Abstract
Autophagy is a lysosomal degradation pathway essential for cell homeostasis, function and differentiation. Under stress conditions, autophagy is induced and targets various cargos, such as bulk cytosol, damaged organelles and misfolded proteins, for degradation in lysosomes. Resulting nutrient molecules are recycled back to the cytosol for new protein synthesis and ATP production. Upregulation of autophagy has beneficial effects against the pathogenesis of many diseases, and pharmacological and physiological strategies to activate autophagy have been reported. Aerobic exercise is recently identified as an efficient autophagy inducer in multiple organs in mice, including muscle, liver, heart and brain. Here we show procedures to induce autophagy in vivo by either forced treadmill exercise or voluntary wheel running. We also demonstrate microscopic and biochemical methods to quantitatively analyze autophagy levels in mouse tissues, using the marker proteins LC3 and p62 that are transported to and degraded in lysosomes along with autophagosomes.
Keywords: Cellular Biology, Issue 120, autophagy, exercise, LC3, brain, muscle, chloroquine, microscopy, mouse
Introduction
Autophagy is an evolutionarily conserved degradation pathway, which is induced in response to various stress conditions such as starvation and hypoxia1,2. During autophagy, double-membrane vesicles, called autophagosomes, incorporate unnecessary or damaged subcellular components and transport them into lysosomes for degradation3. Basal autophagy is essential for cellular function and organism development, and impaired basal autophagy is implicated in many disorders, including neurodegeneration, tumorigenesis and type 2 diabetes4,5,6.
The best-known physiological autophagy inducer is starvation. However, it has two major limitations. First, starvation takes a long period to effectively induce autophagy in animals, e.g., 48 hr of food restriction in mice in most organs. Second, starvation barely induces brain autophagy, due to a relatively stable nutrient supply in the brain. In fact, it is also difficult to detect autophagy induction by small-molecule inducers, as many drugs cannot pass the blood brain barrier. Thus, to better analyze the function of autophagy activation in disease pathogenesis, we recently discovered that exercise is a more potent physiological method to induce autophagy in a short period of time7,8,9. Compared with starvation, autophagy is effectively induced by treadmill running as fast as 30 min. Thus, exercise is a convenient and potent physiological approach to study the mechanism of autophagy in mediating health benefits and preventing diseases.
There are several protein markers for the detection of autophagy activity, including LC3 and p62. LC3 (microtubule-associated protein 1A/1B-light chain 3) is a cytosolic protein (LC3-I form) that is conjugated to PE (phosphatidylethanolamine) upon autophagy induction. PE-lipidated LC3 (LC3-II form) is recruited onto autophagosomal membranes and can be used to visualize autophagosomes when labeled with GFP. Its translocation from the cytosol to punctate structures of autophagosomes under microscopy is an indication of autophagy induction. p62 is a cargo receptor for autophagy substrates (such as ubiquitination proteins), and is incorporated into autophagosomes as well. Since the protein is degraded in lysosomes along with autophagosomes, its levels can be used to measure the autophagy flux. Here we show how to use these markers to quantify autophagy in different mouse tissues induced by aerobic exercise, including forced exercise (treadmill) and voluntary exercise (running wheels). The same procedures can also be applied to in vivo measurement of autophagy after treatment of other inducers.
Protocol
All procedures involving animals were performed according to guidelines approved by the Northwestern University Institutional Animal Care and Use Committee (IACUC).
1. Mouse Models
Use 8-12 week-old mice in the exercise training. To detect exercised-induced autophagy in vivo, use the GFP-LC3 transgenic mice (C57BL/6 background) for imaging studies and C57BL/6 mice for biochemical analyses.
2. Exercise-induced Autophagy
- Treadmill setup - forced exercise
- Acclimate and train the mice on a 10° uphill open treadmill for 2 days. On day 1, exercise the mice for 5 min at 8 m/min, and on day 2, exercise the mice for 5 min at 8 m/min followed by another 5 min at 10 m/min10. Encourage the mice to run by gentle hand nudge.
- On the 3rd day, let the mice undergo a single bout of running for 90 min on the 10° uphill treadmill, starting at the speed of 10 m/min. After 40 min, increase the treadmill speed at a rate of 1 m/min every 10 min for a total of 30 min, and then increase the speed at the rate of 1 m/min every 5 min until it reaches 17 m/min, for a total of 90 min of exercise time and 1,070 meters of running distance.
- Set the electric stimulus at a low intensity range (1.2 Hz), and encourage the mice to run to avoid repeated foot shock.
- For tissue collection, euthanize the mice by isoflurane inhalation, followed by cervical dislocation immediately after exercise. Other approved euthanasia methods can also be used, such as CO2 or I.P. injection of other anesthetics. The level of autophagy does not seem to be affected by methods of euthanasia.
- Running wheel setup - voluntary exercise
- Place a mouse-running wheel (11.4 cm diameter) inside a cage with a single-housed mouse.
- Verify the running capacity using a bike odometer. Set up the wheel parameter [distance (in mm) per full rotation of the wheel] at 358 (11.4*π). Measure the running distance after 24 hr.
- Let the mouse run voluntarily for 2 weeks with the running wheel. For tissue collection, sacrifice the mouse in the morning after the running period.
3. Autophagy Flux Evaluation
- To measure the autophagic flux under resting and exercise conditions, treat mice with the autophagy inhibitor chloroquine for three days and compare them to mice treated with PBS.
- Dissolve chloroquine in PBS (5 mg/ml), and inject chloroquine into mice intraperitoneally at the dose of 50 mg/kg/day for three consecutive days. Sacrifice mice and collect tissues 3 hr after the last injection.
- For exercised mice, pretreat them with chloroquine for two consecutive days at the dose of 50 mg/kg/day. On the third day, inject the mice with the same concentration of chloroquine, and after 90 min let them run on the treadmill for another 90 min.
- Euthanize the mice by isoflurane inhalation, followed by cervical dislocation. Collect tissues immediately after running, so that the total incubation time of chloroquine is the same as control (resting) mice (3 hr). NOTE: Alternatively, use a single injection of chloroquine to determine the autophagy flux in mouse tissues.
4. Tissue Harvest and Fixation
Prior to tissue harvest prepare phosphate buffered saline (PBS) and fresh 4% paraformaldehyde (PFA, caution: personal protection equipment required) in PBS at a final pH of 7.4. Store both solutions at 4 °C.
Fill 30 ml syringe with 15 - 20 ml of 4% PFA (for GFP-LC3 mice). Connect the syringe with a 20 G catheter needle.
After the exercise, euthanize the mice by isoflurane inhalation, followed by cervical dislocation.
Place the animal on a clean work surface on its back. Cut and open the thoracic cavity by cutting the diaphragm to expose the heart.
Make a small incision on the liver to let the blood come out, and inject a total of 15 ml 4% PFA slowly into the right ventricle of the heart via the catheter, until the lung and liver turn completely pale. Perfuse the mouse immediately after the exercise session, using a syringe pump with a constant flow rate of 90 ml/hr.
After perfusion, remove the needle and collect tissues of interest (e.g. brain11 and skeletal muscle). For skeletal muscle, dissect the vastus lateralis. Briefly, pull the skin of the leg backwards to expose the muscles, localize the quadriceps femoris12, and dissect out vastus lateralis, which is the external muscle attached to the upper portion of the femoral bone.
For further fixation and dehydration, place the tissues of GFP-LC3 mice in 4% PFA for 24 hr at 4 °C, and then transfer them to 15% sucrose-PBS at 4 °C overnight or until the tissues have settled, and then to 30% sucrose-PBS at 4 °C overnight or for longer storage. Keep the samples in the dark as much as possible as they may be sensitive to strong light.
For western blot analysis snap freeze the tissues from C57BL/6 mice directly in liquid nitrogen, and store them at -80 °C.
5. Imaging Analysis of GFP-LC3 Puncta
- Tissue process
- Pre-treat the tissues (previously fixed in PFA and stored in 30% sucrose) in embedding medium such as "Optimal Cutting Temperature compound" (OCT) for a few min in a labeled small petri dish.
- Transfer the tissue in a labeled cryomold containing enough fresh OCT to cover the tissue. Orient the sectioning surface toward the bottom of the cryomold (cross section for the vastus lateralis muscle and sagittal section for the hemi-brain). Avoid formation of bubbles.
- Freeze the samples by placing the cryomold for a few min in a covered foam cooler filled with dry ice, until the OCT turns white. Wrap individual samples in labeled foils, seal them in a plastic bag, and store them at -80 °C overnight or longer.
- Use a cryostat to cut sample sections at a thickness of 10 µm, and mount the sections on the slides. Store the slides at -20 °C, and always keep the slides away from light whenever possible.
- Slide preparation
- Remove slides from the freezer and thaw them at room temperature. Cover the tissue section with a drop of mounting media with DAPI. Place a coverslip of appropriate size on top and avoid bubble formation.
- Use nail polish to seal the edges of the coverslip, allow nail polish to dry in the dark at room temperature, and then store the slides in a light-tight container at 4 °C, or proceed with imaging capture of GFP-LC3 puncta.
- Quantification of GFP - LC3 puncta by fluorescence microscopy
- Perform epifluorescence microscopy, capturing pictures in the same condition (exposure and settings) for all the samples. For GFP, use an emission wavelength of 525 nm; for DAPI, use 490 nm. Acquire at least ten images per sample using a 60X objective for vastus lateralis and the frontal cortex region of the brain.
- Quantify the number of GFP-LC3 puncta in a tissue area of 2,500 µm2 in each image for statistical analysis, blinded for experimental groups. Calculate the average number from 10 images of each sample, using the automatic measurement function of "Object Count". Settings for the vastus lateralis muscle are: Threshold L = 10, H = 200; 2X smooth; 1X clean, and settings for the brain frontal cortex are: Threshold L = 11, H = 200; 2X smooth; 1X clean.
6. Western Blot Analysis on Autophagy Markers
- Tissue process
- Prepare the lysis buffer: 50 mM Tris-HCl; 150 mM NaCl; 1 mM EDTA; 1% Triton X-100; protease inhibitor and phosphatase inhibitor (freshly added).
- Remove the tissue of C57BL/6 mice from -80 °C storage. Cut the muscle tissue into small pieces with a razor blade prior to the following process. Add 800 µl (for hemi-brain) or 500 µl (for vastus lateralis) of cold lysis buffer to the sample. Homogenize the sample at 4 °C with a homogenizer at a medium speed.
- Fully lyse the homogenized mixture for another 30 min to 1 hr at 4 °C with rotation.
- Spin down the homogenate at 12,000 x g for 10 min at 4 °C, discard the pellet, and collect the supernatant in a new tube.
- Autophagy analysis
- Determine the protein concentration of tissue lysates using the BCA Protein assay kit according to the manufacturer's specifications. Normalize each sample to the same concentration.
- Combine the sample with the 2x Laemmli sample buffer at a 1:1 ratio, boil it at 95 °C for 5 - 10 min, and proceed with western blot analysis on autophagy markers13,14, such as p62 (anti-p62 antibody, 1:500 dilution) and LC3 (anti-LC3 antibody, 1:500 dilution). Boil the samples immediately after addition of the sample buffer, as longer incubation on ice may generate multiple degraded bands of LC3.
- Quantify p62 and LC3 bands by densitometry analysis using ImageJ, and normalize the value to the corresponding actin band.
Representative Results
This protocol describes two different methods to induce autophagy in mouse tissues by aerobic exercise: a total of 90 min of forced exercise on a multi-lane treadmill proceeded by two days of acclimation; or two weeks of voluntary exercise on a running wheel used by single-housed mice. In each exercise protocol, we can measure the autophagy flux by fluorescence microscopy and western blot analysis in various organs.
We used a transgenic mouse line expressing GFP-tagged LC3 as a reporter system to monitor autophagy by exercise1. Upon autophagy induction, LC3 translocates from the cytosol to the autophagosome in punctate structures. After sectioning, formation of GFP-LC3 puncta can be directly visualized by fluorescence microscopy (Figure 1A). Alternatively, autophagosome structures can also be immunostained by an LC3 antibody for imaging. Either 90 min of treadmill exercise or 2 weeks of voluntary running increased the number of GFP-LC3 puncta in both skeletal muscle (vastus lateralis) and cerebral cortex, compared to the resting condition (Figure 1B). It should be noted that the frontal cortex region has been the major region in the brain where autophagy is clearly induced by either method so far. Exercise also induced the conversion of LC3 from the cytosolic form (LC3-I) to the lipid-conjugated form (LC3-II), which can be detected by western blot analysis (Figure 1C).
Exercise-induced increment of autophagosomes (represented by LC3-II and GFP-LC3 puncta) is due to an elevated autophagic flux, rather than a block in autophagosome degradation, assessed by the use of inhibitors of lysosomal degradation, such as bafilomycin A1 or chloroquine. Here we measured the degradation of the autophagic cargo receptor p62 as an example. Exercise (90 min of treadmill) caused a higher degradation of p62 in skeletal muscle than the resting condition, which was rescued by injection with chloroquine prior to exercise (Figure 2). The similar results are also observed with voluntary exercise by running wheels. Thus, aerobic exercise by treadmill or running wheels induces autophagy in vivo, measured by the steady-state level of LC3 and the degradation of p62.
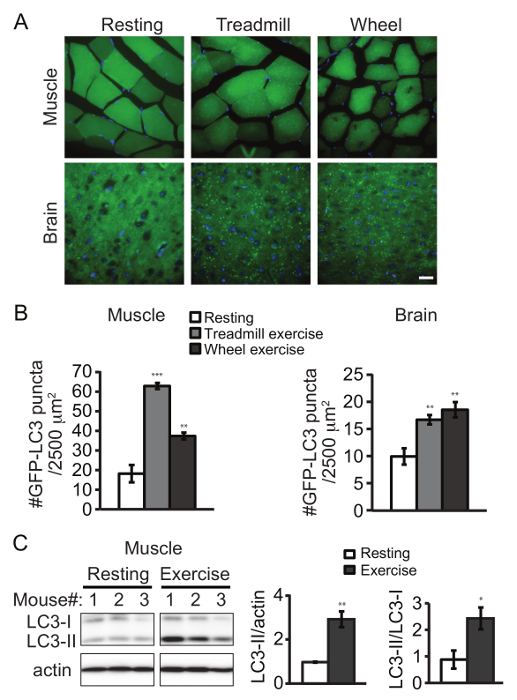 Figure 1. Aerobic Exercise Induces Autophagy in Mouse Tissues. Representative images (A) and quantification (B) of GFP-LC3 puncta in skeletal muscle (vastus lateralis) and brain (frontal cortex) of GFP-LC3 transgenic mice under the control condition (resting), after 90 min of forced exercise (treadmill) or after 2 weeks of voluntary exercise (wheel). Results represent mean ± s.e.m of 10 pictures per mouse. N = 5 mice. The following emission wavelengths were used: GFP - 525 nm; DAPI - 490 nm. (C) Western blot detection of LC3 in skeletal muscle (vastus lateralis) from rested and exercised (by treadmill) mice. Quantification data represent the level of LC3-II normalized to actin (left) and the ratio of LC3-II to LC3-I (right). N = 3 mice. Statistic is comparing each value to the control sample. Results represent mean ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001, t-test. Scale bar, 25 µm. Please click here to view a larger version of this figure.
Figure 1. Aerobic Exercise Induces Autophagy in Mouse Tissues. Representative images (A) and quantification (B) of GFP-LC3 puncta in skeletal muscle (vastus lateralis) and brain (frontal cortex) of GFP-LC3 transgenic mice under the control condition (resting), after 90 min of forced exercise (treadmill) or after 2 weeks of voluntary exercise (wheel). Results represent mean ± s.e.m of 10 pictures per mouse. N = 5 mice. The following emission wavelengths were used: GFP - 525 nm; DAPI - 490 nm. (C) Western blot detection of LC3 in skeletal muscle (vastus lateralis) from rested and exercised (by treadmill) mice. Quantification data represent the level of LC3-II normalized to actin (left) and the ratio of LC3-II to LC3-I (right). N = 3 mice. Statistic is comparing each value to the control sample. Results represent mean ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001, t-test. Scale bar, 25 µm. Please click here to view a larger version of this figure.
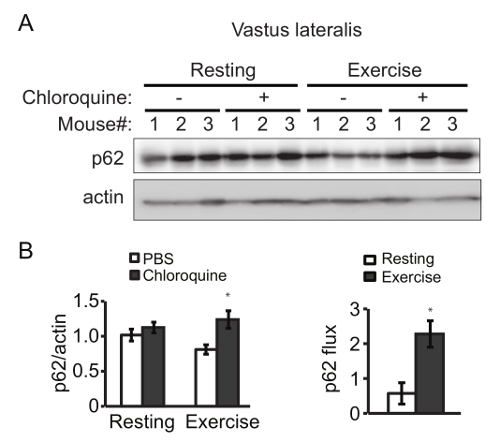 Figure 2. Exercise Increases Autophagy Flux in Mouse Skeletal Muscle. (A) Western blot detection of p62 in vastus lateralis from rested and exercised mice in the presence or absence of the lysosomal inhibitors chloroquine. (B) (Left) Quantification analysis of p62 normalized to the corresponding actin band. (Right) The p62 flux is determined by subtracting the normalized densitometric value of PBS-treated p62 from that of chloroquine-treated p62. Results represent mean ± s.e.m. N = 3 mice. *, P < 0.05, t-test. Please click here to view a larger version of this figure.
Figure 2. Exercise Increases Autophagy Flux in Mouse Skeletal Muscle. (A) Western blot detection of p62 in vastus lateralis from rested and exercised mice in the presence or absence of the lysosomal inhibitors chloroquine. (B) (Left) Quantification analysis of p62 normalized to the corresponding actin band. (Right) The p62 flux is determined by subtracting the normalized densitometric value of PBS-treated p62 from that of chloroquine-treated p62. Results represent mean ± s.e.m. N = 3 mice. *, P < 0.05, t-test. Please click here to view a larger version of this figure.
Discussion
Autophagy is a catabolic process that provides energy and reduces cytotoxicity by lysosomal degradation of cytoplasm components or damaged organelles. Studying autophagy is important to understand the regulation of cellular homeostasis and the mechanisms of stress response. New models and methodologies are emerging in the research field15, to study how impaired autophagy contributes to numerous pathological processes16,17.
Nutrient deprivation (starvation) and pharmacological induction are commonly used approaches to induce autophagy in vitro and in vivo. These methods, especially for animal models, may show adverse effects that can influence the overall results. For example, since it requires at least 48 hr of starvation to induce a detectable level of autophagy, the animals may not have energy needed for regular motor activity, which affects the outcome of many subsequent behavioral studies. Yet pharmacological inducers may also lead to side effects due to their lack of specificity to the autophagy pathway. The major autophagy inducer rapamycin and its derivatives suppress mTOR activity and cause metabolic dysfunction and immunosuppression18,19,20, which should be taken into consideration in the experimental design for long-term treatment. Thus, we have been working on more physiological and robust ways for autophagy activation in animal models.
Recently we and others have identified exercise as an effective, faster and safer autophagy inducer in vivo7,8,9. Both forced exercise by treadmill and voluntary exercise by running wheel have been used to analyze the effects of exercise on autophagy activation, with variations in exercise duration and intensity21,22. For example, an hour of treadmill running (starting at speed of 10 m/min to a maximum of 40 m/min) induces abundant LC3 lipidation in skeletal muscle21, and longer-term voluntary wheel running for 3 months or 4 - 5 weeks also increases basal autophagy and enhances the expression of autophagy proteins in skeletal muscle21,22.
Here we described and compared the two methods (treadmill and running wheel) to exercise mice, and presented optimized shorter protocols with high efficiency in autophagy induction. Each approach has pros and cons. A single bout of 90 min of forced exercise on treadmill is sufficient to induce autophagy in skeletal muscle and brain. We have done a time course of treadmill exercise7, and found that the exercise conditions described here induce the maximal level of autophagy in mouse skeletal muscle, which is also validated by others23. Under these conditions, mice undergo aerobic exercise; as previously reported, aerobic exercise is induced by prolonged running with gradual increases in speeds on a treadmill 24,25,26, whereas the anaerobic condition is obtained by a short duration of high intensity exercise, which increases muscle mass but does not necessarily enhance exercise endurance27,28. Furthermore, the running speed reported here in this protocol positively correlates with oxygen consumption capacity, by indirect methods to assess the aerobic capacity29. Therefore, aerobic exercise on a treadmill is a fast and effective way to induce autophagy.
However, forced running in an enclosed space may exert stress to mice. Thus, to avoid giving additional stress to animals, we use finger nudges or wire tassels as a method to keep mice running, instead of the built-in electric shock. Compared to the treadmill, the use of running wheel has many advantages. It is less stressful, does not require researchers' observation time, and is convenient to study long-term effects of autophagy activation. Yet exercise on a running wheel is voluntary and thus generates variability in terms of running distance and speed among different mice or the same mouse on different days. Therefore, it is necessary to run mice on a wheel for an extended period of time (e.g., several weeks) to minimize the individual variability. Importantly, the approach also requires using an odometer at night to verify that the mouse is actually running. We measured the average running distance of wild-type C57BL/6 mice over a period of 2 weeks, and found that they run approximately 1 km/night at the beginning of training (day 1) and can run up to 8 - 10 km/night at day 14. Overall, the running wheel is an effective method to stimulate autophagy in most organs, although, it is harder to capture autophagosome accumulation in the skeletal muscle compared to using the treadmill. The reason is that skeletal muscle has a high autophagic flux and a fast autophagosome degradation rate, which requires immediate tissue harvesting after running to detect the autophagy induction by GFP-LC3 puncta measurement. In either protocol, rapid PFA perfusion and fixation of the tissues after animal euthanasia is a critical step of the procedure to preserve the autophagosome structures that are otherwise easily degraded during dissection.
This protocol demonstrates how to use aerobic exercise to stimulate autophagy in mouse tissues, including brain and skeletal muscle. These methods can be used to study the mechanisms of the autophagy pathway in the maintenance of tissue function, such as mitochondrial maintenance23, and to study the long-term effects of autophagy induction on the regulation of behavior and health of animal disease models, such as enhancing the analgesic effects of cannabinoids9. Both the treadmill and running wheel methods induce a good level of autophagy; yet it is important to consider their differences when choosing the best approach to meet different research goals.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
We thank the Northwestern University Mouse Histology and Phenotyping Laboratoryfor technical support and assistance, and Noboru Mizushima (University of Tokyo) for providing GFP-LC3 transgenic mice. A. R. and C. H. were supported by the startup funds from Northwestern University and the grant from National Institutes of Health (DK094980).
References
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy K, et al. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Molecular and cellular biology. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Nikoletopoulou V, Papandreou ME, Tavernarakis N. Autophagy in the physiology and pathology of the central nervous system. Cell Death Differ. 2015;22:398–407. doi: 10.1038/cdd.2014.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi A, He C. Emerging roles of autophagy in metabolism and metabolic disorders. Frontiers in biology. 2015;10:154–164. doi: 10.1007/s11515-015-1354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nature reviews. Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Sumpter RJ, Levine B. Exercise induces autophagy in peripheral tissues and in the brain. Autophagy. 2012;8:4. doi: 10.4161/auto.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto K, et al. Autophagy activation by novel inducers prevents BECN2-mediated drug tolerance to cannabinoids. Autophagy. 2016;12:1460–1471. doi: 10.1080/15548627.2016.1187367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JP, Springer DA, Gershengorn MC. The Treadmill Fatigue Test: A Simple, High-throughput Assay of Fatigue-like Behavior for the Mouse. JoVE. 2016. [DOI] [PMC free article] [PubMed]
- Navone SE, et al. Isolation and expansion of human and mouse brain microvascular endothelial cells. Nature protocols. 2013;8:1680–1693. doi: 10.1038/nprot.2013.107. [DOI] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Rando TA. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nature protocols. 2015;10:1612–1624. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslami A, Lujan J. Western blotting: sample preparation to detection. Journal of visualized experiments : JoVE. 2010. [DOI] [PMC free article] [PubMed]
- Bjørkøy G, et al. Chapter 12 Monitoring Autophagic Degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the Pathogenesis of Disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, et al. Duration of rapamycin treatment has differential effects on metabolism in mice. Cell Metab. 2013;17:456–462. doi: 10.1016/j.cmet.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nature reviews. Immunology. 2009;9:324–337. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell. 2014;13:10. doi: 10.1111/acel.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, et al. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI-deficient muscles. Autophagy. 2011;7:1415–1423. doi: 10.4161/auto.7.12.17877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira VA, et al. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:4184–4193. doi: 10.1096/fj.13-228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Verso F, Carnio S, Vainshtein A, Sandri M. Autophagy is not required to sustain exercise and PRKAA1/AMPK activity but is important to prevent mitochondrial damage during physical activity. Autophagy. 2014;10:1883–1894. doi: 10.4161/auto.32154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kregel KC, et al. Resource book for the design of animal exercise protocols. American Physiological Society. 2006. p. 152.
- Lightfoot JT, Turner MJ, Debated KS, Kleeberg SR. Interstrain variation in murine aerobic capacity. Med Sci Sports Exerc. 2001;33:5. doi: 10.1097/00005768-200112000-00012. [DOI] [PubMed] [Google Scholar]
- Rezende EL, Chappell MA, Gomes FR, Malisch JL, Garland T. Maximal metabolic rates during voluntary exercise, forced exercise, and cold exposure in house mice selectively bred for high wheel-running. The Journal of experimental biology. 2005;208:2447–2458. doi: 10.1242/jeb.01631. [DOI] [PubMed] [Google Scholar]
- Kayatekin BM, Gonenc S, Acikgoz O, Uysal N, Dayi A. Effects of sprint exercise on oxidative stress in skeletal muscle and liver. European journal of applied physiology. 2002;87:141–144. doi: 10.1007/s00421-002-0607-3. [DOI] [PubMed] [Google Scholar]
- Kawanaka K, Tabata I, Tanaka A, Higuchi M. Effects of high-intensity intermittent swimming on glucose transport in rat epitrochlearis muscle. J Appl Physiol. 1998;84:4. doi: 10.1152/jappl.1998.84.6.1852. [DOI] [PubMed] [Google Scholar]
- Fernando P, Bonen A, Hoffman-Goetz L. Predicting submaximal oxygen consumption during treadmill running in mice. Can J Physiol Pharmacol. 1993;71:4. doi: 10.1139/y93-128. [DOI] [PubMed] [Google Scholar]


