Abstract
Motivation
Pseudotime analyses of single-cell RNA-seq data have become increasingly common. Typically, a latent trajectory corresponding to a biological process of interest—such as differentiation or cell cycle—is discovered. However, relatively little attention has been paid to modelling the differential expression of genes along such trajectories.
Results
We present switchde, a statistical framework and accompanying R package for identifying switch-like differential expression of genes along pseudotemporal trajectories. Our method includes fast model fitting that provides interpretable parameter estimates corresponding to how quickly a gene is up or down regulated as well as where in the trajectory such regulation occurs. It also reports a P-value in favour of rejecting a constant-expression model for switch-like differential expression and optionally models the zero-inflation prevalent in single-cell data.
Availability and Implementation
The R package switchde is available through the Bioconductor project at https://bioconductor.org/packages/switchde.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Single-cell RNA-sequencing (scRNA-seq) has transformed biology by providing high-throughput quantification of mRNA abundance in individual cells allowing, amongst other things, the identification of novel cell types and gene expression heterogeneity (Trapnell, 2015). Single-cell pseudotime estimation (Ji and Ji, 2016; Reid and Wernisch, 2016; Shin et al., 2015; Trapnell et al., 2014) has also enabled gene expression profiles to be mapped to a unique value known as the pseudotime—a surrogate measure of the cellular state in temporally evolving biological process such as differentiation or cell-cycle.
Once a pseudotime has been assigned to each cell it is possible to identify genes that exhibit a strong pseudotemporal dependence through differential expression testing. An approach first introduced in Trapnell et al. (2014) was to regress gene expression on pseudotime using cubic B-spline basis functions with a Tobit likelihood. However, the flexible nonparametric nature of such models may lead to overfitting and may also be difficult to interpret. To our knowledge no other differential-expression-along-pseudotime models have been proposed.
As a solution to these issues we present switchde, a statistical model and accompanying R package for identifying switch-like differential expression analysis along single-cell trajectories. We model sigmoidal expression changes along pseudotime that provides interpretable parameter estimates corresponding to gene regulation strength and timing along with hypothesis testing for differential expression. Our model optionally incorporates zero-inflation for datasets that exhibit high numbers of missing measurements.
2 Materials and methods
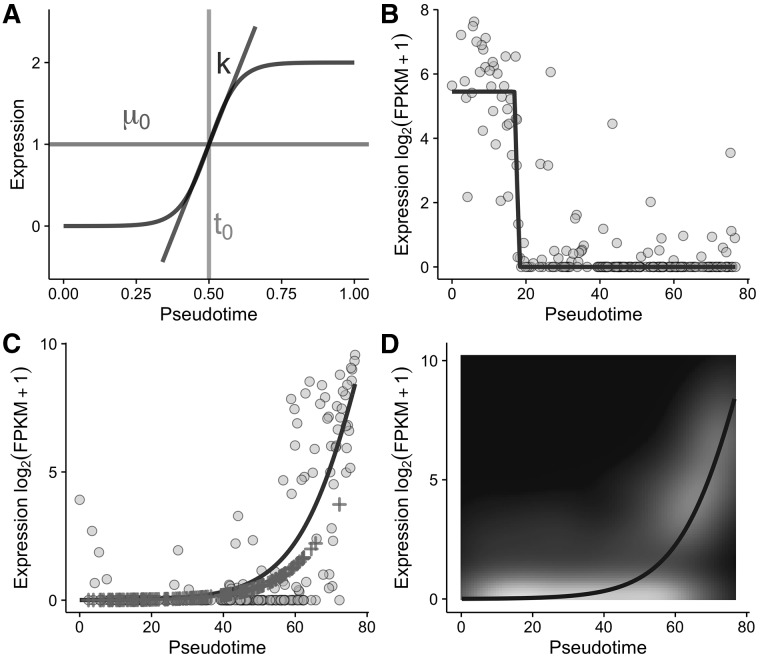
We begin with a C × G expression matrix for G genes and C cells with column vector , that is non-negative and represents gene expression in a form comparable to . We define the sigmoid function as where is the latent pseudotime of cell c. The parameters (Fig. 1A) may be interpreted as the average peak expression level (), the activation strength kg or how quickly a gene is up-or-down regulated and the activation time (), or where in the trajectory the gene regulation occurs.
Fig. 1.
Sigmoidal expression across pseudotime. (A) The sigmoid curve as a model of gene expression along single-cell trajectories, parametrized by the average peak expression μ0, the activation strength k and the activation time t0. (B) An example using the NDC80 gene from the Trapnell dataset (Trapnell et al. (2014)), which had the lowest P-value of all genes tested. Gene expression measurements are shown as the grey points with the maximum likelihood sigmoid fit denoted by the dark line. The maximum likelihood parameter estimates were and . (C) Zero-inflated differential expression for the transcription factor MYOG. Solid line shows the MLE sigmoidal mean while crosses show imputed gene expression measured as zeroes. (D) Posterior predictive density for the zero-inflated model with the solid line denoting MLE sigmoidal mean.
We fit the model using gradient-based L-BFGS-B optimization to find maximum likelihood estimates (MLEs) of the parameters (Supplementary Methods). By setting kg = 0 we identify a nested constant-expression model where and so can perform a likelihood ratio test for differential expression, where twice the difference in the log-likelihood MLE between the constant and sigmoidal models asymptotically follows a distribution with two degrees of freedom.
scRNA-seq data is also known to exhibit a large number of dropouts where the expression measurements of low abundance transcripts are zero (Kharchenko et al. (2014)). This leads to sparse input matrices for downstream analysis which may violate assumptions of statistical models, such as the Gaussian likelihood above. Therefore, we have also developed an extension for datasets with high dropout rates that incorporates a zero-inflated likelihood similar to Pierson and Yau (2015).
3 Results and discussion
We applied switchde to the set of differentiating myoblasts from Trapnell et al. (2014). Using the originally published pseudotimes, we removed cells corresponding to contaminating mesenchymal cells and fitted switch-like models for the 11 253 genes expressed in at least 20% of cells with a mean expression of 0.1 FPKM, which took less than a minute on a laptop computer. 2336 genes were found to be significantly differentially expressed at 5% FDR after Benjamini-Hochberg multiple testing correction. The gene with the lowest reported P-value was NDC80 whose expression is plotted in Figure 1B along with the MLE sigmoid fit. The maximum likelihood parameter estimates were , indicating strong down-regulation and , which given the pseudotimes range from 0 to 77 indicates this down-regulation occurs within the first quarter of the trajectory.
We next applied switchde in zero-inflated mode to a subset of genes from the same dataset. While zero-inflated mode accounts for dropout and is thus a less mis-specified model, the Expectation-Maximization algorithm required for inference takes on average an order of magnitude longer. The resulting fit for the transcription factor MYOG can be seen in Figure 1C. One advantage of the zero-inflated model is that transcripts that exhibit dropout may be imputed given the pseudotemporal trend, shown by the crosses in the figure. Finally, since switchde specifies a fully generative probabilistic model we can generate a posterior predictive distribution of gene expression over pseudotime. This distribution for MYOG is shown in Figure 1D, demonstrating the model is well calibrated with the overall pseudotemporal trend. Further data examples are given in Supplementary Material.
In this paper we have introduced switchde, the first dedicated statistical framework for modelling differential expression over pseudotime. By assuming a parametric model of gene expression along trajectories our model provides interpretable parameter estimates corresponding to gene regulation strength and timing, incorporating zero-inflation that is prevalent in many scRNA-seq datasets. Finally, our model provides hypothesis testing for switch-like differential expression, though in practice this may lead to an inflated false discovery rate due to the assumption that pseudotimes are fixed (Campbell and Yau (2016)).
Supplementary Material
Funding
KRC is supported by a UK Medical Research Council funded doctoral studentship. C.Y. is supported by a UK Medical Research Council New Investigator Research Grant (Ref. No. MR/L001411/1), the Wellcome Trust Core Award Grant Number 090532/Z/09/Z, the John Fell Oxford University Press (OUP) Research Fund and the Li Ka Shing Foundation via a Oxford-Stanford Big Data in Human Health Seed Grant.
Conflict of Interest: none declared.
References
- Campbell K., Yau C. (2016) Order under uncertainty: robust differential expression analysis using probabilistic models for pseudotime inference. PLoS. Comput. Biol.., 12, e1005212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Ji H. (2016) TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic Acids Res., 44, e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P.V. et al. (2014) Bayesian approach to single-cell differential expression analysis. Nat. Methods, 11, 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson E., Yau C. (2015) ZIFA: Dimensionality reduction for zero-inflated single-cell gene expression analysis. Genome Biol., 16, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J.E., Wernisch L. (2016) Pseudotime estimation: deconfounding single cell time series. Bioinformatics, 32, 2973–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. et al. (2015) Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell, 17, 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. (2015) Defining cell types and states with single-cell genomics. Genome Res., 25, 1491–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. (2014) The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol., 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.