Abstract
Motivation
Flow, hyperspectral and mass cytometry are experimental techniques measuring cell marker expressions at the single cell level. The recent increase of the number of markers simultaneously measurable has led to the development of new automatic gating algorithms. Especially, the SPADE algorithm has been proposed as a novel way to identify clusters of cells having similar phenotypes in high-dimensional cytometry data. While SPADE or other cell clustering algorithms are powerful approaches, complementary analysis features are needed to better characterize the identified cell clusters.
Results
We have developed SPADEVizR, an R package designed for the visualization, analysis and integration of cell clustering results. The available statistical methods allow highlighting cell clusters with relevant biological behaviors or integrating them with additional biological variables. Moreover, several visualization methods are available to better characterize the cell clusters, such as volcano plots, streamgraphs, parallel coordinates, heatmaps, or distograms. SPADEVizR can also generate linear, Cox or random forest models to predict biological outcomes, based on the cell cluster abundances. Additionally, SPADEVizR has several features allowing to quantify and to visualize the quality of the cell clustering results. These analysis features are essential to better interpret the behaviors and phenotypes of the identified cell clusters. Importantly, SPADEVizR can handle clustering results from other algorithms than SPADE.
Availability and Implementation
SPADEVizR is distributed under the GPL-3 license and is available at https://github.com/tchitchek-lab/SPADEVizR.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Cytometry is an experimental technique used to characterize cell properties at the single cell level. Flow cytometry is the most common technique and allows measuring simultaneously up to 18 cell markers. Thanks to mass and hyperspectral cytometry techniques, the number of simultaneously measurable cell markers has increased up to 50 (Bendall et al., 2011; Grégori et al., 2014). This increase of measurable cell markers has led to the development of new automatic gating algorithms, such as SPADE (Qiu et al., 2011) or ACCENSE. The aim of these algorithms is to identify group of cells, also named cell clusters, having similar expressions in the whole dataset for selected markers. Cell cluster behaviors are then analyzed in terms of variations of associated cells among the different biological samples and conditions.
The SPADE algorithm, which stands for Spanning-tree Progression Analysis of Density-normalized Events, was developed to identify cell clusters in the context of mass cytometry data. In summary, SPADE is a hierarchical clustering-based algorithm combined to a density-based down-sampling procedure. SPADE results can be mainly summarized by two matrices: the cluster abundance matrix which contains the number of cells associated to each cluster for each sample and the cluster phenotype matrix which contains the marker median expressions for each cluster of each sample.
While SPADE is a powerful approach, the interpretation of the behaviors or phenotypes of the identified cell clusters can be challenging, in particular in the scope of a whole dataset. For instance, SPADE has no methods allowing to highlight cell clusters with a cell abundance statistically different between two biological conditions or associated with an additional biological variable. Moreover, SPADE lacks of visualization methods to deeply characterize the phenotypes of the cell clusters in the whole dataset.
We have developed SPADEVizR, an R package to visualize, analyze and integrate results provided by SPADE. This package extends the original SPADE outputs with techniques such as volcano plots, streamgraphs, parallel coordinates, heatmaps, or distograms. Moreover, several statistical methods allow the identification of clusters with important biological behaviors. SPADEVizR also has features allowing the quantification and the visualization of the quality of clustering results and can be used with results generated by algorithms different from SPADE. We illustrate the capabilities of our package using a mass cytometry dataset (Pejoski et al., 2016), obtained in a macaque vaccination study (Supplementary Fig. S1, and user tutorial).
2 Statistical methods
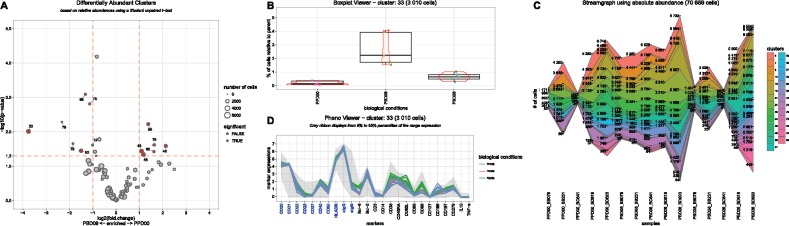
SPADEVizR allows the identification of three types of relevant cell clusters. Abundant Clusters correspond to clusters having a cell abundance statistically greater than a specific threshold for a given set of samples, identified by one sample t-tests. Differentially Abundant Clusters correspond to clusters having a cell abundance statistically different between two biological conditions, identified by two sample t-tests. Correlated Clusters correspond to clusters having a cell abundance correlated with an additional biological variable, identified by Pearson or Spearman correlations. These clusters with important behaviors can be visualized using scatter plot or volcano plot representations (Supplementary Fig. S1A, B and Fig. 1A).
Fig. 1.
Selected visualization representations available in SPADEVizR. (A) Volcano plot showing Differentially Abundant Clusters (DAC) between baseline and 8 days post-boost. DACs are labeled and colored in red. The size of the dots is proportional to the cell cluster sizes. (B) Dotplot and boxplot showing the cell abundances for the cluster 33 in each sample and each condition. A red violon indicates the distribution of cell abundances. (C) Streamgraph showing absolute and relative abundances for a set of clusters across all the samples. (D) Parallel coordinates showing the phenotype of the cluster 33. Grey ribbon represents the marker expression ranges in the entire dataset. More details about this dataset and analysis are provided in Supplementary Figure S1 and in the user manual
Statistical tests can be easily parametrized and corrected for multiple comparisons. Clusters having similar cell abundance profiles can be classified, using various methods such as k-means (user manual and tutorial), and can be visualized using colored circle packing representations (Supplementary Fig. S1C). SPADEVizR can also generate linear, Cox and random forest models to predict biological outcomes, based on the cluster abundances (Supplementary Fig. S2).
3 Visualization methods
Boxplot (Fig. 1B) and kinetic (Supplementary Fig. S1D) representations available in SPADEVizR allows efficient visualizations and comparisons of cluster abundances between different samples and conditions. Moreover, streamgraph representations can display simultaneously absolute and relative cell abundances for a set of clusters (Fig. 1C). While the original SPADE tree representations display data for one single sample, SPADEVizR can display trees based on multiple samples (Supplementary Fig. S1E). Nodes can also be gradient-colored based on marker expression and relevant cell clusters previously identified can be highlighted. The number of cells associated to each cluster for each sample can also be displayed using a dot plot representation to visualize the heterogeneity of cluster sizes (Supplementary Fig. S1F).
In SPADEVizR, phenotypical characterization of the cell clusters can be performed using categorical heatmaps or parallel coordinates (Supplementary Fig. S1G and D). While heatmaps provide global overviews, parallel coordinates provide more details by highlighting the homogeneity of marker expressions between the samples. SPADEVizR can generate multidimensional scaling representations to visualize the similarities between samples or clusters, based on theirs abundance profiles (Supplementary Fig. S1H and I). In such representations, each dot corresponds to a cluster or sample and the distances between the dots are proportional to the Euclidean distances computed based on the cell abundance profiles. Biplot representations can be generated to visualize the co-expressions between two cell markers for one or multiple samples and can be restricted to one or multiple clusters (Supplementary Fig. S1J). Additionally, all pairwise marker co-expressions for selected samples or selected clusters can be visualized using distograms (Supplementary Fig. S1K).
SPADEVizR can generate PDF reports, which gather all these statistical results and visual representations, to make easier the analysis of cell clustering results (user tutorial and user manual). Finally, clusters having uniform phenotypes can be quantified using the Hartigan's Dip test and the interquartile range (IQR). Uniform clusters are defined as clusters having unimodal expression distributions, estimated by the Dip test, and having low spread of expressions, estimated by the IQR, for all their clustering markers. Clusters having low number of associated cells can also be quantified. Reports with heatmaps and expression densities can be generated to visualize the quality of the clustering results (Supplementary Fig. S3).
4 Conclusion
SPADEVizR constitutes a powerful approach for interpreting clustering results from the SPADE algorithm or other automatic gating algorithms. The available statistical and visualization methods are very valuable to analyze high-dimensional cytometry data.
Funding
This work has been supported the grants ‘Investissement d'avenir: Equipements d'Excellence’ – 2010 FlowCyTech (ANR-10-EQPX-02-01) and ‘Infrastructures Nationales en Biologie et Santé’ – 2011 IDMIT (ANR-11-INBS-0008).
Conflict of Interest: none declared.
Supplementary Material
References
- Bendall S.C. et al. (2011) Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science, 332, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grégori G. et al. (2014) Hyperspectral cytometry. Curr. Top. Microbiol. Immunol., 377, 191–210. [DOI] [PubMed] [Google Scholar]
- Pejoski D. et al. (2016) Identification of vaccine-altered circulating B cell phenotypes using mass cytometry and a two-step clustering analysis. J. Immunol., 196, 4814–4831. [DOI] [PubMed] [Google Scholar]
- Qiu P. et al. (2011) Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol., 29, 886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.