Abstract
Summary
Homorepeats are low complexity regions consisting of repetitions of a single amino acid residue. There is no current consensus on the minimum number of residues needed to define a functional homorepeat, nor even if mismatches are allowed. Here we present dAPE, a web server that helps following the evolution of homorepeats based on orthology information, using a sensitive but tunable cutoff to help in the identification of emerging homorepeats.
Availability and Implementation
dAPE can be accessed from http://cbdm-01.zdv.uni-mainz.de/∼munoz/polyx.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Low Complexity (LC) is a general term used to describe regions in protein sequences with little diversity in their amino acid composition, such as tandem repeats and compositionally biased regions (CBR). Homorepeats, or polyX regions, are CBRs composed of runs of a single amino acid residue. There are slight variations in the calculated proportions of human homorepeat-containing proteins, from 5.7% (Jorda and Kajava, 2010) to 7.7% (around 4000 proteins with homorepeats from 51 778 human sequences) (Lobanov and Galzitskaya, 2012). The reason for this diversity is the lack of a standardized cutoff to consider a biased region as a homorepeat, and whether it may contain a mismatch within such region or not. Several cutoffs are described in the literature to define homorepeats: at least five identical residues (Alba and Guigó, 2004), at least six (Lobanov and Galzitskaya, 2012), from at least five to at least seven (Jorda and Kajava, 2010), eight in a window of ten (Schaefer et al., 2012) and four in five (UniProt, http://www.uniprot.org/help/compbias). In fact, cut-offs for homorepeat detection are likely to be dependent not only on amino acid type but also on the species (Lobanov et al., 2016).
Homorepeats have been individually characterized in previous works (Bhattacharyya et al, 2006; Muralidharan and Goldberg, 2013; Salichs et al., 2009; Schaefer et al., 2012), or studied proteome-wise (Lobanov and Galzitskaya, 2012; Lobanov et al., 2016) and stored in databases (Lobanov et al., 2014). Although they have been described as more abundant in eukaryotic proteomes than in prokaryotes (Faux et al, 2005; Jorda and Kajava, 2010), it is not clear how homorepeats evolve. As their development over time has not been yet thoroughly described, it is not known if they increase, decrease or randomly change their length and functions along evolution.
We have tried to tackle these issues by developing dAPE, a web server and database that helps assessing the evolution of homorepeats and their protein context. It uses by default a weak cutoff to help in the identification of emerging or disappearing homorepeats, and allows users to study their own sequences in an easy-to-use setup.
2 Implementation
We used as reference the set of human protein-coding genes from Ensembl (GRCh38.p7, 102147 entries), as well as Ensembl entries from protein coding genes from 13 organisms restricted to proteins with orthologous human genes (Supplementary Table S1). Most of the selected proteomes belong to vertebrates, as homorepeats are more prevalent in them and automatically-obtained orthology information for these organisms is more reliable; orthologous sequences from more distant organisms can also be computed when uploaded as input. We computed for each sequence its homorepeats, using a low cutoff of four identical residues out of six amino acids (4/6). When a homorepeat was found, its neighboring region was extended to ± 30 amino acids to evaluate and report a possible composition bias in the amino acid corresponding to the detected homorepeat in the regions surrounding it. In this extended region, we calculated the maximum number of occurrences of the repeated amino acid using a variable sliding window (length: 4–22). This procedure was followed to generate the dAPE database of homorepeats evolution, and is performed on-the-fly if the user uses as input a set of protein sequences.
3 Application
dAPE is organized both as a database and as a web server. Input to it are either one EnsemblProteinID (or UniProt AC or UniProt ID), already processed in the database, or a set of protein sequences to process from scratch. These sequences must be in FASTA format, and ideally should be part of a cluster of orthologs, to add an evolutionary perspective to the homorepeat detection. The input is entered in a simple form, and each sequence is then processed to show its homorepeats and sequence context (see Implementation). The context of a polyX region may be useful to detect compositionally biased regions surrounding the homorepeat. If sequences are provided, homorepeats are identified and the sequences are aligned using Clustal Omega with default parameters (Sievers et al., 2011). They are also clustered to group similar sequences (threshold of a minimum of 25% identity), using the algorithm FastaHerder2 (Mier and Andrade-Navarro, 2016).
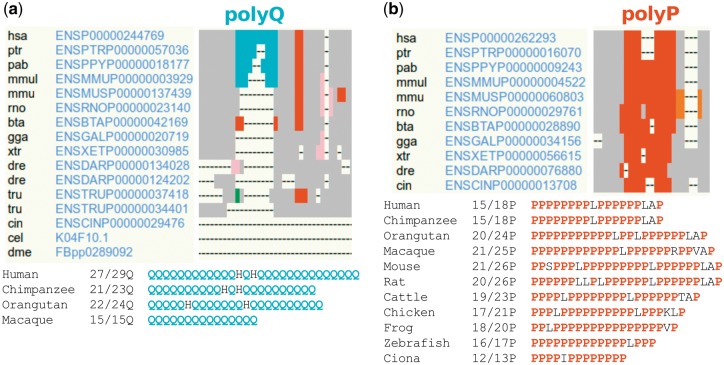
An alignment simplification is displayed, scaled depending on the alignment length, and sorted by clusters (if any). Gaps are represented by dashes, aligned regions in grey and homorepeats are colored according to the repeated amino acid (Fig. 1). One graph per amino acid is created to represent the maximum number of such residue in a variable window size, computed for each homorepeat. This information is also gathered in a table beside each graph, to facilitate the study of the evolution of each individual homorepeat. For example, the distinct appearance and growth in primates of a polyQ in the N-terminal region of Ataxin-1 (Fig. 1a), or the unstable growth and shrink of a polyP in the central part of PRR11 (Fig. 1b). The simplified view of the alignment allows visualizing the variability of length and coexistence of homorepeats within different proteins.
Fig. 1.
dAPE representation of the evolution of two different homorepeats. Gaps are represented by dashes, and aligned regions in grey. Only fragments of the full alignments are shown, for simplicity. Abbreviations of the organisms' names can be found in Supplementary Table S1. (a) Evolution of a polyQ region (in blue, human coordinates: 197-225), using default parameters and ′ENSP00000244769′ (human Ataxin-1) as query. The polyQ run of Ataxin-1 appeared in primates and seems to have grown in length during its evolution. (b) Evolution of a polyP region (in red, human coordinates: 185-202), using default parameters and ′ENSP00000262293′ (human PRR11) as query. The polyP run of PRR11 seems to have appeared before the emergence of tunicates, and displays great length variability
We use a 4/6 homorepeat threshold (see Implementation) as default to facilitate the detection of emerging and disappearing homorepeats. The user can try more restrictive thresholds to visualize only those (computed with the default 4/6 threshold) which are also above the selected cutoff.
dAPE has been developed to change the way homorepeats are studied, from a single protein point of view to an evolutionary one. The simple yet powerful visual alignment simplification shows at a glance the distribution of all the polyX regions in the query and its orthologs, giving information about their context both along evolution and in the sequence.
Multiple sequence alignment of compositionally biased protein sequences is a difficult problem for which, to the best of our knowledge, no standard solution exists. By providing visual means to evaluate the coherence of alignments respect to homorepeat evolution, dAPE facilitates the analysis of compositionally biased proteins and may even help the development of methods to align them.
Supplementary Material
Funding
This work has been supported with funds from the Center for Computational Sciences Mainz (CSM, Johannes Gutenberg University of Mainz, Germany).
Conflict of Interest: none declared.
References
- Alba M.M., Guigó R. (2004) Comparative analysis of amino acid repeats in rodents and humans. Genome Res., 14, 549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A. et al. (2006) Oligoproline effects on polyglutamine conformation and aggregation. J. Mol. Biol., 355, 524–535. [DOI] [PubMed] [Google Scholar]
- Faux N.G. et al. (2005) Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res., 15, 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorda J., Kajava A.V. (2010) Protein homorepeats: sequences, structures, evolution, and functions. Adv. Protein Chem. Struct. Biol., 79, 59–88. [DOI] [PubMed] [Google Scholar]
- Lobanov M.Y., Galzitskaya O.V. (2012) Occurrence of disordered patterns and homorepeats in eukaryotic and bacterial proteomes. Mol. Biosyst., 8, 327–337. [DOI] [PubMed] [Google Scholar]
- Lobanov M.Y. et al. (2014) HRaP: database of occurrence of HomoRepeats and patterns in proteomes. Nucleic Acids Res., 42, D273–D278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov M.Y. et al. (2016) Non-random distribution of homo-repeats: links with biological functions and human diseases. Sci. Rep., 6, 26941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mier P., Andrade-Navarro M. (2016) FastaHerder2: four ways to research protein function and evolution with clustering and clustered databases. J. Comput. Biol., 23, 270–278. [DOI] [PubMed] [Google Scholar]
- Muralidharan V., Goldberg D.E. (2013) Asparagine repeats in Plasmodium falciparum proteins: good for nothing? PLoS Pathog., 9, e1003488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichs E. et al. (2009) Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet., 5, e1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M.H. et al. (2012) Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res., 40, 4273–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F. et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol., 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.