Abstract
Characterization of molecular interactions in terms of basic binding parameters such as binding affinity, stoichiometry, and thermodynamics is an essential step in basic and applied science. MicroScale Thermophoresis (MST) is a sensitive biophysical method to obtain this important information. Relying on a physical effect called thermophoresis, which describes the movement of molecules through temperature gradients, this technology allows for the fast and precise determination of binding parameters in solution and allows the free choice of buffer conditions (from buffer to lysates/sera). MST uses the fact that an unbound molecule displays a different thermophoretic movement than a molecule that is in complex with a binding partner. The thermophoretic movement is altered in the moment of molecular interaction due to changes in size, charge, and hydration shell. By comparing the movement profiles of different molecular ratios of the two binding partners, quantitative information such as binding affinity (pM to mM) can be determined. Even challenging interactions between molecules of small sizes, such as aptamers and small compounds, can be studied by MST. Using the well-studied model interaction between the DH25.42 DNA aptamer and ATP, this manuscript provides a protocol to characterize aptamer-small molecule interactions. This study demonstrates that MST is highly sensitive and permits the mapping of the binding site of the 7.9 kDa DNA aptamer to the adenine of ATP.
Keywords: Biochemistry, Issue 119, Small molecule-aptamer interaction, binding parameters, MicroScale Thermophoresis, binding affinity, stoichiometry, thermodynamics, nucleic acids, binding site mapping
Introduction
Interaction between molecules is the basis of nature. Hence, scientists in many fields of basic and applied research try to understand the fundamental principles of molecular interactions of different kinds. MicroScale Thermophoresis (MST) enables scientists to perform the fast, precise, cost-efficient, and quality-controlled characterization of molecular interactions in solution, with a free choice of buffers. There are already more than 1,000 publications using MST, from 2016 alone, describing different kinds of analyses, including library screenings, binding event validations, competition assays, and experiments with multiple binding partners1-8. In general, MST permits the study of the classical binding parameters, such as binding affinity (pM to mM), stoichiometry, and thermodynamics, of any kind of molecular interaction. A great advantage of MST is the ability to study binding events independent of the size of the interaction partners. Even challenging interactions between small nucleic acid aptamers (15-30 nt) and targets such as small molecules, drugs, antibiotics, or metabolites can be quantified.
Current state-of-the-art technologies to characterize aptamer-target interactions are either lab-intense and highly complex or fail to quantify aptamer-small molecule interactions9,10. Surface Plasmon Resonance (SPR)-based assays11,12 and truly label-free calorimetric approaches, such as Isothermal Titration Calorimetry (ITC)13-15, isocratic elution16, equilibrium filtration17,18, in-line probing19, gel-shift assays, stopped-flow fluorescence spectroscopy20,21, fluorescence anisotropy (FA)22,23, single-molecule fluorescence imaging24,25, and Bio-layer interferometry (BLI)26 are also either imprecise or incompatible with aptamer-small molecule interactions. Other principal issues of these methods are low sensitivity, high sample consumption, immobilization, mass transport limitations on surfaces, and/or buffer restrictions. Only a few of these technologies provide integrated controls for aggregation and adsorption effects.
MST represents a powerful tool for scientists to overcome this limitation to study the interactions between aptamers and small molecules27-29, as well as other targets such as proteins30-33. The technology relies on the movement of molecules through temperature gradients. This directed movement, called "thermophoresis," depends on the size, charge, and hydration shell of the molecule34,35. The binding of a ligand to the molecule will directly alter at least one of these parameters, resulting in a changed thermophoretic mobility. Ligands with small sizes may not have considerable impact in terms of size change from the unbound to the bound state, but they can have dramatic effects on the hydration shell and/or charge. The changes in the thermophoretic movement of molecules after interactions with the binding partner enables the quantification of basic binding parameters2,7,34,36,37.
As depicted in Figure 1A, the MST device consists of an infrared laser focused onto the sample within the glass capillaries using the same optics as for fluorescence detection. The thermophoretic movement of proteins via the intrinsic fluorescence of tryptophans6 or of a fluorescently labeled interaction partner3,8 can be monitored while the laser establishes a temperature gradient (ΔT of 2-6 °C). The resulting temperature difference in space, ΔT, leads to the depletion or accumulation of molecules in the area of elevated temperature, which can be quantified by the Soret coefficient (ST):
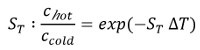
chot represents the concentration in the heated region, and ccold is the concentration in the initial cold region.
As shown in Figure 1B, a typical MST experiment results in an MST movement profile (time trace), consisting of different phases, which can be separated by their respective timescales. The initial fluorescence is measured in the first 5 s in absence of the temperature gradient to define the precise starting fluorescence and to check for photobleaching or photoenhancement. The Temperature Jump (T-Jump) represents the phase in which the fluorescence changes before thermophoretic movement. This initial decrease in fluorescence depends on heat-dependent changes of fluorophore quantum yield. The thermophoresis phase follows, in which the fluorescence decreases (or increases) due to the thermophoretic movement of the molecules until the steady-state distribution is reached. The reverse TJump and concomitant back diffusion of fluorescent molecules can be observed as indicated in Figure 1B after the laser is switched off. In order to access basic binding parameters, different molar ratios of the interaction partners are analyzed and compared. Typically, 16 different ratios are studied in one MST experiment, whereas the optical visible molecule is kept constant and is supplied with an increasing amount of the unlabeled ligand. The interaction between the two binding partners induces changes in the thermophoresis, and thus in the normalized fluorescence, Fnorm, which is calculated as following:
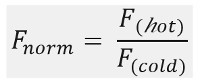
Fhot and Fcold represent averaged fluorescence intensities at defined time points of the MST traces. Binding affinities (Kd or EC50 values) can be calculated by curve fitting (Figure 1C).
Overall, MST is a powerful tool to study molecular interactions of any kind. This manuscript offers a protocol to characterize the challenging interaction between the small molecule adenosine triphosphate (ATP; 0.5 kDa) and the 25-nt short ssDNA aptamer DH25.42 (7.9 kDa). Over the course of the manuscript, the binding site of the aptamer on the ATP molecule is mapped down to the adenine group of the ATP.
Protocol
1. Preparation of the Aptamer Working Stock
Follow the manufacturer's instructions and dissolve the oligonucleotide (5-Cy5-CCTG GGGGAGTATTGCGGAGGAAGG-3, sequence from reference18) in water, reaching a 100-µM final concentration.
Prepare the aptamer working solution by diluting the oligonucleotide stock to 200 nM with binding buffer (20 mM Tris, pH 7.6; 300 mM NaCl; 5 mM MgCl2; 0.01% Tween20).
Incubate the mixture for 2 min at 90 °C, let the sample immediately cool down on ice, and use the sample at room temperature.
2. Preparation of the Ligand Dilution Series
For each ligand (adenosine triphosphate (ATP), adenosine diphosphate (ADP), adenosine monophosphate (AMP), adenine, guanosine triphosphate (GTP), cytosine triphosphate (CTP), deoxyadenosine triphosphate (dATP), and S-adenosyl methionine (SAM); 10 mM stock each), prepare a 16-step serial dilution in 200 µl micro reaction tubes. NOTE: Centrifugation of ligand stocks for 5 min at 14,000 x g may help to remove aggregates. Low volume, low binding reaction tubes are recommended to avoid adsorption of molecules to the tube walls.
Start with a maximum concentration of at least 50 times higher than the estimated affinity and reduce the ligand concentration by 50% in each dilution step. NOTE: The concentration finder tool implemented in the control software simulates binding data and helps with finding the right concentration range for the dilution series.
Fill 20 µl of the ligand stock (10 mM) in tube 1. Add 10 µl of aptamer binding buffer into micro reaction tubes 2 to 16.
Transfer 10 µl of tube 1 to tube 2 and mix properly by pipetting up and down several times. Transfer 10 µL to the next tube and repeat this dilution for the remaining tubes.
Discard the 10 µl excess from the last tube. Avoid any buffer dilution effects. The buffer in tube 1 and in tubes 2-16 must be identical.
3. Preparation of the Final Reaction Mix
Prepare the individual binding reactions with a volume of 20 µl (10 µl of aptamer working solution + 10 µl of the respective ligand dilution) to minimize pipetting errors. A volume of only 4 µl is sufficient to fill the capillary.
Add 10 µl of the 200 nM aptamer working solution to 10 µl of each ligand dilution and mix properly by pipetting up and down several times.
Incubate the samples for 5 min at room temperature and fill the samples into standard capillaries by dipping the capillaries into the sample. Longer incubation times may be necessary for some interactions; however, 5 min is adequate for most. Touch the capillaries only on the sides, NOT on the middle part, where the optical measurement will be taken.
Place the capillaries onto the capillary tray and start the MST device.
4. Starting the MST Device
NOTE: The device provides two pre-installed software packages, the ''control" software for the technical setup of the experimental conditions and the ''analysis" software for the interpretation of the produced data.
Before placing the capillary tray into the MST device, start the control software and adjust the overall desired temperature by selecting ''enable manual temperature control" in the ''temperature control" dropdown menu. Adjust the temperature to 25 °C in this way. NOTE: The MST instruments can be temperature-controlled from 22 to 45 °C.
Wait for the temperature to reach the expected level and then place the capillary tray into the MST device.
Set the LED channel to ''red" for Cy5 dyes and adjust the LED power to gain a fluorescence signal of 300 to 1,000 fluorescence units at the MST device with a standard sensor. 25% LED power is used in this study. NOTE: 6,000 to 18,000 fluorescence units are recommended for the MST with a high-sensitivity sensor.
5. Capillary Scan
Carry out a capillary scan to check different quality aspects of the sample by choosing the capillary position on the "control" software and clicking on "start cap scan"before starting the MST measurement.
Inspect the capillary scan for fluorescence enhancement/quenching and sticking effects (U-shaped or flattened peaks) in the software. NOTE: More details on the detection and handling of fluorescence and sticking effects can be found in the discussion.
6. MST Measurement
NOTE: Before starting the MST measurement, make sure to exclude sticking effects, enhancement/quenching effects, or pipetting errors, and ensure that the capillary scan indicates that the fluorescence signal is sufficient. For more details, see the discussion.
Assign the ligand concentrations from the dilution series to the respective capillary position in the ''control" software. Consider the dilution step of mixing the aptamer and ligand (1:1).
Enter the highest concentration of ligand (5 mM) for capillary #1, select the correct dilution type (here, 1:1), click on the maximum concentration, and use the drag function to automatically assign the remaining concentrations in capillaries #2-16. The lowest concentration is 152.6 nM.
Enter the concentration of the fluorescent aptamer (here, 100 nM) in the respective section of the control software.
Use the default settings, which detect the fluorescence for 5 sec, record the MST for 30 sec, and record the fluorescence for a further 5 sec after the inactivation of the laser to monitor the back diffusion of molecules.
Adjust the laser power to 20% in the respective section of the control software. NOTE: In order to receive the best signal-to-noise ratio and to avoid unspecific effects, a laser power of 20-40% is recommended. In specific cases, a higher laser power may be required to get a good separation of unbound and bound molecules.
Save the experiment after selecting the destination folder and start the MST measurement by pressing the ''Start MST measurement" button. NOTE: The .ntp file will be generated in the destination folder. Using this setup, one measurement lasts 10-15 min.
Repeat the experimental procedure at least twice for a more accurate determination of the EC50 value. NOTE: In order to test the technical reproducibility, the same capillaries can be scanned several times (technical repeats).
7. MST Data Analysis
NOTE: The analysis software enables the analysis of data on the fly during the measurement. The analysis software plots the MST time traces and changes in the normalized fluorescence (Fnorm) versus the ligand concentration37.
Start the MST analysis software (MO.Affinity Analysis) and load the .ntp file from the destination folder. Select "MST" as the analysis type in the data selection menu. NOTE: In case of ligand-dependent fluorescence effects, the initial fluorescence can be chosen for analysis.
Add the respective technical or biological run(s) to a new analysis by drag-and-drop or by pressing the "+" button below the respective experimental run.
Press the information button below the respective experimental run to obtain information on the properties of the experiment, MST traces, capillary scan, capillary shape, initial fluorescence, and bleaching rate. NOTE: These raw data can also be inspected in later steps of the analysis.
Visually inspect the MST traces for aggregation and precipitation effects, visible as bumps and spikes. NOTE: For more information on the detection and handling of aggregation effects, read the discussion.
Visually inspect the capillary scan and the capillary shape overlay for adsorption effects, visible as flattened or U-shaped peaks. Visually inspect the capillary scan and the initial fluorescence for fluorescence effects. Visually inspect the bleaching rate for photobleaching effects.
Switch to the dose-response mode and change the analysis setting to "expert" mode by pressing the respective button. Select "T-Jump" as the MST evaluation strategy.
Select the "Hill" model for curve fitting. The binding parameters will automatically be calculated. Normalize the data by choosing the respective type of normalization in the "compare results" menu. Export the data either as an .xls or .pdf. NOTE: The table below the binding graph summarizes the calculated binding parameters.
Representative Results
In this study, MST was applied to characterize the binding site of the DH25.42 DNA aptamer18 on ATP. In contrast to other studies characterizing the interaction of ATP or ATP-mimicking small molecules with proteins randomly labeled with one or more fluorophores38-40, this study includes a labeled version of the 7.9 kDa ssDNA aptamer with one Cy5 molecule on the 5´ end. Different ATP derivatives and related molecules, all differing from ATP in various positions, were used to map the binding site on the ATP molecule. Dilution series (in 16 steps, reducing the ligand concentration by 50% each) of the different ligands were prepared and mixed with a constant amount of Cy5-labeled aptamer. Samples were analyzed in standard capillaries at 25% LED and 20% laser power. Movement profiles (MST time traces) were recorded and fluorescence units, derived from the T-Jump phase, were plotted versus the ligand concentration (compare Figure 1C). Curve fitting was performed applying the Hill equation, resulting in EC50 values. In 5 independent biological repeats (4 different operators, 2 different aptamer stocks, 2 different ATP stocks), the ATP-aptamer interaction shows a negative binding amplitude of 6 to 13 units and an average EC50 value of 52.3 ± 5.0 µM (Figure 2A). Error bars in the binding graphs and "±" presented in the affinity data represent the standard deviation of 5 biological repeats (5 independent measurements in a different capillary). Previously reported affinities for the interaction of the DH25.42 aptamer with [2,8,5'-3H]-adenosine and ATP agarose were comparable. The interaction of radiolabeled adenosine with the aptamer was quantified by a centrifugal filter assay, and an affinity (Kd) of 6 ± 3 µM was determined. Isocratic elution experiments with ATP immobilized to agarose resulted in an affinity (Kd) of 13 µM18,41.
For a better side-by-side comparison, the data can be normalized to the fraction of complexed molecules (fraction bound, FB) using the following equation: ![]() where value(c) is the MST value measured for the concentration c, free is the MST value for the unbound state (lowest concentration), and complexed is the MST value for the fully bound state (Figure 2B). This normalization is ideal to compare the results of different ligands in one graph, as shown in Figure 2C. The detected affinities of ADP (63.6 ± 5.9 µM in biological duplicates), AMP (91.6 ± 9.1 µM in biological duplicates), and SAM (44.4 ± 3.2 µM in biological duplicates), which differ from ATP in the number of phosphate groups, imply that this position has no or only minor influence on the binding behavior of the aptamer (Figure 2C and D). The OH group at the C2 carbon of the ribose of ATP could also be excluded from being the major binding site, as dATP was also bound by the aptamer with a slightly reduced affinity (64.4 ± 6.1 µM in biological duplicates). Changing the purine group of ATP to the pyrimidine group CTP resulted in non-binding of the aptamer, demonstrating the importance of this group for the interaction. The aptamer bound to adenine with an affinity of 141.7 ± 9.4 µM (in biological duplicates), showing that the binding site must be in this part of the ATP molecule. The purine molecules GTP and ATP differ in the green shaded area represented in Figure 2D, which represents the main binding site of the aptamer on ATP. Another study used non-quantitative elution experiments with different ATP derivatives to elute a radiolabeled aptamer from ATP agarose, which showed comparable results to this MST study18.
where value(c) is the MST value measured for the concentration c, free is the MST value for the unbound state (lowest concentration), and complexed is the MST value for the fully bound state (Figure 2B). This normalization is ideal to compare the results of different ligands in one graph, as shown in Figure 2C. The detected affinities of ADP (63.6 ± 5.9 µM in biological duplicates), AMP (91.6 ± 9.1 µM in biological duplicates), and SAM (44.4 ± 3.2 µM in biological duplicates), which differ from ATP in the number of phosphate groups, imply that this position has no or only minor influence on the binding behavior of the aptamer (Figure 2C and D). The OH group at the C2 carbon of the ribose of ATP could also be excluded from being the major binding site, as dATP was also bound by the aptamer with a slightly reduced affinity (64.4 ± 6.1 µM in biological duplicates). Changing the purine group of ATP to the pyrimidine group CTP resulted in non-binding of the aptamer, demonstrating the importance of this group for the interaction. The aptamer bound to adenine with an affinity of 141.7 ± 9.4 µM (in biological duplicates), showing that the binding site must be in this part of the ATP molecule. The purine molecules GTP and ATP differ in the green shaded area represented in Figure 2D, which represents the main binding site of the aptamer on ATP. Another study used non-quantitative elution experiments with different ATP derivatives to elute a radiolabeled aptamer from ATP agarose, which showed comparable results to this MST study18.
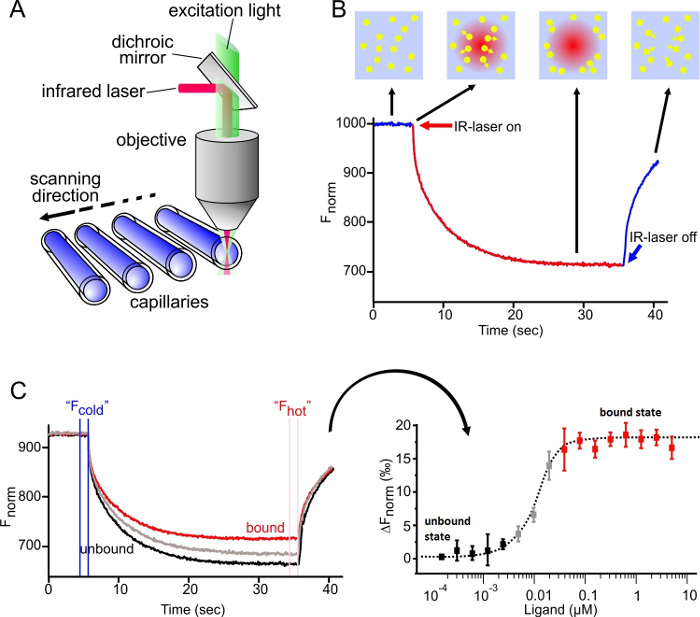 Figure 1: MicroScale Thermophoresis. (A) The technical setup of the MST technology is shown. The optics focus on the center of glass capillaries, thereby detecting the fluorescence signal of the optically visible molecule. An IR laser is utilized to establish a temperature gradient in the observation window of the optical system. Changes in fluorescence can be utilized to monitor the thermophoretic movement of the molecules in solution (B) MST time trace-movement profile of molecules in a temperature gradient. The initial fluorescence is measured for 5 sec while the laser is off. Switching on the laser generates a temperature gradient. After the immediate T-Jump phase, in which the fluorescent dye decreases its signal yield upon heat induction, the thermophoretic movement takes place and is observed for 30 sec. After the laser is turned off, the molecules diffuse back. (C) Results of a typical MST experiment: (Left) 16 capillaries containing the same concentration of fluorescent molecule and an increasing concentration of the unlabeled ligand; the MST time traces are recorded and normalized to their initial fluorescence. (Right) The normalized fluorescence; the difference between Fcold and Fhot is plotted against the concentration of the ligand. A curve fit of this data yields binding parameters, such as the binding affinity. Re-print with permission from Elsevier, Methods28; license number 3890230800113. Please click here to view a larger version of this figure.
Figure 1: MicroScale Thermophoresis. (A) The technical setup of the MST technology is shown. The optics focus on the center of glass capillaries, thereby detecting the fluorescence signal of the optically visible molecule. An IR laser is utilized to establish a temperature gradient in the observation window of the optical system. Changes in fluorescence can be utilized to monitor the thermophoretic movement of the molecules in solution (B) MST time trace-movement profile of molecules in a temperature gradient. The initial fluorescence is measured for 5 sec while the laser is off. Switching on the laser generates a temperature gradient. After the immediate T-Jump phase, in which the fluorescent dye decreases its signal yield upon heat induction, the thermophoretic movement takes place and is observed for 30 sec. After the laser is turned off, the molecules diffuse back. (C) Results of a typical MST experiment: (Left) 16 capillaries containing the same concentration of fluorescent molecule and an increasing concentration of the unlabeled ligand; the MST time traces are recorded and normalized to their initial fluorescence. (Right) The normalized fluorescence; the difference between Fcold and Fhot is plotted against the concentration of the ligand. A curve fit of this data yields binding parameters, such as the binding affinity. Re-print with permission from Elsevier, Methods28; license number 3890230800113. Please click here to view a larger version of this figure.
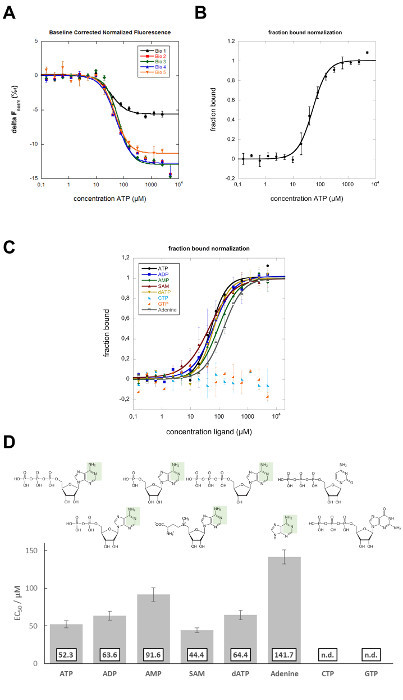 Figure 2:MST data analysis. (A) The baseline corrected normalized fluorescence ΔFnorm (‰), derived from the MST TJump signal, is plotted against the ATP concentration (in µM). The Hill equation (EC50) was applied for curve fitting. The error bars represent the standard deviation from five biological repeats. (B) Fraction-bound plot of the data shown in A. The respective data sets were normalized to the fraction bound, and the average of these normalized data is presented in the binding graph. The error bars indicate the standard deviation of 5 biological repeats. (C) The fraction-bound graph shows a quantitative comparison (Hill fit) of the different ligands to the aptamer. The error bars represent the standard deviation of two biological repeats. (D) Binding affinities (EC50) of different ligands to the aptamer. The green shaded area indicates the binding site of the aptamer on the adenine group of ATP. This figure is modified from Elsevier, Methods28; license number 3890230800113. Data sets from the previous study are reanalyzed and expanded within this study. Please click here to view a larger version of this figure.
Figure 2:MST data analysis. (A) The baseline corrected normalized fluorescence ΔFnorm (‰), derived from the MST TJump signal, is plotted against the ATP concentration (in µM). The Hill equation (EC50) was applied for curve fitting. The error bars represent the standard deviation from five biological repeats. (B) Fraction-bound plot of the data shown in A. The respective data sets were normalized to the fraction bound, and the average of these normalized data is presented in the binding graph. The error bars indicate the standard deviation of 5 biological repeats. (C) The fraction-bound graph shows a quantitative comparison (Hill fit) of the different ligands to the aptamer. The error bars represent the standard deviation of two biological repeats. (D) Binding affinities (EC50) of different ligands to the aptamer. The green shaded area indicates the binding site of the aptamer on the adenine group of ATP. This figure is modified from Elsevier, Methods28; license number 3890230800113. Data sets from the previous study are reanalyzed and expanded within this study. Please click here to view a larger version of this figure.
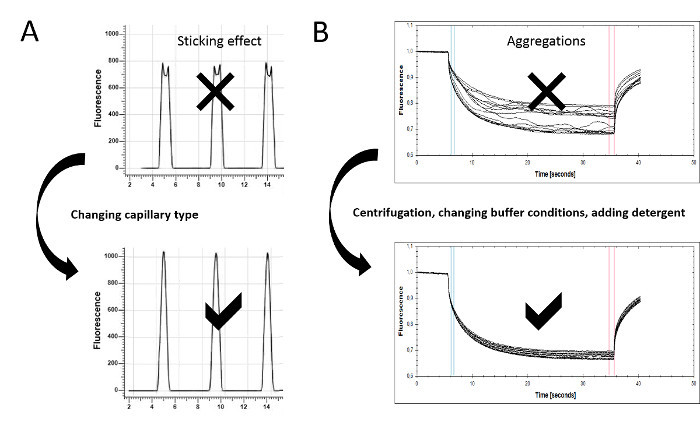 Figure 3:Assay optimization for MST experiments. (A) Protein adsorption and sticking effects can be detected in the capillary scan. Irregular peak shapes, such as flattened or U-shaped peaks, indicate the sticking of the sample material to the glass surface. Changing the capillary type (standard, premium, or hydrophobic) can prevent molecule adsorption, resulting in a regular peak shape. (B) The MST time traces also serve as a quality control, since aggregates can be detected there as bumps and spikes. The experimental conditions can be optimized by improving the solubility of the molecules (e.g., including detergents such as Pluronic F-127 or varying pH values or salt concentrations). Centrifugation may help to remove larger aggregates. To ensure optimal data quality, the MST time traces should resemble the given example. Please click here to view a larger version of this figure.
Figure 3:Assay optimization for MST experiments. (A) Protein adsorption and sticking effects can be detected in the capillary scan. Irregular peak shapes, such as flattened or U-shaped peaks, indicate the sticking of the sample material to the glass surface. Changing the capillary type (standard, premium, or hydrophobic) can prevent molecule adsorption, resulting in a regular peak shape. (B) The MST time traces also serve as a quality control, since aggregates can be detected there as bumps and spikes. The experimental conditions can be optimized by improving the solubility of the molecules (e.g., including detergents such as Pluronic F-127 or varying pH values or salt concentrations). Centrifugation may help to remove larger aggregates. To ensure optimal data quality, the MST time traces should resemble the given example. Please click here to view a larger version of this figure.
Discussion
Quality controls:
Unspecific sticking/adsorption of sample material to surfaces, as well as aggregation effects, have a dramatic influence on the quality of the affinity data. However, only a few state-of-the-art technologies offer accurate and rapid options to monitor and avoid these effects. MST offers integrated quality controls that detect and help to overcome these issues, allowing for the stepwise optimization of the technical setup. Important information on sticking and fluorescence effects can be extracted from the capillary scan and capillary shape (steps 5.2 and 7.5), whereas aggregation/precipitation effects (step 7.7) can be monitored in the movement profiles. These controls enable for the constant improvement of data quality and represent critical steps in the protocol.
Detection of adsorption effects and troubleshooting:
The capillary scan is primarily conducted as an initial step to determine the position of the capillaries on the tray holder by scanning the fluorescence over it. Each peak represents one capillary, indicating the starting point for the MST measurement. Visual inspection of the peak shape enables the determination of the adsorption of molecules to the glass surface, which is an essential quality control that should be performed before starting the MST experiment. The capillary shape overlay allows for the easy detection of flattened, bumpy peak shapes, or even of peaks that resemble a U-form, indicating the adsorption/sticking of molecules to the inner glass surface of the capillary (Figure 3A, upper panel). Differently coated capillary types (standard, premium, and hydrophobic) help to ensure the solubility and minimal adsorption of the molecules to the capillaries. Capillaries should be tested for their suitability prior to the binding experiments. Detergents such as Tween (0.005-0.1%) or Pluronic F127 (0.01-0.1%) may also prevent unspecific adsorption effects. Optimization at this stage of the experiment is essential for high data quality (compare Figure 3A, lower panel).
Detection of fluorescence effects and troubleshooting:
Variations in the peak height of the scanned capillaries offer another layer of information on sample characteristics. Random differences in capillary fluorescence may, on the one hand, be due to pipetting errors or, on the other hand, originate from large sample aggregates in the scanning region that may increase the fluorescence drastically. High pipetting accuracy is mandatory to avoid dilution effects. Mixing all solutions by pipetting up and down increases the accuracy further. In addition, it is essential to keep the buffer consistent in each capillary. Strategies to handle aggregation effects are presented in a later paragraph.
Systemic changes of fluorescence with increasing ligand concentration often indicate ligand dependent quenching/enhancement effects. The "SD Test" is carried out in order to discriminate binding-induced fluorescence changes from unspecific fluorescence loss; this test monitors fluorescence intensities under denaturing conditions. In case of ligand-dependent fluorescence effects, the fluorescence intensities under denaturing condition should be identical, independent of the concentration of titrant. If the difference in fluorescence intensities is still present under these conditions, material was either lost due to unspecific adsorption to tube walls or due to aggregation and subsequent centrifugation.
In order to perform the SD Test, 10 µl of the first and 10 µl of the last ligand dilution step are each carefully transferred to a fresh tube containing 10 µl of a 2x SD Mix (4% SDS and 40 mM DTT). The samples are mixed and the molecules are denatured for 5 min at 95 °C. After filling the samples into capillaries, the fluorescence intensities are measured. If a ligand-dependent fluorescence effect is detected, the data can be analyzed directly by the binding information deduced from the fluorescence intensity changes.
Detection of aggregation effects and troubleshooting:
Bumpy, uneven MST time traces indicate aggregation and/or precipitation effects (Figure 3B, upper panel). Non-aggregated sample material displays a clean and smooth thermophoretic movement profile (Figure 3B, lower panel). The centrifugation of the sample material prior to use (5-10 min at 14,000 x g), the addition of detergents (0.005-0.1% Tween-20, 0.01-0.1% Pluronic F-127, or similar), the use of BSA (>0.5 mg/ml), or the change of buffer conditions (pH and ionic strength) are recommended to minimize the aggregation effects and to optimize the data quality. In order to obtain high-quality data, it is essential to minimize the aggregation effects.
Data analysis and curve fitting:
A thermophoretic movement profile is divided into different phases, which can be inspected either separately or concomitantly. The T-Jump describes the sudden change in fluorescence yield upon temperature change, representing an intrinsic characteristic of fluorophores. Direct binding in the close surroundings of the fluorophore or conformation changes upon binding highly affect the T-Jump. The slower thermophoresis refers to the movement of molecules in the temperature field, offering information on the overall structure of the formed complex. The default type of data analysis employs the "Thermophoresis + T-Jump" setting, exploiting both phenomena to determine the binding parameters. In case the two phases have opposite directions, it is recommended to analyze the phases separately. Please note that time-effects and different species of molecules might lead to differences in the affinity of the thermophoresis and T-Jump. Another type of analysis option is the manual setting, which enables the investigation of specific regions of the movement profile. Disturbances due to aggregation or convection can be excluded in this way. In order to improve data quality, cursors may be set before aggregation signals. Examining the early thermophoresis commonly produces data with less noise, since convection is a time-dependent process. This early manual setting is highly recommended for experiments with high laser power (80%). Upon choosing the data analysis setting, two fitting models integrated in the analysis software can be applied to fit the binding curve. The fit function for Kd from the law of mass action is suited for 1:1 binding modes or for multiple binding sites possessing the same affinity. The concentration-independent dissociation constant Kd describes the equilibrium between the bound and unbound state; thus, the affinity of a binding site to a ligand is measured. The concentration-dependent EC50 value derived from the Hill equation represents the effective dose of a ligand at which half of the labeled molecules are in the bound state. The Hill fit should be applied to multivalent binding models, especially if they are cooperative.
The integrated quality controls represent one great advantage of MST over technologies such as SPR or ITC. These controls allow for the fast and easy optimization of the technical setup to ensure optimal data quality. In addition to the time-efficient measurements (Kd in 15 min) and the immobilization-free setup, MST offers the free choice of buffers, permitting the assessment of molecular interactions, even in lysates and sera5,42,43. Due to the fact that molecular thermophoresis depends on the size, charge, and hydration shell of molecules, there are no limitations in the size of the measured interaction partners during MST measurements. The dynamic affinity range, from pM to mM, together with the low sample consumption, completes the strengths of the MST technology. It is worth mentioning that MST generates precise data on basic binding parameters, such as binding affinity, stoichiometry, and thermodynamics. However, MST does not allow for the measurement of kon and koff rates. In addition, one has to consider that, for most MST experiments, a fluorescent modification must be added to one of the interaction partners. The generated data are in good agreement with state-of-the-art technologies, such as SPR and ITC32,43-46. However, these data are quality-controlled, the experiments are faster, and they consume less material. Overall, MST represents a powerful technology orthogonal to state-of-the-art technologies, with some additional advantages.
Disclosures
C.E. and T.S. are employees of 2bind GmbH, which provides biophysical analytical services. Publication fees for this video-article are paid for by 2bind GmbH.
Acknowledgments
The authors have no acknowledgements.
References
- Linke P, et al. An Automated Microscale Thermophoresis Screening Approach for Fragment-Based Lead Discovery. J Biomol Screen. 2015;21(4):414–421. doi: 10.1177/1087057115618347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerabek-Willemsen M, et al. MicroScale Thermophoresis: Interaction analysis and beyond. Journal of Molecular Structure. 2014;1077:101–113. [Google Scholar]
- Zillner K, et al. Microscale thermophoresis as a sensitive method to quantify protein: nucleic acid interactions in solution. Methods Mol Biol. 2012;815:241–252. doi: 10.1007/978-1-61779-424-7_18. [DOI] [PubMed] [Google Scholar]
- Zhang W, Duhr S, Baaske P, Laue E. Microscale thermophoresis for the assessment of nuclear protein-binding affinities. Methods Mol Biol. 2014;1094:269–276. doi: 10.1007/978-1-62703-706-8_21. [DOI] [PubMed] [Google Scholar]
- Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- Seidel SA, et al. Label-free microscale thermophoresis discriminates sites and affinity of protein-ligand binding. Angew Chem Int Ed Engl. 2012;51(42):10656–10659. doi: 10.1002/anie.201204268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel SA, et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59(3):301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert T, et al. Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin. Mol Cell. 2012;48(3):434–444. doi: 10.1016/j.molcel.2012.08.021. [DOI] [PubMed] [Google Scholar]
- McKeague M, Derosa MC. Challenges and opportunities for small molecule aptamer development. J Nucleic Acids. 2012;2012:748913. doi: 10.1155/2012/748913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscito A, DeRosa MC. Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications. Front Chem. 2016;4:14. doi: 10.3389/fchem.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AL, McKeague M, Liang JC, Smolke CD. Kinetic and equilibrium binding characterization of aptamers to small molecules using a label-free, sensitive, and scalable platform. Anal Chem. 2014;86(7):3273–3278. doi: 10.1021/ac5001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AL, McKeague M, Smolke CD. Facile characterization of aptamer kinetic and equilibrium binding properties using surface plasmon resonance. Methods Enzymol. 2014;549:451–466. doi: 10.1016/B978-0-12-801122-5.00019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing M, Bowser MT. Methods for measuring aptamer-protein equilibria: a review. Anal Chim Acta. 2011;686(1-2):9–18. doi: 10.1016/j.aca.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloski JE, Dombrowski SE, Bevilacqua PC. Thermodynamics of ligand binding to a heterogeneous RNA population in the malachite green aptamer. Biochemistry. 2012;51(1):565–572. doi: 10.1021/bi201642p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnouf D, et al. kinITC: a new method for obtaining joint thermodynamic and kinetic data by isothermal titration calorimetry. J Am Chem Soc. 2012;134(1):559–565. doi: 10.1021/ja209057d. [DOI] [PubMed] [Google Scholar]
- Mannironi C, Scerch C, Fruscoloni P, Tocchini-Valentini GP. Molecular recognition of amino acids by RNA aptamers: the evolution into an L-tyrosine binder of a dopamine-binding RNA motif. RNA. 2000;6(4):520–527. doi: 10.1017/s1355838200991763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263(5152):1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34(2):656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329(5993):845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44(40):13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- Jucker FM, Phillips RM, McCallum SA, Pardi A. Role of a heterogeneous free state in the formation of a specific RNA-theophylline complex. Biochemistry. 2003;42(9):2560–2567. doi: 10.1021/bi027103+. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Lv Q, Wang H. Aptamer fluorescence anisotropy sensors for adenosine triphosphate by comprehensive screening tetramethylrhodamine labeled nucleotides. Biosens Bioelectron. 2015;70:188–193. doi: 10.1016/j.bios.2015.03.031. [DOI] [PubMed] [Google Scholar]
- Zhang D, et al. A sensitive fluorescence anisotropy method for detection of lead (II) ion by a G-quadruplex-inducible DNA aptamer. Anal Chim Acta. 2014;812:161–167. doi: 10.1016/j.aca.2013.12.029. [DOI] [PubMed] [Google Scholar]
- Elenko MP, Szostak JW, van Oijen AM. Single-molecule imaging of an in vitro-evolved RNA aptamer reveals homogeneous ligand binding kinetics. J Am Chem Soc. 2009;131(29):9866–9867. doi: 10.1021/ja901880v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenko MP, Szostak JW, van Oijen AM. Single-molecule binding experiments on long time scales. Rev Sci Instrum. 2010;81(8):083705. doi: 10.1063/1.3473936. [DOI] [PubMed] [Google Scholar]
- Zichel R, Chearwae W, Pandey GS, Golding B, Sauna ZE. Aptamers as a sensitive tool to detect subtle modifications in therapeutic proteins. PLoS One. 2012;7(2):31948. doi: 10.1371/journal.pone.0031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baaske P, Wienken CJ, Reineck P, Duhr S, Braun D. Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew Chem Int Ed Engl. 2010;49(12):2238–2241. doi: 10.1002/anie.200903998. [DOI] [PubMed] [Google Scholar]
- Entzian C, Schubert T. Studying small molecule-aptamer interactions using MicroScale Thermophoresis (MST) Methods. 2016;97:27–34. doi: 10.1016/j.ymeth.2015.08.023. [DOI] [PubMed] [Google Scholar]
- Valenzano S, et al. Screening and Identification of DNA Aptamers to Tyramine Using in Vitro Selection and High-Throughput Sequencing. ACS Comb Sci. 2016;18(6):302–313. doi: 10.1021/acscombsci.5b00163. [DOI] [PubMed] [Google Scholar]
- Jauset Rubio M, et al. beta-Conglutin dual aptamers binding distinct aptatopes. Anal Bioanal Chem. 2016;408(3):875–884. doi: 10.1007/s00216-015-9179-z. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, et al. Aptamer Binding Studies Using MicroScale Thermophoresis. Methods Mol Biol. 2016;1380:99–111. doi: 10.1007/978-1-4939-3197-2_8. [DOI] [PubMed] [Google Scholar]
- Stoltenburg R, Schubert T, Strehlitz B. In vitro Selection and Interaction Studies of a DNA Aptamer Targeting Protein A. PLoS One. 2015;10(7):0134403. doi: 10.1371/journal.pone.0134403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinghorn AB, et al. Aptamer Affinity Maturation by Resampling and Microarray Selection. Anal Chem. 2016;88(14):6981–6985. doi: 10.1021/acs.analchem.6b01635. [DOI] [PubMed] [Google Scholar]
- Duhr S, Braun D. Why molecules move along a temperature gradient. Proc Natl Acad Sci U S A. 2006;103(52):19678–19682. doi: 10.1073/pnas.0603873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun D, Libchaber A. Trapping of DNA by thermophoretic depletion and convection. Phys Rev Lett. 2002;89(18):188103. doi: 10.1103/PhysRevLett.89.188103. [DOI] [PubMed] [Google Scholar]
- Duhr S, Arduini S, Braun D. Thermophoresis of DNA determined by microfluidic fluorescence. Eur Phys J E Soft Matter. 2004;15(3):277–286. doi: 10.1140/epje/i2004-10073-5. [DOI] [PubMed] [Google Scholar]
- Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 2011;9(4):342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Dragnea V, Bauer CE. Adenylate Charge Regulates Sensor Kinase CheS3 To Control Cyst Formation in Rhodospirillum centenum. MBio. 2015;6(3):00546. doi: 10.1128/mBio.00546-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brvar M, et al. Structure-based discovery of substituted 4,5'-bithiazoles as novel DNA gyrase inhibitors. J Med Chem. 2012;55(14):6413–6426. doi: 10.1021/jm300395d. [DOI] [PubMed] [Google Scholar]
- Pogorelcnik B, et al. 4,6-Substituted-1,3,5-triazin-2(1H)-ones as monocyclic catalytic inhibitors of human DNA topoisomerase IIalpha targeting the ATP binding site. Bioorg Med Chem. 2015;23(15):4218–4229. doi: 10.1016/j.bmc.2015.06.049. [DOI] [PubMed] [Google Scholar]
- Jhaveri S, Rajendran M, Ellington AD. In vitro selection of signaling aptamers. Nat Biotechnol. 2000;18(12):1293–1297. doi: 10.1038/82414. [DOI] [PubMed] [Google Scholar]
- Khavrutskii L, et al. Protein purification-free method of binding affinity determination by microscale thermophoresis. J Vis Exp. 2013. [DOI] [PMC free article] [PubMed]
- Ramakrishnan M, et al. Probing cocaine-antibody interactions in buffer and human serum. PLoS One. 2012;7(7):40518. doi: 10.1371/journal.pone.0040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, et al. Antiviral activity and interaction mechanisms study of novel glucopyranoside derivatives. Bioorg Med Chem Lett. 2015;25(18):3840–3844. doi: 10.1016/j.bmcl.2015.07.068. [DOI] [PubMed] [Google Scholar]
- Wan C, et al. Insights into the molecular recognition of the granuphilin C2A domain with PI(4,5)P2. Chem Phys Lipids. 2015;186(4,5):61–67. doi: 10.1016/j.chemphyslip.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Harazi A, et al. The Interaction of UDP-N-Acetylglucosamine 2-Epimerase/N-Acetylmannosamine Kinase (GNE) and Alpha-Actinin 2 Is Altered in GNE Myopathy M743T Mutant. Mol Neurobiol. 2016. [DOI] [PubMed]


