SUMMARY
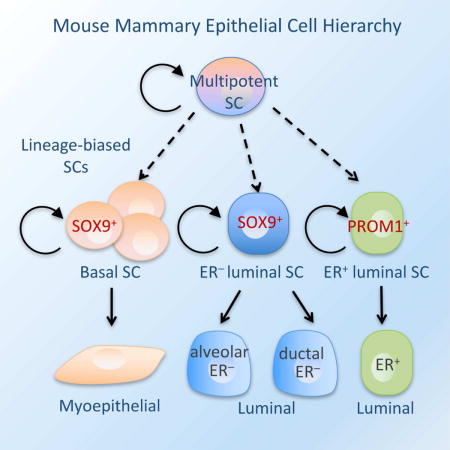
Delineating the mammary differentiation hierarchy is important for studying mammary gland development and tumorigenesis. Mammary luminal cells are considered a major origin of human breast cancers. However, how ER+ and ER− luminal cells are developed and maintained remains poorly understood. The prevailing model suggests that a common stem/progenitor cell generate both cell types. Through genetic lineage tracing in mice, we find that SOX9-expressing cells specifically contribute to the development and maintenance of ER− luminal cells and, to a lesser degree, basal cells. In parallel, PROM1-expressing cells only give rise to ER+ luminal cells. Both SOX9+ and PROM1+ cells specifically sustain their respective lineages even after pregnancy-caused tissue remodeling or serial transplantation, demonstrating characteristic properties of long-term repopulating stem cells. Thus, our data reveal that mouse mammary ER+ and ER− luminal cells are two independent lineages that are maintained by distinct stem cells, providing a revised mammary epithelial cell hierarchy.
eTOC Blurb
Wang et al. discovered two distinct lineage-biased stem cells in the mouse mammary gland: one contributes to the development of estrogen receptor negative luminal cells; the other maintain the development of estrogen receptor positive luminal cells. These findings provide a new framework for studying mammary differentiation and breast cancer etiology.
INTRODUCTION
Mammary gland development and homeostasis involves extensive postnatal growth and tissue remodeling. While the mouse mammary epithelium is specified during embryogenesis, it remains largely quiescent as a rudimentary ductal structure until puberty (Cowin and Wysolmerski, 2010; Watson and Khaled, 2008). During puberty, the ductal rudiment undergoes extensive growth and branching morphogenesis to form a fully developed mammary ductal tree, which then goes through constant turnover during each estrous cycle (Khokha and Werb, 2011; Watson and Khaled, 2008). At pregnancy, the ductal tree massively expands to form milk-secreting alveoli, which are then cleared by apoptosis after lactation through a process called involution. Each mammary gland can sustain repeated rounds of alveologenesis and involution during the reproductive period of the organism. This remarkable tissue remodeling demands robust stem/progenitor activities, and identifying the stem/progenitor cells involved in mammary development and homeostasis is a major focus of the mammary gland field (Makarem et al., 2013; Visvader and Stingl, 2014).
The mammary epithelium is composed of heterogeneous cell types classified into two lineages: basal and luminal. The basal lineage, consisting mostly myoepithelial cells, forms the outer layer of the ducts adjacent to the basement membrane. The luminal lineage includes ductal and alveolar luminal cells, which constitute the inner layer of the ducts and the milk-secreting alveoli, respectively. Luminal cells are also classified by their expression of hormone receptors, particularly estrogen receptor (ER). While ducts contain both ER− and ER+ luminal cells, alveolar luminal cells are mainly ER− (Visvader and Smith, 2011; Visvader and Stingl, 2014). Previous studies of transplanted cell populations have identified multipotent stem cells capable of regenerating the entire mammary ductal tree (Plaks et al., 2013; Shackleton et al., 2006; Sleeman et al., 2006; Spike et al., 2012; Stingl et al., 2006; Zeng and Nusse, 2010). However, subsequent lineage-tracing studies have revealed that basal- or luminal-restricted unipotent stem cells, as well as multipotent stem cells, can all contribute to postnatal mammary gland development and maintenance, suggesting the existence of heterogeneous stem cell populations in the mammary gland (Rios et al., 2014; van Amerongen et al., 2012; Van Keymeulen et al., 2011; Wang et al., 2015).
Despite the substantial progress, the interrelationship of various luminal cell types and the identity of their stem/progenitor cells remains poorly understood (Sreekumar et al., 2015; Visvader and Stingl, 2014). It has been widely believed that a common luminal stem/progenitor cell produces all luminal cell types, including both ER+ and ER− cells (Visvader and Stingl, 2014). This common luminal stem/progenitor cell is thought to be ER−, and the ER+ cells are considered mature cell types, as they lack substantial proliferative potential in vitro (Shehata et al., 2012; Sleeman et al., 2007). However, recent studies found that NOTCH1-expressing progenitors generates ER− but not ER+ luminal cells and that ER+ cells can undergo significant proliferation in vivo (Giraddi et al., 2015; Rodilla et al., 2015). Mathematical modeling of adult mammary cell division kinetics suggests that ER+ and ER− luminal cells may be sustained by progenitors within each population in the resting adult gland (Giraddi et al., 2015). These findings raise questions of the common luminal stem/progenitor model. However, it remains unclear whether the proliferating ER+ cells are long-term repopulating stem cells or only short-term, rapidly dividing progenitors that must be replenished by more primitive stem cells. It is also unclear which luminal stem/progenitor cells produce ER+ cells during mammary ductal tree development and alveologenesis. Thus, long-term in vivo fate mapping studies are required to elucidate the differentiation hierarchy of luminal cells.
We have recently identified SOX9 as a key transcription factor regulating mammary stem/progenitor cell fate (Guo et al., 2012). To further define mammary stem/progenitor cell populations, we carried out lineage tracing for SOX9-expressing cells. Surprisingly, we found that postnatal SOX9+ cells generated basal and ER− luminal cells but not ER+ cells, whereas Prominin 1 (PROM1)-expressing cells specifically maintained ER+ luminal cells during mammary gland development and homeostasis. Both SOX9+ and PROM1+ cells were long-term repopulating and specifically regenerated their respective lineages even after repeated rounds of pregnancy or serial transplantation. These results revealed that ER+ and ER− luminal cells are two independent lineages that are maintained by distinct long-lived stem cells in the postnatal mouse mammary gland.
RESULTS
SOX9+ cells are long-term repopulating and contribute to the ER− luminal and basal lineages in postnatal mammary ducts
To determine the role of SOX9+ cells in mammary gland development and homeostasis, we carried out lineage-tracing experiments that allow faithful assessment of stem and progenitor cell fates in their native tissue microenvironment. For this, we crossed Sox9-CreERT2 mice with R26R-tdTomato reporter mice (Figure 1A) (Kopp et al., 2011; Madisen et al., 2010), and treated the animals with tamoxifen at postnatal day 28 (P28). To minimize any interference with mammary gland development, we opted for a low dose tamoxifen (1.5 mg/mouse) as used in other studies, which allows ~7% of SOX9+ cells to be labelled (Rios et al., 2014; Shehata et al., 2014). We found that ~95% of tdTomato+ cells are SOX9+ 24 hours after the tamoxifen treatment (Figure S1A). The small numbers of tdTomato+/SOX9− cells are likely due to the downregulation of SOX9 expression after the tamoxifen pulse in these cells, as SOX9 is a highly unstable protein (~5-hour half-life) (Luanpitpong et al., 2016). This result confirms the specificity of Sox9-CreERT2 in the mammary gland, consistent with findings in other tissue types (Kopp et al., 2011).
Figure 1. SOX9+ cells are long-term repopulating and maintain the ER− luminal and basal lineages in the postnatal mammary gland.
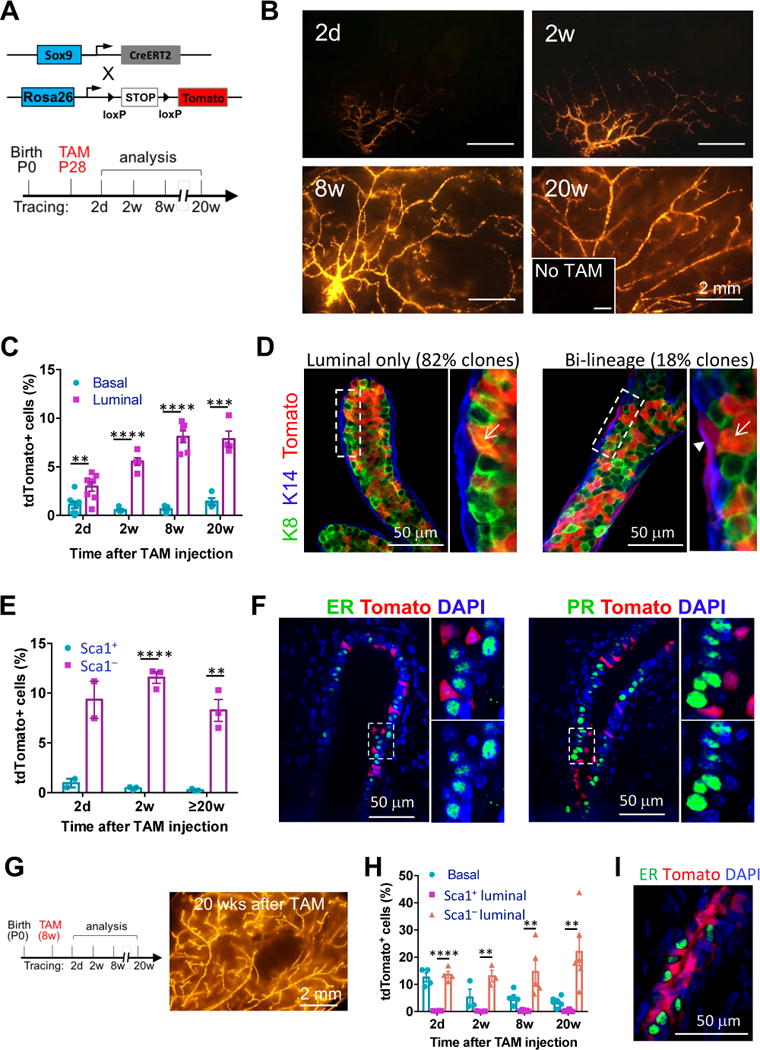
(A) The mouse models (upper) and the experimental schedule (lower) for lineage-tracing experiments shown in Figure 1B to 1F.
(B) Representative tdTomato whole-mount images of Sox9-CreERT2; R26R-tdTomato mammary glands at the indicated time points after tamoxifen treatment. Mice without tamoxifen treatment (No TAM) at 24 weeks of age were used as the negative control (inset). Minimum 3 animals were examined for each time point.
(C) Percentages of tdTomato+ cells in the basal and luminal populations at the indicated time points, as determined by flow cytometry (mean ± SEM, n = 3–8).
(D) K8 and K14 immunostaining of mammary gland sections for characterizing the tdTomato+ cells after 20 weeks of lineage tracing. Representative luminal only and bi-lineage tdTomato+ ducts are shown. The arrow head and arrows point to basal and luminal cells, respectively. Total 57 tdTomato+ cell clusters in 3 mice were counted.
(E) Percentages of tdTomato+ cells in the Sca1+ and Sca1− luminal populations at the indicated time points, as determined by flow cytometry (mean ± SEM, n = 2–3).
(F) ER and PR immunostaining of mammary gland sections 2 weeks after tamoxifen treatment. Magnification of the selected areas is shown on the right of each panel. 98% of tdTomato+ cells were ER− and PR−.
(G) Experimental schedule (left) for adult Sox9-CreERT2 lineage-tracing experiments as shown in Figure 1G to 1I, and a representative tdTomato whole-mount image (right) of Sox9-CreERT2; R26R-tdTomato mammary glands 20 weeks after the tamoxifen treatment.
(H) Percentages of tdTomato+ cells in the basal, Sca1+ and Sca1− luminal populations at indicated time points after the tamoxifen treatment, as determined by flow cytometry (mean ± SEM, n = 3–6).
(I) ER immunostaining of mammary glands after 20 weeks of lineage tracing of adult SOX9+ cells. 99.5% tdTomato+ cells were ER−.
*P < 0.05, **P < 0.01, ***P<0.001, and ****P < 0.0001.
See also Figure S1.
We then carried out long-term lineage tracing of the labeled SOX9+ cells in nulliparous mice (Figure 1A). These cells robustly contributed to pubertal mammary gland development (Figure 1B). The contribution of tdTomato+ cells to mammary ducts was maintained at high levels even after 20 weeks of lineage tracing (Figure 1B). In contrast, mice without tamoxifen treatment displayed no tdTomato signal in the mammary gland (Figure 1B).
We further assessed the contribution of SOX9+ cells to various mammary lineages by flow cytometry using established cell surface markers (Figure S1B) (Shehata et al., 2012). Two days after tamoxifen treatment, both luminal (~3%) and basal (~1%) cells were labeled by tdTomato (Figure 1C). During lineage tracing, the frequency of tdTomato+ luminal cells increased substantially to ~8% in adult mice, whereas the frequency of tdTomato+ basal cells remained steady (Figure 1C). Because the absolute numbers of both luminal and basal cells increase dramatically from puberty to adult (Giraddi et al., 2015), this suggests that SOX9+ cells in both lineages are highly proliferative, although they preferentially contribute to the luminal lineage. Consistent with this notion, immunostaining revealed that 80% of tdTomato+ cell clusters contained only keratin 8 positive (K8+) luminal cells after 20 weeks of lineage tracing (Figures 1D and S1C), whereas the remaining 20% clusters contained both luminal and basal cells (Figure 1D and S1C). These bipotent clusters could be generated by a single bipotent stem/progenitor cell or by adjacent lineage-committed stem/progenitor cells. To distinguish these two scenarios, we reduced the tamoxifen dose to 25 μg/mouse to label SOX9+ cells at clonal density (Figure S1D). In this condition, we observed only luminal- or basal-specific clones, but virtually no bipotent clones (Figure S1D). This indicates that most SOX9+ cells are committed to either the luminal or basal fate, and bipotent SOX9+ cells are rare.
We then asked whether SOX9+ cells give rise to both ER− and ER+ luminal subpopulations using previously defined Sca1 and CD49b markers (Figure S1B) (Shehata et al., 2012). Surprisingly, SOX9+ cells predominantly contributed to the Sca1−CD49b+ population (mostly ER− cells) but not the Sca1+ population (mostly ER+ cells) (Figure 1E). We also confirmed that tdTomato+ cells did not express ER or progesterone receptor (PR) by immunostaining (Figure 1F).
We then examined whether the ER− luminal lineage is already separated from the ER+ cells before the onset of puberty. For this, we started lineage tracing at postnatal day 14 (Figure S1E). The prepubescent SOX9+ cells robustly contributed to mammary gland development and homeostasis, even after 25 weeks of lineage tracing (Figure S1E). Similar to cells labeled during puberty, the prepubescent cells were biased toward the luminal fate, and specifically gave rise to ER− luminal cells (Figures S1F and S1G). We also assessed the fate of adult SOX9+ cells by administering tamoxifen at 8 weeks of age. These cells were also long-term repopulating (Figure 1G) and gave rise virtually only to ER− luminal and basal cells but not ER+ cells (Figures 1H and 1I). Together, these results demonstrate that mouse ductal ER− luminal cells are developed and maintained independently from ER+ luminal cells by SOX9+ long-term repopulating cells. Importantly, SOX9 is similarly expressed in ER− luminal and basal cells in human breast as in different mouse strains, suggesting a conserved role of SOX9 between human and mouse (Figure S1H).
SOX9+ cells contribute to alveologenesis
During pregnancy, the mammary gland undergoes massive expansion to generate milk-secreting alveoli, which are mainly composed of ER− luminal cells and a layer of sparse basal cells. Whether ductal and lobular structures are generated by common or distinct precursor cells is still under debate (Visvader and Smith, 2011; Visvader and Stingl, 2014). Therefore, we examined whether SOX9+ cells also contribute to the development of alveoli (Figure 2A). Indeed, the labeled SOX9+ cells efficiently contributed to alveologenesis (Figure 2B). Like in nulliparous glands, these cells preferentially contributed to the luminal lineage in pregnant glands (Figure 2C). Interestingly, nearly all tdTomato+ alveoli had only either labeled luminal or labeled basal cells, and rarely did a single alveolus contain both labeled luminal and basal cells (Figure 2D and S2A). Furthermore, these labeled cells were virtually all ER− (Figure 2E). This supports the notion that two distinct types of SOX9+ cells are involved in postnatal mammary gland development, one contributing to ER− luminal and the other to basal cells. This also suggests that basal and luminal cells within each alveolus are derived from different stem/progenitor cells.
Figure 2. SOX9+ cells contribute to alveologenesis and persist after repeated pregnancy.
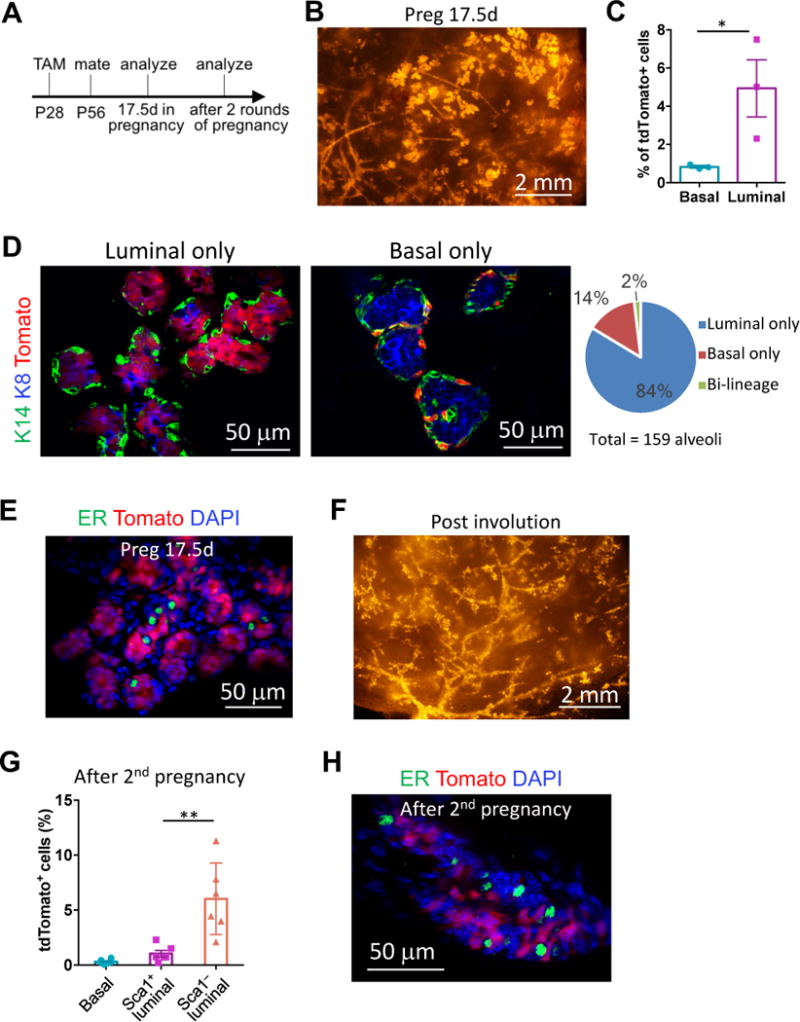
(A) The experimental schedule for lineage-tracing studies shown in Figure 2.
(B) A representative tdTomato whole-mount image of Sox9-CreERT2; R26R-tdTomato mammary glands on day 17.5 of pregnancy (Preg17.5d).
(C) Percentages of tdTomato+ cells in the basal and luminal populations of Preg17.5d mice, as determined by flow cytometry (mean ± SEM, n = 3, paired t-test).
(D) K8 and K14 immunostaining of Preg17.5d mammary gland sections for characterizing the tdTomato+ cells. Examples of alveoli with either tdTomato-labeled luminal or basal cells were shown. The graph on the right shows percentage of alveoli containing the indicated types of tdTomato+ cells.
(E) ER immunostaining of Preg17.5d mammary gland sections. 99% tdTomato+ cells were ER−.
(F) A representative tdTomato whole-mount image of Sox9-CreERT2; R26R-tdTomato mammary glands at least 3 weeks after weaning of the second litter.
(G) Percentage of tdTomato+ cells in the basal, Sca1+ and Sca1− luminal populations 3 weeks after weaning of the second litter (mean ± SEM, n = 6).
(H) ER immunostaining of mammary glands after the 2nd pregnancy. 99.5% of tdTomato+ cells were ER−.
*P < 0.05 and **P < 0.01.
See also Figure S2.
We further assessed the long-term repopulating ability of SOX9+ cells by subjecting the animals to repeated pregnancy and involution, which causes massive turnover of the mammary epithelium. We found that the mammary glands, including both ducts and the residual alveoli, remained robustly populated by tdTomato+ cells even after at least two rounds of pregnancy (Figure 2F). Furthermore, these cells preferentially committed to the ER− luminal fate (Figures 2G, 2H and S2B). Interestingly, although SOX9+ cells generated a small number of Sca1+ cells (Figure 2G and S2B), these cells remained as ER− (Figure 2H), consistent with the previous report that Sca1 expression becomes somewhat promiscuous after pregnancy (Shehata et al., 2012).
PROM1+ cells specifically maintain the ER+ lineage
Our unexpected finding that SOX9+ cells repopulate ER− cells but not ER+ cells questions the prevailing model that ER− and ER+ luminal cells are sustained by a common stem or progenitor cell population. Thus, we sought to identify the cells that maintain the ER+ lineage. We first evaluated the expression patterns of reported ER+ cell markers in the mammary gland, in order to identify a specific Cre driver for ER+ cells. Although Sca1 is an effective marker for separating ER+ and ER− luminal cells (Shehata et al., 2012), it is also expressed in significant numbers of basal and stromal cells (Figure S3A). However, we confirmed that another ER+ cell marker PROM1 (Prominin-1, also known as CD133) was exclusively expressed in ER+ luminal cells in the mammary gland of different mouse strains (Figures S3A and S3B) (Sleeman et al., 2007). Thus, we crossed Prom1-CreERT2 knock-in mice with R26R-tdTomato mice to lineage trace ER+ cells (Figure 3A) (Zhu et al., 2009). We verified that 48 hours post tamoxifen treatment in P28 Prom1-CreERT2; R26R-tdTomato mice, only PROM1+ cells were labeled with tdTomato (Figure S3C).
Figure 3. PROM1+ cells are long-term populating and specifically maintain the ER+ lineage.
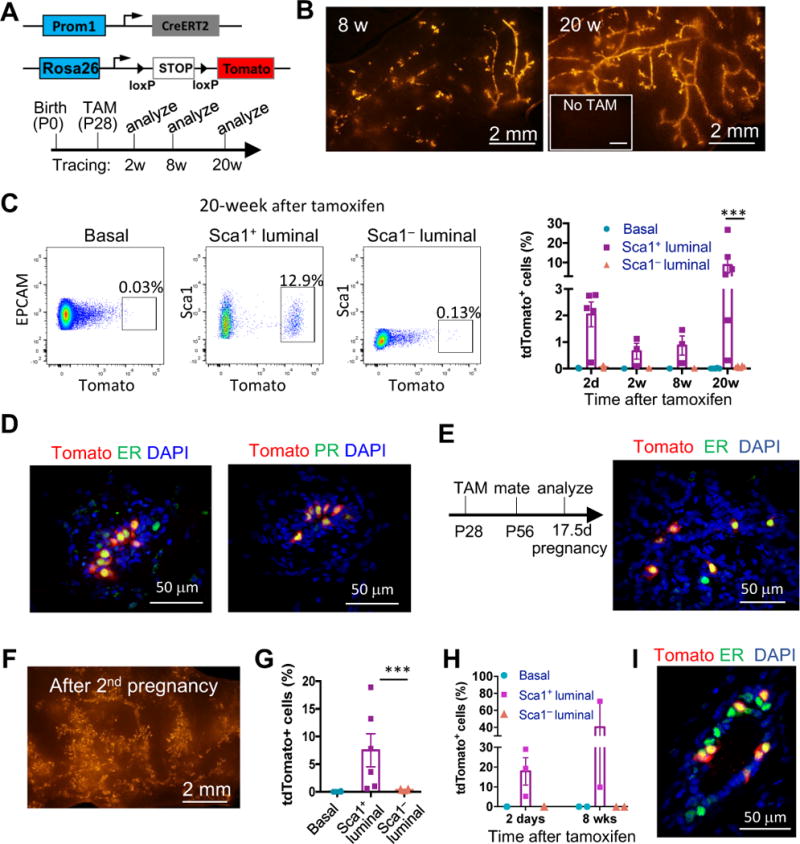
(A) The mouse models (upper) and the experimental schedule (lower) for PROM1+ cell lineage-tracing experiments as shown in Figure 3A–3D.
(B) Representative tdTomato whole-mount images of Prom1-CreERT2; R26R-tdTomato mammary glands at the indicated time points after tamoxifen treatment. Mice without tamoxifen treatment (No TAM) at 24 weeks of age were used as the negative control. Minimum 3 animals were examined for each time point.
(C) Representative flow cytometric profiles of the basal, Sca1+ and Sca1− luminal populations showing percentages of tdTomato+ cells after 20 weeks of lineage tracing. Percentages of tdTomato+ cells in specific cell populations are quantified on the right (mean ± SEM, n = 3–6, paired t-test).
(D) ER and PR immunostaining of mammary gland sections after 20 weeks of lineage tracing. 97% and 100% of tdTomato+ cells are ER+ and PR+, respectively.
(E) The experimental schedule for lineage-tracing studies of Prom1-CreERT2; R26R-tdTomato mice during pregnancy (left), and ER immunostaining of mammary gland sections at Preg17.5d (right). All tdTomato+ cells were ER+.
(F) A representative tdTomato whole-mount image of Prom1-CreERT2; R26R-tdTomato mammary glands 3 weeks after 2 rounds of pregnancy and lactation.
(G) Percentages of tdTomato+ cells in the indicated mammary cell types 3 weeks after 2 rounds of pregnancy and lactation, as determined by flow cytometry (mean ± SEM, n = 6, paired t-test).
(H) Percentages of tdTomato+ cells in the indicated mammary cell types at the indicated time points of adult PROM1+ cell lineage tracing (n = 2–3).
(I) ER immunostaining of mammary gland sections after 8 weeks of adult PROM1+ cell lineage tracing. 96% tdTomato+ cells were ER+.
***P<0.001.
See also Figure S3.
We then carried out long-term lineage tracing of PROM1+ cells starting from early puberty (Figure 3A). The labeled cells efficiently contributed to mammary ductal tree development and maintenance, even after 20 weeks of lineage tracing (Figure 3B). Furthermore, these cells specifically gave rise to Sca1+ and ER+PR+ luminal cells (Figure 3C and 3D). Interestingly, although only a small fraction (2%) of the Sca1+ cells were labeled by tdTomato initially, the frequency of tdTomato+ cells increased substantially after long-term tracing to average 9% at 20 weeks, suggesting that labeled PROM1+ cells have a high proliferative potential (Figure 3C).
We further examined the fate of PROM1+ cells during and after pregnancy. Although a relatively small percentage of cells are ER+ in pregnant glands due to expansion of ER− alveolar cells, all cells derived from the labeled PROM1+ cells were ER+ (Figure 3E). Furthermore, after two rounds of pregnancy, these cells still robustly contributed to the mammary ductal tree and remained as Sca1+ (Figure 3F and 3G). We also examined the fate of adult PROM1+ cells by initiating lineage tracing in 7 weeks old mice. These cells also specifically gave rise to ER+ cells (Figure 3H and 3I). Thus, our data demonstrates that the ER+ luminal lineage is indeed an independent lineage maintained by PROM1+ cells.
ER− and ER+ luminal cells are regenerated independently by distinct stem cells during serial transplantation
The ability to regenerate in serial transplantation has been considered a cardinal feature of long-term repopulating stem cells (Eaves, 2015; Visvader and Clevers, 2016). Therefore, we asked whether SOX9+ and PROM1+ cells can contribute to serial mammary gland regeneration and maintain their lineage commitment. Previously studies have shown that purified luminal cells, when transplanted by themselves, cannot regenerate mammary ductal trees (Shackleton et al., 2006; Stingl et al., 2006). However, when mixed with basal cells, they can contribute to the luminal lineage in the mammary outgrowths (Van Keymeulen et al., 2011). This suggests that for unipotent stem cells to regenerate complex epithelial tissue, they need to act in concert with stem cells of complementary lineages. We reasoned that the proper ratio and spatial allocation of distinct stem cells can be best maintained by transplanting small fragments of mammary ducts, therefore allowing unipotent stem cells to contribute to tissue regeneration.
To fate map SOX9+ cells during transplantation, we used fragments from Sox9-CreERT2; R26R-tdTomato mice that had been pre-treated with tamoxifen (see Figure 4A for the schematic diagram). In these fragments, a subset of SOX9+ cells and their progeny were labeled with tdTomato. Eight weeks after transplantation, we observed that the tdTomato+ cells robustly contributed to the regenerated mammary ductal trees (Figure 4B). Importantly, these cells specifically gave rise to ER− luminal cells (Figure 4C). To further test their long-term repopulating ability, we performed secondary transplantation using fragments of the primary outgrowths. Similarly, the progeny of labeled SOX9+ cells efficiently contributed to the outgrowths of secondary transplantation (Figure 4B) and remained committed to the ER− luminal cell fate (Figure 4D and S4A).
Figure 4. ER− and ER+ luminal lineages are regenerated by different unipotent stem cells during serial mammary gland transplantation.
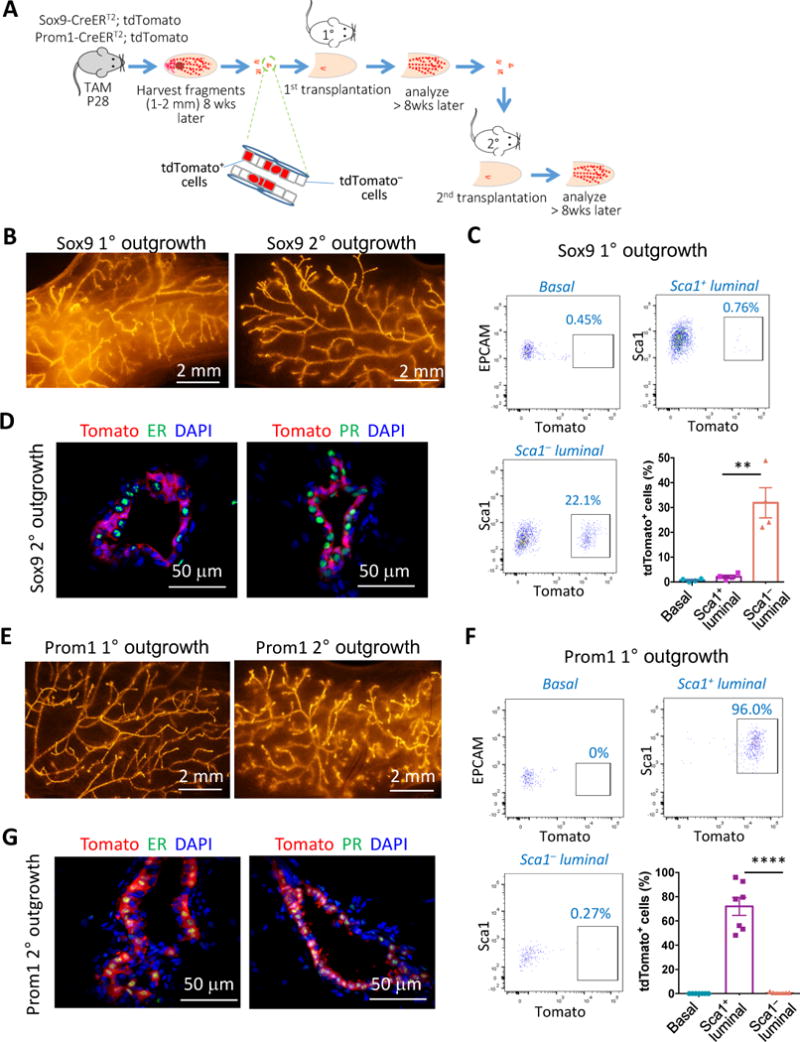
(A) A schematic diagram of the serial transplantation experiments. Sox9-CreERT2; R26R-tdTomato or Prom1-CreERT2; R26R-tdTomato mice were treated with tamoxifen at P28. Small mammary ductal fragments were isolated ≥8 weeks later and used for cleared mammary fat pad transplantation. The fragments of primary outgrowths were then used for secondary transplantation.
(B) Representative tdTomato whole-mount images of the primary and secondary outgrowths regenerated by Sox9-CreERT2; R26R-tdTomato mammary ductal fragments.
(C) Representative flow cytometric profiles and quantification of percentages of tdTomato+ cells in the indicated cell types of Sox9-CreERT2; R26R-tdTomato primary outgrowths (mean ± SEM, n = 4).
(D) ER and PR immunostaining of the secondary outgrowths from Sox9-CreERT2; R26R-tdTomato ductal fragments. 97% and 99% of tdTomato+ cells were ER− and PR−, respectively.
(E) Representative tdTomato whole-mount images of the primary and secondary outgrowths derived from Prom1-CreERT2; R26R-tdTomato mammary ductal fragments.
(F) Representative flow cytometric profiles and quantification of frequency of tdTomato+ cells in the indicated cell types of Prom1-CreERT2; R26R-tdTomato primary outgrowths (mean ± SEM, n = 7).
(G) ER and PR immunostaining of the secondary outgrowths from Prom1-CreERT2; R26R-tdTomato ductal fragments. 97% and 98% of tdTomato+ cells were ER+ and PR+, respectively. **P < 0.01 and ****P < 0.0001.
See also Figure S4.
We next investigated whether PROM1+ cells can regenerate ER+ cells during serial transplantation using similar approaches (Figure 4A). As expected, PROM1+ cells efficiently contributed to the mammary outgrowths of serial transplantation (Figure 4E). And in parallel to the SOX9+ cells, they only gave rise to ER+ cells in both primary and secondary outgrowths (Figures 4F, 4G and S4B). Of note, in some outgrowths, nearly all ER+ cells were derived from the labeled PROM1+ cells, suggesting that ER+ cells can be almost entirely regenerated by PROM1+ cells (Figure 4F and 4G). Together, these results strongly support the conclusion that SOX9+ and PROM1+ cells are long-term repopulating stem cells for the ER− and ER+ luminal cell lineages, respectively.
DISCUSSION
Although much progress has been made in elucidating the mammary epithelial cell hierarchy, how various luminal cell types are developed remains an unanswered question in the mammary gland field (Makarem et al., 2013; Sreekumar et al., 2015; Visvader and Stingl, 2014). It has been widely assumed that a common stem/progenitor cell population generates both ER+ and ER− luminal cells, although direct experimental evidence for this is still lacking. By combining long-term lineage tracing and serial transplantation approaches in this study, we provide clear evidence demonstrating that postnatal ER+ and ER− luminal cells are instead two independent lineages that are maintained by distinct long-lived stem cells. We showed that SOX9+ cells repopulate ER− luminal and basal cells but not ER+ cells, whereas PROM1+ cells specifically repopulate ER+ luminal cells during various stages of mammary gland development and regeneration. Together with recent studies (Giraddi et al., 2015; Rodilla et al., 2015), our findings provide an important revision of the mammary epithelial differentiation hierarchy (Figure S4C).
We used rigorous assays to distinguish long-term repopulating stem cells versus short-term progenitors, an important distinction which have not been resolved in the previous studies (Giraddi et al., 2015; Rodilla et al., 2015). It has been shown that certain progenitors can sustain mammary gland growth for an extended period of time (e.g. 8 weeks) and are only depleted after long-term lineage tracing (e.g. 20 weeks) (Rios et al., 2014). Thus, to determine stem cell capability, we carried out lineage tracing for at least 20 weeks and subjected the mice to multiple rounds of pregnancy. Furthermore, we tested the ability of these cells to regenerate specific lineages following serial transplantation as another rigorous criterion of stemness. Under all these stringent conditions, SOX9+ and PROM1+ cells remain long-term repopulating and robustly contribute to their respective lineages. Thus, our findings provide key experimental evidence supporting the conclusion that ER+ and ER− luminal cells are maintained by separate lineage-biased stem cells.
Our data suggests that SOX9+ cells contribute to either ER− luminal cells or basal cells, and rarely did a single SOX9+ cell contribute to both lineages. Based on the recent report that basal and luminal cells are mainly self-maintaining and there is little “flux” of cells between these two compartments (Wuidart et al., 2016), we think there are two types of SOX9+ cells, one in the luminal and the other in the basal compartment (Figure S4C). In addition to SOX9+ cells, other basal stem cells, such as LGR5+ and AXIN2+ cells, are likely to contribute to the basal lineage as well (de Visser et al., 2012; Koren et al., 2015; Plaks et al., 2013; van Amerongen et al., 2012). The exact relationship of these cells remains to be defined in future. Interestingly, the luminal-committed SOX9+ cells are similar to recently characterized NOTCH1+ cells that are restricted to the ER− luminal cell fate in the postnatal mammary gland (Rodilla et al., 2015). It is likely that these SOX9+ and NOTCH1+ cells largely overlap, as SOX9 is a key downstream target of NOTCH (Shih et al., 2012; Yanger et al., 2013). Of note, our previous work showed that SOX9 is a key regulator in transplantable multipotent stem cells (Guo et al., 2012). The limited multipotent repopulating activity of SOX9+ cells in lineage tracing is likely due to the difference of cell fates in transplantation versus endogenous gland development, as shown in recent studies (van Amerongen et al., 2012; Van Keymeulen et al., 2011).
The finding that ER+ cells are capable of self-maintenance is unexpected, as the prevailing view considers these cells as mature cells derived from ER− common luminal stem/progenitor cells (Tornillo and Smalley, 2015; Visvader and Stingl, 2014). We showed that PROM1+ cells specifically repopulate ER+ cells in mice. Whether PROM1 can be applied to identify ER+ luminal cells in human has not been directly tested, although it seems to be more broadly expressed in human breast (Lim et al., 2009; Raouf et al., 2008). Since it is not uncommon for human and mouse stem cells to have different cell surface markers, future work will be needed to identify human equivalent of mouse PROM1+ cells. Furthermore, although we have shown a clear role of lineage-biased stem cells in luminal cell development and regeneration, it is important to point out that our results do not rule out the contribution of multipotent or bipotent stem cells in these processes. Future work is need to demonstrate the relationship of various stem cell types. It also remains to be determined whether the functions of various multipotent and lineage-biased stem cells are influenced by mouse stain background. Nevertheless, the discovery of lineage-restricted stem cells raises important questions as to whether they serve as cells-of-origin for distinct breast cancer subtypes. It is particularly intriguing to speculate whether ER+ stem cells are a preferred origin of ER+ cancers, which represent the majority of human breast cancers.
EXPERIMENTL PROCEDURES
Mouse reagents
Sox9-CreERT2 (JAX #018829) and Prom1-CreERT2 mice (JAX #017743) were bred with R26R-tdTomato mice (JAX #007914) to generate inducible lineage-tracing models. Female mice at indicated ages were used for experiments. All experimental procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine.
Tamoxifen-induced lineage tracing
Female mice were treated by a single intraperitoneal injection of 1.5 mg tamoxifen/mouse (Sigma T5648; diluted in 100 μl corn oil) for lineage tracing from P28 days and 8 weeks, except for lineage tracing from P14 days (2 mg/mouse) and clonal analysis of SOX9+ cells (25 μg/mouse).
Cleared mammary fat pad transplantation
Mammary ductal fragments (1–2 mm) containing tdTomato+ cells were dissected from the endogenous glands or primary outgrowths and transplanted into cleared mammary fat pads of 3–4 weeks old female NOD-SCID mice as previously described (Guo et al. 2012). One fragment was transplanted into each cleared fat pad. The fat pads were analyzed 8–12 weeks post-transplantation. Only the outgrowth with branched ductal trees and terminal end buds was scored as positive reconstitution and included in the analysis.
Mammary cell flow cytometry, mammary whole-mount and immunostaining
Mammary cell flow cytometric analysis was performed as previously described (Zhang et al., 2016). Detailed protocols for flow cytometry, whole-mount and immunostaining are described in the Supplemental Experimental Procedure.
Statistical analysis
All data are represented as mean ± standard error of the mean (SEM). Comparisons between two groups were done using the unpaired t test unless stated otherwise, and P < 0.05 is considered significant. Statistical analyses were performed with GraphPad Prism 6.
Supplementary Material
HIGHLIGHTS.
SOX9+ cells maintain the development and long-term homeostasis of ER− luminal cells
SOX9+ cells contribute to alveologenesis
PROM1+ cells sustain the development and long-term homeostasis of ER+ luminal cells
ER− and ER+ luminal cells are regenerated by distinct cells during transplantation
Acknowledgments
We are grateful to Dr. Pamela Stanley for critical reading of the manuscript. We thank the Flow Cytometry, Histopathology, Analytical Imaging core facilities of Albert Einstein College of Medicine for technical assistance (supported by Einstein Cancer Center Support Grant P30 CA013330), and the New York State Department of Health (NYSTEM Program) for shared facility grant support (C029154). This work is supported by grants from NYSTEM (C028109 and C029571), Susan Komen for the Cure (CCR12224440), and the V Foundation for Cancer Research. W. G. is a V Scholar. J. R. C. was supported by the 2T32GM007491-36 Training Program in Cellular and Molecular Biology and Genetics and the NYSTEM Training Grant (C30392GG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
C.W. and W.G. designed the study. C.W. and J.R.C. performed the experiments, acquired and analyzed data. M.H.O and W.G. analyzed data. C.W. and W.G. wrote the manuscript with inputs from all other authors. W.G. supervised the study.
References
- Cowin P, Wysolmerski J. Molecular mechanisms guiding embryonic mammary gland development. Cold Spring Harb Perspect Biol. 2010;2:a003251. doi: 10.1101/cshperspect.a003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser KE, Ciampricotti M, Michalak EM, Tan DW, Speksnijder EN, Hau CS, Clevers H, Barker N, Jonkers J. Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol. 2012;228:300–309. doi: 10.1002/path.4096. [DOI] [PubMed] [Google Scholar]
- Eaves CJ. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood. 2015;125:2605–2613. doi: 10.1182/blood-2014-12-570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraddi RR, Shehata M, Gallardo M, Blasco MA, Simons BD, Stingl J. Stem and progenitor cell division kinetics during postnatal mouse mammary gland development. Nat Commun. 2015;6:8487. doi: 10.1038/ncomms9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha R, Werb Z. Mammary gland reprogramming: metalloproteinases couple form with function. Cold Spring Harb Perspect Biol. 2011;3:a004333. doi: 10.1101/cshperspect.a004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Schaffer AE, Hao E, Shih HP, Seymour PA, Ma J, Sander M. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011;138:653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Reavie L, do Couto JP, De Silva D, Stadler MB, Roloff T, Britschgi A, Eichlisberger T, Kohler H, Aina O, et al. PIK3CA induces multipotency and multi-lineage mammary tumours. Nature. 2015;525:114–118. doi: 10.1038/nature14669. [DOI] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Luanpitpong S, Li J, Manke A, Brundage K, Ellis E, McLaughlin SL, Angsutararux P, Chanthra N, Voronkova M, Chen YC, et al. SLUG is required for SOX9 stabilization and functions to promote cancer stem cells and metastasis in human lung carcinoma. Oncogene. 2016;35:2824–2833. doi: 10.1038/onc.2015.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarem M, Spike BT, Dravis C, Kannan N, Wahl GM, Eaves CJ. Stem cells and the developing mammary gland. J Mammary Gland Biol Neoplasia. 2013;18:209–219. doi: 10.1007/s10911-013-9284-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC, de Sauvage F, Klein OD, Werb Z. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell reports. 2013;3:70–78. doi: 10.1016/j.celrep.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- Rodilla V, Dasti A, Huyghe M, Lafkas D, Laurent C, Reyal F, Fre S. Luminal progenitors restrict their lineage potential during mammary gland development. PLoS Biol. 2015;13:e1002069. doi: 10.1371/journal.pbio.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shehata M, Teschendorff A, Sharp G, Novcic N, Russell A, Avril S, Prater M, Eirew P, Caldas C, Watson CJ, et al. Phenotypic and functional characterization of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M, van Amerongen R, Zeeman AL, Giraddi RR, Stingl J. The influence of tamoxifen on normal mouse mammary gland homeostasis. Breast Cancer Res. 2014;16:411. doi: 10.1186/s13058-014-0411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Kopp JL, Sandhu M, Dubois CL, Seymour PA, Grapin-Botton A, Sander M. A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Development. 2012;139:2488–2499. doi: 10.1242/dev.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman KE, Kendrick H, Robertson D, Isacke CM, Ashworth A, Smalley MJ. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176:19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike BT, Engle DD, Lin JC, Cheung SK, La J, Wahl GM. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell. 2012;10:183–197. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A, Roarty K, Rosen JM. The mammary stem cell hierarchy: a looking glass into heterogeneous breast cancer landscapes. Endocr Relat Cancer. 2015;22:T161–176. doi: 10.1530/ERC-15-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Tornillo G, Smalley MJ. ERrrr…where are the progenitors? Hormone receptors and mammary cell heterogeneity. J Mammary Gland Biol Neoplasia. 2015;20:63–73. doi: 10.1007/s10911-015-9336-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Bowman AN, Nusse R. Developmental Stage and Time Dictate the Fate of Wnt/beta-Catenin-Responsive Stem Cells in the Mammary Gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Clevers H. Tissue-specific designs of stem cell hierarchies. Nat Cell Biol. 2016;18:349–355. doi: 10.1038/ncb3332. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Smith GH. Murine mammary epithelial stem cells: discovery, function, and current status. Cold Spring Harb Perspect Biol. 2011;3:a004879. doi: 10.1101/cshperspect.a004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Cai C, Dong X, Yu QC, Zhang XO, Yang L, Zeng YA. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015;517:81–84. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008;135:995–1003. doi: 10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- Wuidart A, Ousset M, Rulands S, Simons BD, Van Keymeulen A, Blanpain C. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 2016;30:1261–1277. doi: 10.1101/gad.280057.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, Stanger BZ. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Christin JR, Wang C, Ge K, Oktay MH, Guo W. Mammary-Stem-Cell-Based Somatic Mouse Models Reveal Breast Cancer Drivers Causing Cell Fate Dysregulation. Cell reports. 2016;16:3146–3156. doi: 10.1016/j.celrep.2016.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


