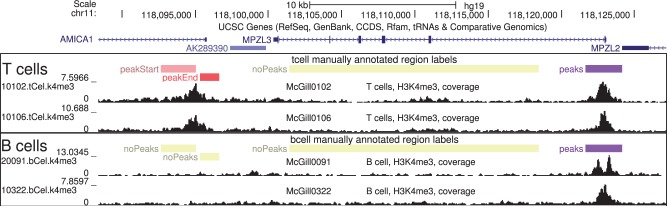
Fig. 1.
Labels indicate some genomic regions with and without peaks. Visual inspection of ChIP-seq normalized coverage plots was used to create labels that encode where peaks should and should not be detected in these H3K4me3 profiles for T cell and B cell samples. Exactly 1 peak start/end should be detected in each peakStart/peakEnd region. There should be no overlapping peaks in each noPeaks region, and at least 1 overlapping peak in each peaks region