Abstract
Motivation
A genome-scale reconstruction of human metabolism, Recon 2, is available but no interface exists to interactively visualize its content integrated with omics data and simulation results.
Results
We manually drew a comprehensive map, ReconMap 2.0, that is consistent with the content of Recon 2. We present it within a web interface that allows content query, visualization of custom datasets and submission of feedback to manual curators.
Availability and Implementation
ReconMap can be accessed via http://vmh.uni.lu, with network export in a Systems Biology Graphical Notation compliant format released under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License. A Constraint-Based Reconstruction and Analysis (COBRA) Toolbox extension to interact with ReconMap is available via https://github.com/opencobra/cobratoolbox.
1 Introduction
A genome-scale metabolic reconstruction represents the full portfolio of metabolic and transport reactions that can occur in a given organism. A mathematical model can be derived from such a reconstruction, allowing one to simulate of an organism’s phenotypic behaviour under a particular condition (Palsson, 2006). Recon 2 (Thiele et al., 2013) is a very comprehensive knowledge-base of human metabolism and has been applied for numerous biomedical studies, including the mapping and analysis of omics datasets (Aurich and Thiele, 2016). However, despite numerous visualization efforts using automated layouts (Jensen and Papin, 2014), there is no genome-scale and biochemically intuitive human metabolic map available for visualization of omic data in its network context. Here, we release ReconMap, a comprehensive, manually curated map of human metabolism presented utilizing the Google Maps Application Programming Interface (API) for highly responsive interactive navigation within a platform that facilitates queries and custom data visualization.
2 Features
ReconMap content was derived from Recon 2.04, obtained from the Virtual Metabolic Human database (VMH, http://vmh.uni.lu). Reactions (hyperedges) were manually laid out using the biochemical network editor CellDesigner (Funahashi et al., 2008). Each metabolite (node) was designated by its abbreviation and a letter corresponding to the compartment, in which the reaction occurs (e.g. ‘[c]’ for cytosol). Metabolites present in a high number of reactions, e.g. common cofactors, were replicated across the map to minimize hyperedge crossover.
ReconMap is presented using Molecular Interaction NEtwoRk visualization (MINERVA, Gawron et al., 2016), a standalone webservice built on the Google Maps API, that enables low latency web display and interactive navigation of large-scale molecular interaction networks. Each metabolite and reaction in ReconMap links to the corresponding curated content provided by the VMH database. Moreover, MINERVA funtionality connects ReconMap to external databases, such as the CHEMBL database (Bento et al., 2013).
2.1 Overlay of simulation results and multi-omics datasets
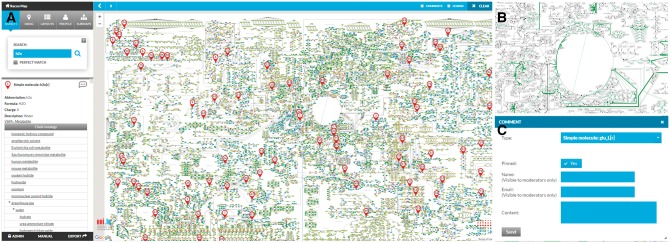
Recon-derived simulation results can be visualized on ReconMap using a new extension to the COBRA Toolbox (Schellenberger et al., 2011). By submitting an account request through the ‘ADMIN’ area of ReconMap, the user can perform a simulation, e.g. Flux Balance Anlalysis, using the COBRA toolbox function ‘optimizeCBmodel’, then call the function ‘buildFluxDistLayout’ to write the input file for a context-specific ReconMap Overlay. This permits the user to translate each flux value into a custom thickness and color within a simple tab-delimited file to highlight certain reactions. Similarly, registered users can display omic data on ReconMap via the ‘Overlay’ menu, by uploading a tab-delimited file assigning a different color and thickness to each node and reaction (Fig. 1).
Fig. 1.
(A) Web interface to ReconMap with search functionality. Information retrieved for a specific molecule are shown, along with external links; (B) overlay of a flux distribution, using differential thickness and color of the edges; (C) Feedback interface that allows users to provide suggestions and corrections to entities of the ReconMap and Recon2
2.2 Community-driven refinement of ReconMap & Recon2
All users may post suggestions for refinement and expansion that are linked to a specific metabolite or reaction in specific locations of the map (right click then select ‘Add comment’). Each suggestion is forwarded to VMH curators for consideration when planning further curation effort. As such, ReconMap enables the community-driven refinement of human metabolic reconstruction and visualization.
2.3 Connecting ReconMap and PDMap
The Parkinson’s disease map (PDMap, Fujita et al., 2016, http://pdmap.uni.lu) displays molecular interactions known to be involved in the pathogenesis of Parkinson’s disease. A total of 168 metabolites connect ReconMap and PDMap via standard identifiers. These connections are available in the metabolites description as well as in their detail pages in the VMH website. This feature is particularly interesting when mapping omics datasets on both maps, thereby allowing the simultaneous investigation of metabolic and non-metabolic pathways relevant for Parkinson’s and other neurodegenerative diseases.
3 Implementation
ReconMap was drawn using CellDesigner, is displayed using the MINERVA platform, built on the Google Map API, using human reconstruction content from the VMH database http://vmh.uni.lu. Matlab scripts for analysis of COBRA Toolbox simulation results using ReconMap are freely available from https://opencobra.github.io/cobratoolbox.
4 Discussion
ReconMap allows for efficient visualization of manually curated human metabolic reactions and metabolites from the VMH database, with numerous connections to complimentary online resources. ReconMap is a generic visualization of human metabolism and serves as a template for generation of cell-, tissue- and organ-specific maps. Moreover, omics data and flux distributions resulting from simulations can be visualized in ReconMap in a network context via an extension to The COBRA Toolbox. ReconMap can be readily connected to disease-specific maps, such as the Parkinson’s disease map, thereby enabling investigations beyond metabolic pathways. Future directions include multiscale visualization, conserved moiety tracing (Haraldsdottir et al., 2016), drug target search and increased synergy with simulation tools.
Funding
This work was supported by the Luxembourg National Research Fund (FNR) through the National Centre of Excellence in Research (NCER) on Parkinson’s disease and the ATTRACT programme (FNR/A12/01), and by the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 668738.
Conflict of Interest: none declared.
References
- Aurich M., Thiele I. (2016) Computational modeling of human metabolism and its application to systems biomedicine. Syst. Med., 253–281. [DOI] [PubMed] [Google Scholar]
- Bento A. et al. (2013) The ChEMBL bioactivity database: an update. Nucleic Acids Res., gkt1031.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K. et al. (2016) Integrating pathways of Parkinson’s disease in a molecular interaction map. Mol. Neurobiol., 49, 253–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi A. et al. (2008) CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc. IEEE, 96, 1254–1265. [Google Scholar]
- Gawron P. et al. (2016) MINERVA – a platform for visualisation and curation of molecular interaction networks. Syst. Biol. Appl, doi:10.1038/npjsba.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraldsdottir H.S. et al. (2016) Identification of conserved moieties in metabolic networks by graph theoretical analysis of atom transition networks. PloS Comput. Biol. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P., Papin J. (2014) MetDraw: automated visualisation of genome-scale metabolic network reconstructions and high-throughput data. Bioinformatics, 30, 1327–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson B. et al. (2006) Systems Biology: Properties of Reconstructed Networks. Cambridge University Press, Cambridge, England. [Google Scholar]
- Schellenberger J. et al. (2011) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2. 0. Nat. Protoc., 6, 1290–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I. et al. (2013) A community-driven global reconstruction of human metabolism. Nat. Biotechnol., 31, 419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]