Abstract
Motivation
Chloroplast genomes are now produced in the hundreds for angiosperm phylogenetics projects, but current methods for annotation, alignment and tree estimation still require some manual intervention reducing throughput and increasing analysis time for large chloroplast systematics projects.
Results
Verdant is a web-based software suite and database built to take advantage a novel annotation program, annoBTD. Using annoBTD, Verdant provides accurate annotation of chloroplast genomes without manual intervention. Subsequent alignment and tree estimation can incorporate newly annotated and publically available plastomes and can accommodate a large number of taxa. Verdant sharply reduces the time required for analysis of assembled chloroplast genomes and removes the need for pipelines and software on personal hardware.
Availability and Implementation
Verdant is available at: http://verdant.iplantcollaborative.org/plastidDB/. It is implemented in PHP, Perl, MySQL, Javascript, HTML and CSS with all major browsers supported.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Chloroplast genomes, or plastomes, are a valuable tool for phylogenetics in angiosperms as they provide a less complex, albeit partial, history relative to nuclear loci. Plastomes are also easily obtainable from low coverage genome sequencing (Soltis et al., 2013; Steele et al., 2012) making them a desirable by-product from multiple sequencing projects. A major hurdle in scalable use of these data is quick and accurate annotation of plastomes and subsequent alignment and phylogenetic estimation. The time and necessary computational resources or skill required to complete these tasks may act as a barrier for novel research in underrepresented flowering plant groups.
Here, we present Verdant, a taxonomically structured, database-driven suite of tools for annotation, alignment and tree estimation of chloroplast genomes in a web-based platform. An exhaustive tutorial is provided, but much of Verdant’s interface is designed to be as intuitive as possible.
Verdant provides a number of key features designed for usability, including:
Automated annotation of both whole and partial plastomes for protein coding genes, tRNAs and rRNAs using our novel software, annoBTD.
Orientation and orthology focused alignments of annotated genes, rRNAs, tRNAs, introns and intergenic regions using MAFFT (Katoh and Standley, 2013).
Phylogeny estimation using RAxML (Stamatakis, 2014).
Annotation visualization using Circos (Krzywinski et al., 2009) and JBrowse (Skinner et al., 2009).
Downloadable datasets of aligned and unaligned plastome regions, both individual gene or concatenated plastome trees, and project metadata including full plastome size, large single copy (LSC), small single copy (SSC), and inverted repeat (IRA or IRB) sizes and locations and total number of annotated features present.
These functions are enabled by an underlying database consisting of high-quality plastomes downloaded from GenBank and newly annotated, secure and user-populated databases for individual projects. Users can then release their data to the public database at their discretion.
2 Implementation
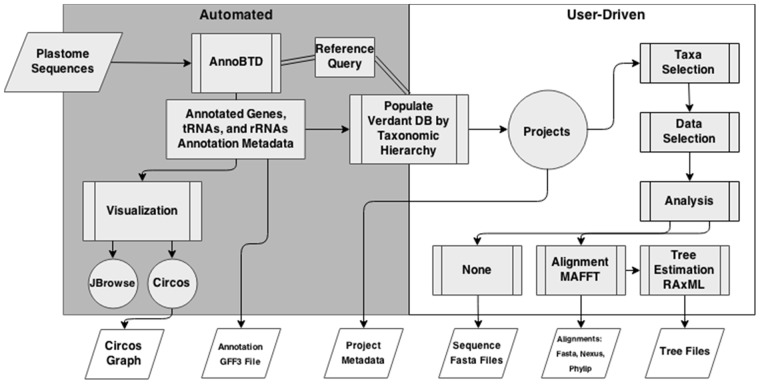
Verdant is broken into two primary workflows (see Fig. 1). The first, which is automatic upon upload, involves the annotation of the plastome sequence(s) using our novel software, annoBTD, the population of the user’s personal and secure data structure, and the creation of Circos and JBrowse visualization for each plastome. The second workflow is completely user-driven and includes project creation, taxon selection, feature selection, alignment and phylogenetic tree reconstruction.
Fig. 1.
Verdant workflow. Workflow diagram depicting the automated (grey box) and user-driven (white box) steps and options available in Verdant. Parallelograms at the bottom of the diagram represent downloadable files available
2.1 Annotation with annoBTD
Our novel annotation software, annoBTD, completely removes the need for manual annotation, although such intervention may occasionally be necessary with some aberrant plastomes. The time for annotation of a full plastome sequence is approximately 10–30 min, and annotations can be downloaded in GFF3 format. AnnoBTD is an alternative to the current standard web-based program DOGMA (Wyman et al., 2004), which is effective and easy to use but requires manual intervention for final and accurate annotations thus limiting throughput.
2.1.1 Protein coding genes
Details of annoBTD are found in Supplementary Information. A novel feature includes de novo ORF identification; ORFs are then identified to reference using the five most closely related species available in the database. Once putative identity is established for an ORF, its position in the plastome informs the final annotation decision. Overlap of two different genes, such as psbD and psbC, is allowed. Start and stop codons for each gene, as well as intron–exon boundaries, are estimated from the sequence by methods that do not require canonical start codons or exact boundary matches. AnnoBTD also finds very small exons that may be missed by other annotation programs.
2.1.2 rRNAs and tRNAs
Because they are conserved in chloroplasts, rRNAs and tRNAs are detected by an optimized blastn and annotated via position and length in the plastome sequence.
2.1.3 LSC, SSC and IR
The LSC, SSC and IR regions of the plastome, if the full sequence is given, are estimated by identifying the repetitive IR sequences and assigning LSC and SSC by size.
2.2 Analyses in Verdant
Verdant’s project management system allows users to create multiple projects adding their own plastome data or publicly available data from the database. Users then choose single genes, ranges of genes, or whole plastomes to include in their analyses. Unaligned or aligned sequences may be downloaded. For alignments, each region of the genome, annotated feature or inter-annotated region, is aligned separately and then concatenated into a single alignment. The MAFFT nucleotide direction option is used to keep all regions properly oriented to each other in order to maintain alignment accuracy over inversion events. In cases where a taxon does not have a specific feature, the region is left as an indel for the taxon in the alignment. Both individual and concatenated alignments are provided to the user for download. Phylogenies are estimated using both individual region alignments and the concatenated alignments with all RAxML files available for download.
3 Conclusion
The annotation and project development features of Verdant provide a high-throughput method for conducting phylogenetic analyses using whole chloroplast sequences, a much needed utility with the glut of plastome data now available. Because of its focus on phylogenetic applications, Verdant complements other tools developed for functional studies of plastome biology. Future additions to Verdant will include more evolutionary analyses to look at plastome structure and function and, ultimately, user created modules.
Supplementary Material
Acknowledgements
Authors thank multiple beta testers during the development of Verdant. We also thank three anonymous reviewers whose comments greatly improved the manuscript.
Funding
This work was supported by the National Science Foundation [DEB 11456884 and DEB-1457748 to E.A.K].
Conflict of Interest: none declared.
References
- Katoh K., Standley D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol., 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. (2009) Circos: an information aesthetic for comparative genomics. Genome Res., 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M. et al. (2009) JBrowse: a next-generation genome browser. Genome Res., 19, 1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E. et al. (2013) The potential of genomics in plant systematics. Taxon, 62, 886–898. [Google Scholar]
- Stamatakis A. (2014) RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele P.R. et al. (2012) Quality and quantity of data recovered from massively parallel sequencing: examples in Asparagales and Poaceae. Am. J. Bot., 99, 330–348. [DOI] [PubMed] [Google Scholar]
- Wyman S.K. et al. (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics, 20, 3252–3255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.