare A. P., Friedman A. E., Bedi G. S., Holtzman D. M., Deane R., and Zlokovic B. V.. 2007. Transport pathways for clearance of human Alzheimer’s amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J. Cereb. Blood Flow Metab. 27: 909–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deane R., Sagare A., Hamm K., Parisi M., Lane S., Finn M. B., Holtzman D. M., and Zlokovic B. V.. 2008. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J. Clin. Invest. 118: 4002–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fryer J. D., Demattos R. B., McCormick L. M., O’Dell M. A., Spinner M. L., Bales K. R., Paul S. M., Sullivan P. M., Parsadanian M., Bu G., et al. 2005. The low density lipoprotein receptor regulates the level of central nervous system human and murine apolipoprotein E but does not modify amyloid plaque pathology in PDAPP mice. J. Biol. Chem. 280: 25754–25759. [DOI] [PubMed] [Google Scholar]
- 123.Kim J., Castellano J. M., Jiang H., Basak J. M., Parsadanian M., Pham V., Mason S. M., Paul S. M., and Holtzman D. M.. 2009. Overexpression of low-density lipoprotein receptor in the brain markedly inhibits amyloid deposition and increases extracellular A beta clearance. Neuron. 64: 632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Basak J. M., Verghese P. B., Yoon H., Kim J., and Holtzman D. M.. 2012. Low-density lipoprotein receptor represents an apolipoprotein E-independent pathway of Abeta uptake and degradation by astrocytes. J. Biol. Chem. 287: 13959–13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bell R. D., Winkler E. A., Singh I., Sagare A. P., Deane R., Wu Z., Holtzman D. M., Betsholtz C., Armulik A., Sallstrom J., et al. 2012. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature. 485: 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Halliday M. R., Rege S. V., Ma Q., Zhao Z., Miller C. A., Winkler E. A., and Zlokovic B. V.. 2016. Accelerated pericyte degeneration and blood-brain barrier breakdown in apolipoprotein E4 carriers with Alzheimer’s disease. J. Cereb. Blood Flow Metab. 36: 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Alata W., Ye Y., St-Amour I., Vandal M., and Calon F.. 2015. Human apolipoprotein E varepsilon4 expression impairs cerebral vascularization and blood-brain barrier function in mice. J. Cereb. Blood Flow Metab. 35: 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bien-Ly N., Boswell C. A., Jeet S., Beach T. G., Hoyte K., Luk W., Shihadeh V., Ulufatu S., Foreman O., Lu Y., et al. 2015. Lack of widespread BBB disruption in Alzheimer’s disease models: focus on therapeutic antibodies. Neuron. 88: 289–297. [DOI] [PubMed] [Google Scholar]
- 129.Koistinaho M., Lin S., Wu X., Esterman M., Koger D., Hanson J., Higgs R., Liu F., Malkani S., Bales K. R., et al. 2004. Apolipoprotein E promotes astrocyte colocalization and degradation of deposited amyloid-beta peptides. Nat. Med. 10: 719–726. [DOI] [PubMed] [Google Scholar]
- 130.Saavedra L., Mohamed A., Ma V., Kar S., and de Chaves E. P.. 2007. Internalization of beta-amyloid peptide by primary neurons in the absence of apolipoprotein E. J. Biol. Chem. 282: 35722–35732. [DOI] [PubMed] [Google Scholar]
- 131.Paresce D. M., Ghosh R. N., and Maxfield F. R.. 1996. Microglial cells internalize aggregates of the Alzheimer’s disease amyloid beta-protein via a scavenger receptor. Neuron. 17: 553–565. [DOI] [PubMed] [Google Scholar]
- 132.Paresce D. M., Chung H., and Maxfield F. R.. 1997. Slow degradation of aggregates of the Alzheimer’s disease amyloid beta-protein by microglial cells. J. Biol. Chem. 272: 29390–29397. [DOI] [PubMed] [Google Scholar]
- 133.Mandrekar S., Jiang Q., Lee C. Y., Koenigsknecht-Talboo J., Holtzman D. M., and Landreth G. E.. 2009. Microglia mediate the clearance of soluble Abeta through fluid phase macropinocytosis. J. Neurosci. 29: 4252–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Beffert U., Aumont N., Dea D., Lussier-Cacan S., Davignon J., and Poirier J.. 1999. Apolipoprotein E isoform-specific reduction of extracellular amyloid in neuronal cultures. Brain Res. Mol. Brain Res. 68: 181–185. [DOI] [PubMed] [Google Scholar]
- 135.Hu X., Crick S. L., Bu G., Frieden C., Pappu R. V., and Lee J. M.. 2009. Amyloid seeds formed by cellular uptake, concentration, and aggregation of the amyloid-beta peptide. Proc. Natl. Acad. Sci. USA. 106: 20324–20329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nielsen H. M., Veerhuis R., Holmqvist B., and Janciauskiene S.. 2009. Binding and uptake of A beta1–42 by primary human astrocytes in vitro. Glia. 57: 978–988. [DOI] [PubMed] [Google Scholar]
- 137.Shaffer L. M., Dority M. D., Gupta-Bansal R., Frederickson R. C., Younkin S. G., and Brunden K. R.. 1995. Amyloid beta protein (A beta) removal by neuroglial cells in culture. Neurobiol. Aging. 16: 737–745. [DOI] [PubMed] [Google Scholar]
- 138.Wyss-Coray T., Loike J. D., Brionne T. C., Lu E., Anankov R., Yan F., Silverstein S. C., and Husemann J.. 2003. Adult mouse astrocytes degrade amyloid-beta in vitro and in situ. Nat. Med. 9: 453–457. [DOI] [PubMed] [Google Scholar]
- 139.Cole G. M., and Ard M. D.. 2000. Influence of lipoproteins on microglial degradation of Alzheimer’s amyloid beta-protein. Microsc. Res. Tech. 50: 316–324. [DOI] [PubMed] [Google Scholar]
- 140.Lee C. Y., Tse W., Smith J. D., and Landreth G. E.. 2012. Apolipoprotein E promotes beta-amyloid trafficking and degradation by modulating microglial cholesterol levels. J. Biol. Chem. 287: 2032–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J. S., Younkin S., et al. 2013. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P. V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A. I., Lah J. J., et al. 2013. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368: 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Atagi Y., Liu C. C., Painter M. M., Chen X. F., Verbeeck C., Zheng H., Li X., Rademakers R., Kang S. S., Xu H., et al. 2015. Apolipoprotein E is a ligand for triggering receptor expressed on myeloid cells 2 (TREM2). J. Biol. Chem. 290: 26043–26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bailey C. C., DeVaux L. B., and Farzan M.. 2015. The triggering receptor expressed on myeloid cells 2 binds apolipoprotein E. J. Biol. Chem. 290: 26033–26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yeh F. L., Wang Y., Tom I., Gonzalez L. C., and Sheng M.. 2016. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron. 91: 328–340. [DOI] [PubMed] [Google Scholar]
- 146.Wang Y., Cella M., Mallinson K., Ulrich J. D., Young K. L., Robinette M. L., Gilfillan S., Krishnan G. M., Sudhakar S., Zinselmeyer B. H., et al. 2015. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 160: 1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yuan P., Condello C., Keene C. D., Wang Y., Bird T. D., Paul S. M., Luo W., Colonna M., Baddeley D., and Grutzendler J.. 2016. TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe axonal dystrophy. Neuron. 92: 252–264. [DOI] [PubMed] [Google Scholar]
- 148.Li X., Montine K. S., Keene C. D., and Montine T. J.. 2015. Different mechanisms of apolipoprotein E isoform-dependent modulation of prostaglandin E2 production and triggering receptor expressed on myeloid cells 2 (TREM2) expression after innate immune activation of microglia. FASEB J. 29: 1754–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li Y. J., Hauser M. A., Scott W. K., Martin E. R., Booze M. W., Qin X. J., Walter J. W., Nance M. A., Hubble J. P., Koller W. C., et al. 2004. Apolipoprotein E controls the risk and age at onset of Parkinson disease. Neurology. 62: 2005–2009. [DOI] [PubMed] [Google Scholar]
- 150.Gao J., Huang X., Park Y., Liu R., Hollenbeck A., Schatzkin A., Mailman R. B., and Chen H.. 2011. Apolipoprotein E genotypes and the risk of Parkinson disease. Neurobiol. Aging. 32: 2106.e1–2106.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McKee A. C., Cantu R. C., Nowinski C. J., Hedley-Whyte E. T., Gavett B. E., Budson A. E., Santini V. E., Lee H. S., Kubilus C. A., and Stern R. A.. 2009. Chronic traumatic encephalopathy in athletes: progressive tauopathy after repetitive head injury. J. Neuropathol. Exp. Neurol. 68: 709–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Panegyres P. K., Beilby J., Bulsara M., Toufexis K., and Wong C.. 2006. A study of potential interactive genetic factors in Huntington’s disease. Eur. Neurol. 55: 189–192. [DOI] [PubMed] [Google Scholar]
- 153.Kehoe P., Krawczak M., Harper P. S., Owen M. J., and Jones A. L.. 1999. Age of onset in Huntington disease: sex specific influence of apolipoprotein E genotype and normal CAG repeat length. J. Med. Genet. 36: 108–111. [PMC free article] [PubMed] [Google Scholar]
- 154.Agosta F., Vossel K. A., Miller B. L., Migliaccio R., Bonasera S. J., Filippi M., Boxer A. L., Karydas A., Possin K. L., and Gorno-Tempini M. L.. 2009. Apolipoprotein E epsilon4 is associated with disease-specific effects on brain atrophy in Alzheimer’s disease and frontotemporal dementia. Proc. Natl. Acad. Sci. USA. 106: 2018–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Praline J., Blasco H., Vourc’h P., Garrigue M. A., Gordon P. H., Camu W., Corcia P., Andres C. R., and French A. L. S. S. G.. 2011. APOE epsilon4 allele is associated with an increased risk of bulbar-onset amyotrophic lateral sclerosis in men. Eur. J. Neurol. 18: 1046–1052. [DOI] [PubMed] [Google Scholar]
- 156.Li Y. J., Pericak-Vance M. A., Haines J. L., Siddique N., McKenna-Yasek D., Hung W. Y., Sapp P., Allen C. I., Chen W., Hosler B., et al. 2004. Apolipoprotein E is associated with age at onset of amyotrophic lateral sclerosis. Neurogenetics. 5: 209–213. [DOI] [PubMed] [Google Scholar]
- 157.Lacomblez L., Doppler V., Beucler I., Costes G., Salachas F., Raisonnier A., Le Forestier N., Pradat P. F., Bruckert E., and Meininger V.. 2002. APOE: a potential marker of disease progression in ALS. Neurology. 58: 1112–1114. [DOI] [PubMed] [Google Scholar]
- 158.Drory V. E., Birnbaum M., Korczyn A. D., and Chapman J.. 2001. Association of APOE epsilon4 allele with survival in amyotrophic lateral sclerosis. J. Neurol. Sci. 190: 17–20. [DOI] [PubMed] [Google Scholar]
- 159.Govone F., Vacca A., Rubino E., Gai A., Boschi S., Gentile S., Orsi L., Pinessi L., and Rainero I.. 2014. Lack of association between APOE gene polymorphisms and amyotrophic lateral sclerosis: a comprehensive meta-analysis. Amyotroph. Lateral Scler. Frontotemporal Degener. 15: 551–556. [DOI] [PubMed] [Google Scholar]
- 160.Mui S., Rebeck G. W., McKenna-Yasek D., Hyman B. T., and Brown R. H. Jr. 1995. Apolipoprotein E epsilon 4 allele is not associated with earlier age at onset in amyotrophic lateral sclerosis. Ann. Neurol. 38: 460–463. [DOI] [PubMed] [Google Scholar]
- 161.Gustafson L., Abrahamson M., Grubb A., Nilsson K., and Fex G.. 1997. Apolipoprotein-E genotyping in Alzheimer’s disease and frontotemporal dementia. Dement. Geriatr. Cogn. Disord. 8: 240–243. [DOI] [PubMed] [Google Scholar]
- 162.Bernardi L., Maletta R. G., Tomaino C., Smirne N., Di Natale M., Perri M., Longo T., Colao R., Curcio S. A., Puccio G., et al. 2006. The effects of APOE and tau gene variability on risk of frontotemporal dementia. Neurobiol. Aging. 27: 702–709. [DOI] [PubMed] [Google Scholar]
- 163.Ji Y., Liu M., Huo Y. R., Liu S., Shi Z., Liu S., Wisniewski T., and Wang J.. 2013. Apolipoprotein Epsilon epsilon4 frequency is increased among Chinese patients with frontotemporal dementia and Alzheimer’s disease. Dement. Geriatr. Cogn. Disord. 36: 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fabre S. F., Forsell C., Viitanen M., Sjogren M., Wallin A., Blennow K., Blomberg M., Andersen C., Wahlund L. O., and Lannfelt L.. 2001. Clinic-based cases with frontotemporal dementia show increased cerebrospinal fluid tau and high apolipoprotein E epsilon4 frequency, but no tau gene mutations. Exp. Neurol. 168: 413–418. [DOI] [PubMed] [Google Scholar]
- 165.Ohm T. G., Kirca M., Bohl J., Scharnagl H., Gross W., and Marz W.. 1995. Apolipoprotein E polymorphism influences not only cerebral senile plaque load but also Alzheimer-type neurofibrillary tangle formation. Neuroscience. 66: 583–587. [DOI] [PubMed] [Google Scholar]
- 166.Gallyas F. 1971. Silver staining of Alzheimer’s neurofibrillary changes by means of physical development. Acta Morphol. Acad. Sci. Hung. 19: 1–8. [PubMed] [Google Scholar]
- 167.Engelborghs S., Dermaut B., Marien P., Symons A., Vloeberghs E., Maertens K., Somers N., Goeman J., Rademakers R., Van den Broeck M., et al. 2006. Dose dependent effect of APOE epsilon4 on behavioral symptoms in frontal lobe dementia. Neurobiol. Aging. 27: 285–292. [DOI] [PubMed] [Google Scholar]
- 168.Riemenschneider M., Diehl J., Muller U., Forstl H., and Kurz A.. 2002. Apolipoprotein E polymorphism in German patients with frontotemporal degeneration. J. Neurol. Neurosurg. Psychiatry. 72: 639–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Srinivasan R., Davidson Y., Gibbons L., Payton A., Richardson A. M., Varma A., Julien C., Stopford C., Thompson J., Horan M. A., et al. 2006. The apolipoprotein E epsilon4 allele selectively increases the risk of frontotemporal lobar degeneration in males. J. Neurol. Neurosurg. Psychiatry. 77: 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Strittmatter W. J., Saunders A. M., Goedert M., Weisgraber K. H., Dong L. M., Jakes R., Huang D. Y., Pericak-Vance M., Schmechel D., and Roses A. D.. 1994. Isoform-specific interactions of apolipoprotein E with microtubule-associated protein tau: implications for Alzheimer disease. Proc. Natl. Acad. Sci. USA. 91: 11183–11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tesseur I., Van Dorpe J., Spittaels K., Van den Haute C., Moechars D., and Van Leuven F.. 2000. Expression of human apolipoprotein E4 in neurons causes hyperphosphorylation of protein tau in the brains of transgenic mice. Am. J. Pathol. 156: 951–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Huang Y., Liu X. Q., Wyss-Coray T., Brecht W. J., Sanan D. A., and Mahley R. W.. 2001. Apolipoprotein E fragments present in Alzheimer’s disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. Proc. Natl. Acad. Sci. USA. 98: 8838–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Harris F. M., Brecht W. J., Xu Q., Tesseur I., Kekonius L., Wyss-Coray T., Fish J. D., Masliah E., Hopkins P. C., Scearce-Levie K., et al. 2003. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer’s disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc. Natl. Acad. Sci. USA. 100: 10966–10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Brecht W. J., Harris F. M., Chang S., Tesseur I., Yu G. Q., Xu Q., Dee Fish J., Wyss-Coray T., Buttini M., Mucke L., et al. 2004. Neuron-specific apolipoprotein e4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. J. Neurosci. 24: 2527–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Harris F. M., Brecht W. J., Xu Q., Mahley R. W., and Huang Y.. 2004. Increased tau phosphorylation in apolipoprotein E4 transgenic mice is associated with activation of extracellular signal-regulated kinase: modulation by zinc. J. Biol. Chem. 279: 44795–44801. [DOI] [PubMed] [Google Scholar]
- 176.Allen B., Ingram E., Takao M., Smith M. J., Jakes R., Virdee K., Yoshida H., Holzer M., Craxton M., Emson P. C., et al. 2002. Abundant tau filaments and nonapoptotic neurodegeneration in transgenic mice expressing human P301S tau protein. J. Neurosci. 22: 9340–9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Iwai A., Masliah E., Yoshimoto M., Ge N., Flanagan L., de Silva H. A., Kittel A., and Saitoh T.. 1995. The precursor protein of non-A beta component of Alzheimer’s disease amyloid is a presynaptic protein of the central nervous system. Neuron. 14: 467–475. [DOI] [PubMed] [Google Scholar]
- 178.Jakes R., Spillantini M. G., and Goedert M.. 1994. Identification of two distinct synucleins from human brain. FEBS Lett. 345: 27–32. [DOI] [PubMed] [Google Scholar]
- 179.Withers G. S., George J. M., Banker G. A., and Clayton D. F.. 1997. Delayed localization of synelfin (synuclein, NACP) to presynaptic terminals in cultured rat hippocampal neurons. Brain Res. Dev. Brain Res. 99: 87–94. [DOI] [PubMed] [Google Scholar]
- 180.Lashuel H. A., Overk C. R., Oueslati A., and Masliah E.. 2013. The many faces of alpha-synuclein: from structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 14: 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Irwin D. J., Lee V. M., and Trojanowski J. Q.. 2013. Parkinson’s disease dementia: convergence of alpha-synuclein, tau and amyloid-beta pathologies. Nat. Rev. Neurosci. 14: 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mikolaenko I., Pletnikova O., Kawas C. H., O’Brien R., Resnick S. M., Crain B., and Troncoso J. C.. 2005. Alpha-synuclein lesions in normal aging, Parkinson disease, and Alzheimer disease: evidence from the Baltimore Longitudinal Study of Aging (BLSA). J. Neuropathol. Exp. Neurol. 64: 156–162. [DOI] [PubMed] [Google Scholar]
- 183.Mattila P. M., Rinne J. O., Helenius H., Dickson D. W., and Roytta M.. 2000. Alpha-synuclein-immunoreactive cortical Lewy bodies are associated with cognitive impairment in Parkinson’s disease. Acta Neuropathol. 100: 285–290. [DOI] [PubMed] [Google Scholar]
- 184.Jellinger K. A. 2007. Morphological substrates of parkinsonism with and without dementia: a retrospective clinico-pathological study. J. Neural Transm. Suppl. 72: 91–104. [DOI] [PubMed] [Google Scholar]
- 185.Hughes A. J., Daniel S. E., Blankson S., and Lees A. J.. 1993. A clinicopathologic study of 100 cases of Parkinson’s disease. Arch. Neurol. 50: 140–148. [DOI] [PubMed] [Google Scholar]
- 186.Sabbagh M. N., Adler C. H., Lahti T. J., Connor D. J., Vedders L., Peterson L. K., Caviness J. N., Shill H. A., Sue L. I., Ziabreva I., et al. 2009. Parkinson disease with dementia: comparing patients with and without Alzheimer pathology. Alzheimer Dis. Assoc. Disord. 23: 295–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kotzbauer P. T., Cairns N. J., Campbell M. C., Willis A. W., Racette B. A., Tabbal S. D., and Perlmutter J. S.. 2012. Pathologic accumulation of alpha-synuclein and Abeta in Parkinson disease patients with dementia. Arch. Neurol. 69: 1326–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Irwin D. J., Grossman M., Weintraub D., Hurtig H. I., Duda J. E., Xie S. X., Lee E. B., Van Deerlin V. M., Lopez O. L., Kofler J. K., et al. 2017. Neuropathological and genetic correlates of survival and dementia onset in synucleinopathies: a retrospective analysis. Lancet Neurol. 16: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wilson R. S., Bennett D. A., Gilley D. W., Beckett L. A., Schneider J. A., and Evans D. A.. 2000. Progression of parkinsonism and loss of cognitive function in Alzheimer disease. Arch. Neurol. 57: 855–860. [DOI] [PubMed] [Google Scholar]
- 190.Marder K., Tang M. X., Cote L., Stern Y., and Mayeux R.. 1995. The frequency and associated risk factors for dementia in patients with Parkinson’s disease. Arch. Neurol. 52: 695–701. [DOI] [PubMed] [Google Scholar]
- 191.Korczyn A. D. 2001. Dementia in Parkinson’s disease. J. Neurol. 248(Suppl 3): III1–III4. [DOI] [PubMed] [Google Scholar]
- 192.Lippa C. F., Schmidt M. L., Lee V. M., and Trojanowski J. Q.. 2001. Alpha-synuclein in familial Alzheimer disease: epitope mapping parallels dementia with Lewy bodies and Parkinson disease. Arch. Neurol. 58: 1817–1820. [DOI] [PubMed] [Google Scholar]
- 193.Li Y. J., Scott W. K., Hedges D. J., Zhang F. Y., Gaskell P. C., Nance M. A., Watts R. L., Hubble J. P., Koller W. C., Pahwa R., et al. 2002. Age at onset in two common neurodegenerative diseases is genetically controlled. Am. J. Hum. Genet. 70: 985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Martin E. R., Scott W. K., Nance M. A., Watts R. L., Hubble J. P., Koller W. C., Lyons K., Pahwa R., Stern M. B., Colcher A., et al. 2001. Association of single-nucleotide polymorphisms of the tau gene with late-onset Parkinson disease. JAMA. 286: 2245–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Huang X., Chen P. C., and Poole C.. 2004. APOE-[epsilon]2 allele associated with higher prevalence of sporadic Parkinson disease. Neurology. 62: 2198–2202. [DOI] [PubMed] [Google Scholar]
- 196.Lill C. M., Roehr J. T., McQueen M. B., Kavvoura F. K., Bagade S., Schjeide B. M., Schjeide L. M., Meissner E., Zauft U., Allen N. C. , et al. 2012. Comprehensive research synopsis and systematic meta-analyses in Parkinson’s disease genetics: the PDGene database. PLoS Genet. 8: e1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Josephs K. A., Tsuboi Y., Cookson N., Watt H., and Dickson D. W.. 2004. Apolipoprotein E epsilon 4 is a determinant for Alzheimer-type pathologic features in tauopathies, synucleinopathies, and frontotemporal degeneration. Arch. Neurol. 61: 1579–1584. [DOI] [PubMed] [Google Scholar]
- 198.Irwin D. J., White M. T., Toledo J. B., Xie S. X., Robinson J. L., Van Deerlin V., Lee V. M., Leverenz J. B., Montine T. J., Duda J. E., et al. 2012. Neuropathologic substrates of Parkinson disease dementia. Ann. Neurol. 72: 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Siderowf A., Xie S. X., Hurtig H., Weintraub D., Duda J., Chen-Plotkin A., Shaw L. M., Van Deerlin V., Trojanowski J. Q., and Clark C.. 2010. CSF amyloid {beta} 1-42 predicts cognitive decline in Parkinson disease. Neurology. 75: 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Saito Y., Kawashima A., Ruberu N. N., Fujiwara H., Koyama S., Sawabe M., Arai T., Nagura H., Yamanouchi H., Hasegawa M., et al. 2003. Accumulation of phosphorylated alpha-synuclein in aging human brain. J. Neuropathol. Exp. Neurol. 62: 644–654. [DOI] [PubMed] [Google Scholar]
- 201.Wakabayashi K., Kakita A., Hayashi S., Okuizumi K., Onodera O., Tanaka H., Ishikawa A., Tsuji S., and Takahashi H.. 1998. Apolipoprotein E epsilon4 allele and progression of cortical Lewy body pathology in Parkinson’s disease. Acta Neuropathol. 95: 450–454. [DOI] [PubMed] [Google Scholar]
- 202.Tsuang D., Leverenz J. B., Lopez O. L., Hamilton R. L., Bennett D. A., Schneider J. A., Buchman A. S., Larson E. B., Crane P. K., Kaye J. A., et al. 2013. APOE epsilon4 increases risk for dementia in pure synucleinopathies. JAMA Neurol. 70: 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gallardo G., Schluter O. M., and Sudhof T. C.. 2008. A molecular pathway of neurodegeneration linking alpha-synuclein to ApoE and Abeta peptides. Nat. Neurosci. 11: 301–308. [DOI] [PubMed] [Google Scholar]
- 204.Emamzadeh F. N., Aojula H., McHugh P. C., and Allsop D.. 2016. Effects of different isoforms of apoE on aggregation of the alpha-synuclein protein implicated in Parkinson’s disease. Neurosci. Lett. 618: 146–151. [DOI] [PubMed] [Google Scholar]
- 205.Theendakara V., Peters-Libeu C. A., Spilman P., Poksay K. S., Bredesen D. E., and Rao R. V.. 2016. Direct Transcriptional Effects of Apolipoprotein E. J. Neurosci. 36: 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]