Abstract
Sample preparation is key for optimal detection and visualization of analytes in Matrix-assisted Laser Desorption/Ionization (MALDI) Imaging Mass Spectrometry (IMS) experiments. Determining the appropriate protocol to follow throughout the sample preparation process can be difficult as each step must be optimized to comply with the unique characteristics of the analytes of interest. This process involves not only finding a compatible matrix that can desorb and ionize the molecules of interest efficiently, but also selecting the appropriate matrix deposition technique. For example, a wet matrix deposition technique, which entails dissolving a matrix in solvent, is superior for desorption of most proteins and peptides, whereas dry matrix deposition techniques are particularly effective for ionization of lipids. Sublimation has been reported as a highly efficient method of dry matrix deposition for the detection of lipids in tissue by MALDI IMS due to the homogeneity of matrix crystal deposition and minimal analyte delocalization as compared to many wet deposition methods 1,2. Broadly, it involves placing a sample and powdered matrix in a vacuum-sealed chamber with the samples pressed against a cold surface. The apparatus is then lowered into a heated bath (sand or oil), resulting in sublimation of the powdered matrix onto the cooled tissue sample surface. Here we describe a sublimation protocol using 1,5-diaminonaphthalene (DAN) matrix for the detection and visualization of gangliosides in the rat brain using MALDI IMS.
Keywords: Neuroscience, Issue 121, Sublimation, MALDI Imaging Mass Spectrometry, Gangliosides, DAN matrix, brain, membrane lipids
Introduction
Matrix-assisted Laser Desorption/Ionization (MALDI) Imaging Mass Spectrometry (IMS) is becoming a highly sought after technique for visualization of the spatial distribution of lipids, peptides and proteins across intact sample surfaces. MALDI IMS was previously known as an analytical technique for pre-purified analytes, but in recent years, it has been drawing attention in many other disciplines because of the ability to combine the accuracy of mass spectrometry with high resolution visual/anatomical reference points without the need for any external labelling. As the scientific pool of researchers utilizing this technique continues to grow, there is increased need for standardized, easy-to-follow protocols to assist in the development and optimization of IMS experiments. Gangliosides, a group of membrane lipids highly abundant in the central nervous system, are ideal for MALDI IMS experiments as their location, embedded within the membrane, makes certain species impossible to detect using conventional Immuno-labelling. Additionally, we have shown, using MALDI IMS, that these lipids, which function as modulators of cell signaling, among other things, have unique anatomical distribution patterns in the healthy rodent brain that are altered after brain injury 3,4,5. Gangliosides are located at a higher mass range compared to most lipid species, and are thus most suited to the MALDI imaging platform.
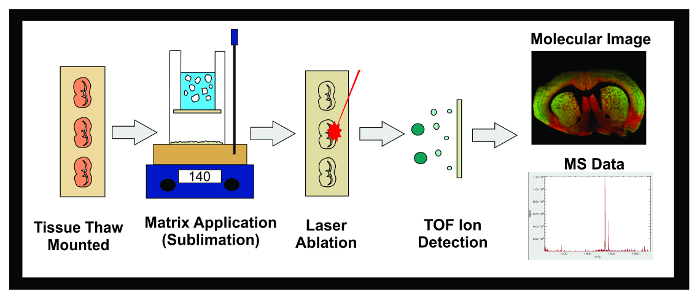 Figure 1: Workflow of MALDI IMS Experiment. Diagram of the general workflow of a MALDI IMS experiment using sublimation. Tissue frozen at -80 °C is sectioned in a cryostat and 10 µm sections are thaw mounted onto conductive ITO slides. The slide is then placed in a desiccator until sublimation. Slides are inserted into the sublimation apparatus and an even layer of matrix is applied to the tissue sample surface. Samples are frozen overnight in a -20 °C freezer then placed in a desiccator for 10 min. Once standards have been applied, the samples are inserted into the MALDI instrument where a laser is directed across the tissue causing desorbed molecules in the matrix to ionize. The ions travel down a flight tube and separate based on their mass (time-of-flight/TOF) until they reach the detector. The information on the ionic abundance of analytes within a predetermined mass-to-charge (m/z) range is displayed as both a molecular image and mass spectrum. This data can be used to both visualize and quantify the ionic abundance of the analyte of interest within the imaged tissue. Please click here to view a larger version of this figure.
Figure 1: Workflow of MALDI IMS Experiment. Diagram of the general workflow of a MALDI IMS experiment using sublimation. Tissue frozen at -80 °C is sectioned in a cryostat and 10 µm sections are thaw mounted onto conductive ITO slides. The slide is then placed in a desiccator until sublimation. Slides are inserted into the sublimation apparatus and an even layer of matrix is applied to the tissue sample surface. Samples are frozen overnight in a -20 °C freezer then placed in a desiccator for 10 min. Once standards have been applied, the samples are inserted into the MALDI instrument where a laser is directed across the tissue causing desorbed molecules in the matrix to ionize. The ions travel down a flight tube and separate based on their mass (time-of-flight/TOF) until they reach the detector. The information on the ionic abundance of analytes within a predetermined mass-to-charge (m/z) range is displayed as both a molecular image and mass spectrum. This data can be used to both visualize and quantify the ionic abundance of the analyte of interest within the imaged tissue. Please click here to view a larger version of this figure.
Sample preparation for MALDI IMS is highly variable as each step of the process must be customized to the analytes of interest. The defining feature of MALDI-based experiments is the use of a matrix coating deposited onto the sample surface prior to analysis. In addition to the role of absorbing and transferring radiation energy from the laser during the ablation process, the matrix also serves to isolate various analytes from the sample, thereby facilitating the analysis of compounds of interest 6,7. Homogenous application of the matrix to the sample surface is the most crucial step in the sample preparation process. Improper matrix deposition can lead to large heterogeneous matrix crystal formations and the development of artifacts, low ion-signal, and poor reproducibility 7.
Due to the affinity of certain matrices to isolate specific analytes, the type of matrix selected for an experiment can significantly alter the outcome. The matrices used for imaging of proteins and peptides often differ from those used for imaging lipids and the process is further complicated by the need for additional procedures such as washing and rehydration steps in order to successfully detect signals from tissue. Although washing steps exist for the enhancement of lipid signals 8, they are not a prerequisite for the detection of most lipid species. When selecting a matrix for a lipid imaging experiment, it is important to consider the polarity of the lipid of interest as this will narrow the range of suitable matrices. For example, gangliosides contain sialic acid residues which give them an overall negative polarity. There are a number of matrices that can effectively desorb and ionize gangliosides from tissue; however, factors such as matrix-derived peaks in the spectrum and stability of the matrix under vacuum must be taken under consideration. 1,5-diaminonapthalene (DAN) matrix is sufficiently stable under instrument vacuum conditions for the majority of imaging applications and has demonstrated a high degree of sensitivity for lipid desorption and can be used for the analysis of lipids in both positive and negative ion modes 2. DAN matrix, when compared to other negative lipid affinity matrices such as dihydroxybenzoic acid (DHB), 9-aminoacridine (9-AA), and 5-chloro-2-mercaptobenzothiazole (CMBT), was able to most efficiently desorb gangliosides from rat brain tissue in negative ion mode (manuscript in preparation).
Selecting the appropriate method of matrix deposition is of equal importance to selecting the matrix itself. Wet matrix deposition methods wherein the solid matrix is dissolved in an organic solvent, and deposited by pneumatic or automated sprayers or spotters, are particularly effective for the desorption of proteins and peptides as the liquid permeates the sample to allow for extraction of compounds and co-crystallization with the matrix. Although these techniques can also be used for lipid applications, analyte delocalization and uneven matrix crystal formations are common occurrences due to the high abundance and solubility of lipids in solvents, particularly in tissue 2,9. Because lipids are readily ionized from tissue, dry matrix deposition techniques, such as sublimation, offer a simple, cost effective alternative to sprayers while circumventing many of the drawback of these techniques. The success of sublimation in MALDI IMS experiments is attributed to features such as microcrystalline matrix morphology which increases the surface area for matrix-analyte binding, increased matrix purity, and homogenous matrix deposition leading to increased reproducibility compared to wet matrix techniques 1,10.
Sublimation involves heating a powdered matrix under vacuum immediately below a cooled sample surface resulting in solid to gas-phase transition of the powdered matrix followed by deposition onto the tissue sample surface. During sublimation, matrix deposition can be controlled by varying factors such as time, temperature and pressure to provide highly reproducible results. A single sublimation experiment can take anywhere from 5 to 20 min depending on the type of matrix selected, which can be re-used several times before disposal. The apparatus can be purchased commercially at a fraction of the price of automated sprayers and is easily taken apart for cleaning and maintenance. The low cost and relative simplicity of this matrix deposition technique make it ideal for researchers beginning or expanding upon lipid imaging applications in MALDI IMS. Although information detailing protocols for sublimation of tissues for IMS have been reported 11, few standardized protocols exist which focus on the basic workflow involved with carrying out a sublimation experiment for imaging high mass lipids in negative ion mode, making it difficult to establish the technique without extensive trial and error. The following is an experimental protocol aiming to fill that gap for the sublimation of DAN matrix onto rat brain sections for high resolution imaging and detection of gangliosides.
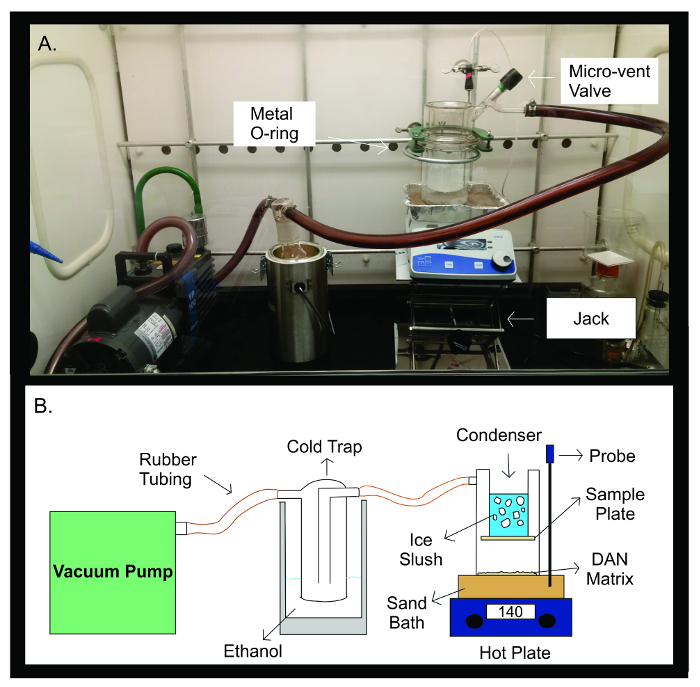 Figure 2: Sublimation Apparatus. Photograph (A) and schematic diagram (B) of the sublimation apparatus. The vacuum pump is connected by rubber tubing to a cold trap filled with 300 mL of ethanol. The cold trap is then connected by rubber tubing to the sublimation apparatus. The apparatus is made up of two separate pieces of glassware that are sealed together with a metal U-joint. The top half of the sublimator contains the condenser which is filled with ice slush. The sample plate is taped onto the bottom of the condenser, inside the sealed glass apparatus. The bottom half of the sublimation apparatus contains the DAN matrix, spread out evenly facing the sample plate. During sublimation, the glass apparatus is placed on a sand bath heated to 140 °C by a hot plate directly below it. The temperature probe helps maintain a stable temperature throughout the sublimation experiment through feedback of the sand bath temperature compared to the preset temperature for the experiment. Please click here to view a larger version of this figure.
Figure 2: Sublimation Apparatus. Photograph (A) and schematic diagram (B) of the sublimation apparatus. The vacuum pump is connected by rubber tubing to a cold trap filled with 300 mL of ethanol. The cold trap is then connected by rubber tubing to the sublimation apparatus. The apparatus is made up of two separate pieces of glassware that are sealed together with a metal U-joint. The top half of the sublimator contains the condenser which is filled with ice slush. The sample plate is taped onto the bottom of the condenser, inside the sealed glass apparatus. The bottom half of the sublimation apparatus contains the DAN matrix, spread out evenly facing the sample plate. During sublimation, the glass apparatus is placed on a sand bath heated to 140 °C by a hot plate directly below it. The temperature probe helps maintain a stable temperature throughout the sublimation experiment through feedback of the sand bath temperature compared to the preset temperature for the experiment. Please click here to view a larger version of this figure.
Protocol
All animal handling procedures described below adhere to the University of Western Ontario's animal care committee (2016-014).
1. Tissue Preparation and Sectioning
- Extract brain tissue from the rat through a process known as "Fresh Frozen Extraction" (FFE).
- Euthanize the rat with an overdose of pentobarbital sodium and monitor reflexes in the limbs. Once all reflexes have ceased, sever the head of the rat using a guillotine.
- Carefully separate the muscles and other tissues from the skull using a scalpel. Use bone-cutting forceps to break the skull to expose the brain, starting from the base of the skull (foramen magnum) to the anterior portion.
- Once the brain is exposed, carefully scoop it out with a surgical spatula and immediately place on crushed dry ice for flash freezing. NOTE: The brain will be very malleable. Therefore, take extreme care when placing the brain on dry ice. If the brain is crushed or pressed against a surface, it will alter its shape and freeze in that position.
Remove fresh frozen tissue (not perfused with formaldehyde or embedded in OCT) from - 80° C freezer and place on dry ice.
Place several drops of water on a cryostat holder and place on either dry ice or cryostat freeze bar. Press tissue lightly onto cryostat holder as it begins to freeze and hold until water freezes around the base of the tissue and anchors it in place. Add additional water to further secure the tissue on the holder if necessary. NOTE: Water is used instead of OCT media for mounting tissue in order to avoid contamination of the tissue with compounds which can affect the detection of the desired signal.
Position tissue in cryostat. Section tissue up to the desired anatomical location. Ensure that the cutting thickness is between 8 - 12 µm. NOTE: When sectioning tissue in communal cryostat devices, it is imperative that separate materials such as blades, brushes, anti-roll bars, and holders be used in order to avoid contamination with embedding compounds. The inside of the cryostat should also be cleaned thoroughly with ethanol before use.
Using the cryostat anti-roll bar, slowly section flattened tissue sections. Holes or markings on the tissue can occur if the roll bar placement is incorrect or blade is dull. Move the tissue to the center of slicing platform using clean paintbrushes.
- Mounting
- Freeze conductive slides or metal plates by placing them in the cryostat while slicing. When the slide is completely frozen, carefully move the sectioned tissue onto the conductive surface of the slide using clean paintbrushes. Once all tissue sections are positioned correctly on the slide, place a finger under the slide, opposite the tissue, and press until the section thaws. NOTE: Sections may fold or curl during the thawing process when using this mounting method. This can be reduced by keeping the slide in the cryostat when thawing the tissue. If these issues become problematic, an alternative, warm-mount method is listed below.
- Alternatively, take a room temperature Indium-tin Oxide (ITO) slide (or metal plate) and lightly press down on frozen tissue section on the slicing platform surface, conductive side down. This will lead to the tissue section thawing evenly onto the surface of the slide with little curling or folding of the tissue. NOTE: Condensation may appear below the section when mounting using this method which may lead to loss of certain proteins and lipids.
Place slides with tissue in a desiccator for 5 - 10 min.
2. Sublimator Apparatus Set-up
NOTE: Perform these steps in a fume hood.
- Place a sand bath in an aluminum container onto a hot plate, with the hot plate on a metal scissor lift of appropriate surface area (i.e. larger than the hot plate). Turn on the hot plate and set the temperature to 140 °C. NOTE: The melting point for DAN matrix is between 187 - 190 °C. Do not exceed this temperature on the hotplate.
- If the hot plate is equipped with a temperature feedback probe, use it to monitor sand temperature throughout the experiment and ensure temperature consistency. This feature can assist with experiment reproducibility across various sublimation experiments. NOTE: The sand bath should be contained within an aluminum container as a glass container may shatter at high temperatures.
Place 300 mg of DAN matrix onto the bottom surface of sublimation apparatus. Place the matrix in the center of the apparatus and spread out in an even layer in the approximate width and length of the slide being sublimated. CAUTION: DAN matrix is toxic. Therefore, it is important wear gloves, masks, and safety goggles at all times when handling the powdered matrix. DAN should be stored in the dark as it is light sensitive.
- Tape a metal plate onto the inner surface of the apparatus with the plate making direct contact with the bottom of the condenser in order to ensure even distribution of temperature cooling across the entire surface of the slide during sublimation. Alternatively, sand the bottom of the condenser to ensure a flat surface.
- Place the tape along the outer edges of the plate and adhere to the sides of the inner glassware. If the tape is placed under the plate and adheres to the bottom of the inner glass surface, the temperature distribution may not be even and could result in uneven matrix distribution across the surface of the slide.
- Tape a blank (test) slide diagonally across the surface of the metal plate with the tape again placed on the outer edges of the slide.
Connect the top and bottom portions of the apparatus, with a rubber O-ring in the middle to ensure a complete seal.
Place a metal U-joint around the center of the apparatus and tighten vices until the top and bottom half of the apparatus are sealed tightly together. Place in the metal O-ring above the sand bath.
Take a handful of crushed ice and place it in the condenser. Fill condenser ¼ to ½ full with cold water to create ice slush. The ice slush will cool the metal plate on the inside of the apparatus and subsequently the slide adhered to it. Wait at least 5 min for the temperature to reach a steady state.
Pour 300 mL of ethanol in the cold trap container and place the glassware into the container. Drop 2 - 3 small pieces of dry ice into the ethanol in the bottom of the cold trap container. The cold trap input has a glass tube running to the bottom of the cylinder, while the output does not.
Connect the vacuum pump to the cold trap output using rubber tubing. Use another piece of rubber tubing to connect the cold trap input to the sublimation apparatus. Ensure that the tubing is tightly secured using metal clamps if available.
Use the vacuum pump to deliver a vacuum of 30 - 50 mT. Allow the pump to run for at least 5 min for pressure equilibration. Vices on U-ring of sublimation apparatus may have to be tightened again once the vacuum pump is turned on because of decreased pressure in the apparatus. NOTE: It is highly recommended that a vacuum gauge be attached to the pump to monitor the pressure of vacuum during the experiment and to test for leaks in pressure in order to achieve the highest possible reproducibility between experiments. However, most pumps are designed to maintain a constant pressure, therefore the gauge may not be essential for experienced users. Additionally, vacuum seal grease can be used to help maintain vacuum pressure.
3. Sublimation
Ensure that the sand bath temperature has stabilized at 140 °C and that the apparatus is secured in the metal O-ring above the sand bath with all tubing connected and the vacuum turned on.
- Set timer for 7 min but do not start timer. Slowly raise the scissor lift and sand bath up to the sublimation apparatus until the U-joint of the sublimator is well above the metal O-ring. This extra space allows for adjustment of the apparatus on the sand.
- Quickly press the sublimator gently on the sand surface to ensure that the apparatus is sitting evenly on the sand, and then immediately start the timer.
- When the timer sounds, turn off the vacuum pump and carefully lower the scissor lift until the sublimator is no longer touching the sand bath and is sitting securely in the metal O-ring.
- Slowly loosen the micro-vent valve to release pressure in the apparatus. Loosen the metal clamp around the rubber tubing of the sublimation apparatus and slowly begin to loosen the tube. Once the rubber tube has been loosened slightly, bend the tube to one side to allow residual pressure to escape.
- When ambient pressure returns, carefully remove the rubber tubing from the sublimation apparatus.
Loosen the vices on the U-joint of the apparatus and remove. Carefully separate the two halves of the sublimation apparatus and pull off the slide from the top of the inner glassware. Examine slide to confirm even matrix distribution. NOTE: If matrix is uneven, the powdered matrix in the bottom of the sublimator can be repositioned or the sublimator apparatus can be repositioned in the sand for the next slide. The sublimation process can be repeated several times using the same matrix, however, the matrix will eventually become darker from repeated heat exposure and the quantity of powder will decrease such that the sublimation time will have to be adjusted slightly to compensate. For this reason, it is important to monitor the amount of matrix being sublimated after each experiment to ensure consistency in matrix deposition between slides
If the distribution and amount of matrix sublimated is sufficient, tape a new slide with tissue onto the inside of the sublimator and repeat Section 3. NOTE: For quality control (QC) purposes, the amount of matrix sublimated can be measured after each experiment by weighing the slide before and after sublimation and dividing the weight of sublimated matrix with the surface area of the slide 2, it should be noted that due to the lack of precision of most scales beyond 4 decimal points and variability between scales, these measurements should be only be used a guide for optimal matrix deposition as opposed to a fully quantifiable means of QC unless a high precision balance is used (see Figure 3).
4. Tissue Storage/Rehydration
After sublimation, store slides in a sealed container or small cassette in sealed plastic bag.
Incubate slides in -20 °C freezer for 2 h or overnight. NOTE: The goal of the freezing process is to act as a rehydration step for the desorption of tissue materials into the matrix. The freezing process has also been shown to prevent degradation of lipid signals for up to 1 week when stored at -80 °C 12. For this reason, the amount of time the samples are stored in the freezer can be varied to a certain degree to suit the needs of the experimenter without significantly altering signal detection (as observed in our lab). However, we have noticed some discoloration of matrix when samples are frozen for longer than 24 h at -20 °C. Thus, we recommend imaging the sublimated tissue before that time or storing tissue at -80 °C for longer incubation periods.
Remove slides from freezer (and container) and place in a desiccator for 5 - 10 min.
5. Imaging and Analysis
Remove slides from desiccator and apply instrument standards to ensure mass accuracy of MALDI instrument during imaging. The type of standards will vary depending on instrumentation. Standards are generally applied evenly across entire slide, surrounding tissue to be imaged (Figure 3D).
Insert slide into MALDI instrument and follow manufacturer's instructions for imaging experiments (instrument methods should be optimized for a 1,000 - 2,000 mass range). Representative result (Figure 4) was acquired in reflectron negative mode with a 70 µm raster and 20 shots/spectrum (acquisition time ~ 2 h).
Representative Results
Upon completion of the sublimation experiment, the two halves of the glass apparatus are separated in the fume hood and the slide, taped to the condenser, can be removed (Figure 3A). At this point, the slide should be examined for uneven matrix distribution and the time, temperature, or placement of the apparatus in the sand bath may have to be adjusted for the next slide. A successful DAN sublimation experiment will result in an even coating of grey/brown matrix along the surface of the slide where the anatomical features of the tissue can be clearly visualized and the amount of matrix on the glass slide is similar to the amount on the tissue (Figure 3B). For example, if too much matrix has been sublimated on the tissue, the thickness of the matrix will cover the features of the tissue and only the general shape will be discernable. However, if too little matrix is sublimated, the tissue will have a darker appearance than the rest of the slide (Figure 3C). Both too much and too little matrix will result in poor signal in the MALDI instrument. For quality control purposes, Thomas et al. weighed the amount of matrix deposited on the slide and divided the weight by the surface area of the slide. They reported an optimal amount of matrix deposition for several matrices including DAN (110 µg/cm²) 2. Although most balances lack the precision needed to replicate such results, we have attempted this method of QC; however, due to a high degree of variability, we can only advise a suitable range of matrix deposition found to be associated successful versus unsuccessful sublimation experiments with DAN matrix as determined through signal detection in the instrument. A matrix measurement of below 100 µg/cm² was associated with "too little" matrix conditions while a measurement of above 140 µg/cm² was associated with "too much". Matrix deposition between 100 and 140 µg/cm² was found to be an optimal quantity for imaging with DAN matrix. Calibrating standards should be applied on the glass slide around the tissue samples to ensure the mass accuracy of the detected IMS signals (Figure 3D)
In a MALDI IMS experiment, the molecular image allows for the visualization of the spatial distribution of the analyte of interest along with any unknown species that may be present in a given mass range. The molecular images of gangliosides in this experiment were overlaid using ImageJ software to show the anatomical distribution of several ganglioside species across the cortical and subcortical regions of the rat brain (Figure 4A). The most abundant gangliosides in the brain are the A-series species GD1a and GM1 but also contain gangliosides GM2 and GM3 which can all be found between the 1,000-2,000 m/z mass range, a higher mass range than most common brain lipids, making these gangliosides easy to identify along the spectrum (Figure 4B). The ionic abundance of each ganglioside species can be used to semi-quantify differences or changes in ganglioside species within the same image. Because a certain amount of variability in sample prep cannot be avoided in IMS, between scan comparisons are considered to be semi-quantitative.
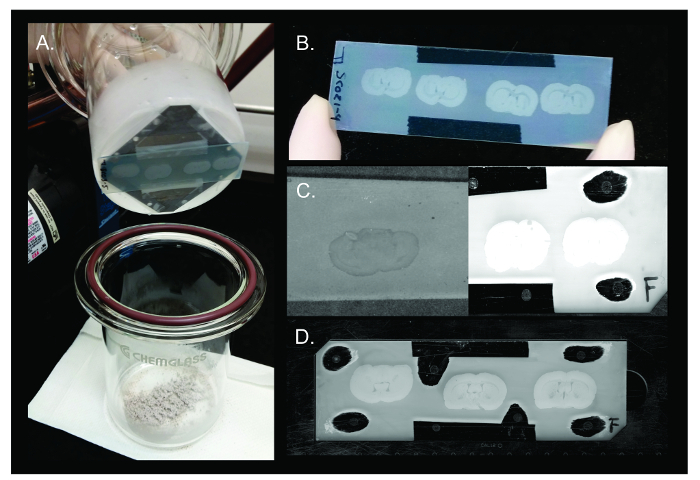 Figure 3. DAN Sublimation. Representative results of sublimation of DAN matrix on rat brain tissue. (A) Upon completion of sublimation, the two halves of the apparatus are separated and the slide is removed from the condenser. (B) DAN matrix spreads evenly across multiple rat brain tissue sections on the surface of the slide to allow for highly reproducible results (between 100 - 150 µg/cm²). (C) Examples of failed sublimation experiments where too little matrix (left - <90 µg/cm²) or too much matrix (right - >150 µg/cm²) was deposited. Too little matrix will result in insufficient desorption of analytes, while too much matrix will not allow the laser to penetrate to the tissue during acquisition, resulting in low ion detection. (D) Slide containing rat brain tissue sections sublimated with DAN matrix and calibration standards applied is ready for insertion into MALDI instrument for imaging. Please click here to view a larger version of this figure.
Figure 3. DAN Sublimation. Representative results of sublimation of DAN matrix on rat brain tissue. (A) Upon completion of sublimation, the two halves of the apparatus are separated and the slide is removed from the condenser. (B) DAN matrix spreads evenly across multiple rat brain tissue sections on the surface of the slide to allow for highly reproducible results (between 100 - 150 µg/cm²). (C) Examples of failed sublimation experiments where too little matrix (left - <90 µg/cm²) or too much matrix (right - >150 µg/cm²) was deposited. Too little matrix will result in insufficient desorption of analytes, while too much matrix will not allow the laser to penetrate to the tissue during acquisition, resulting in low ion detection. (D) Slide containing rat brain tissue sections sublimated with DAN matrix and calibration standards applied is ready for insertion into MALDI instrument for imaging. Please click here to view a larger version of this figure.
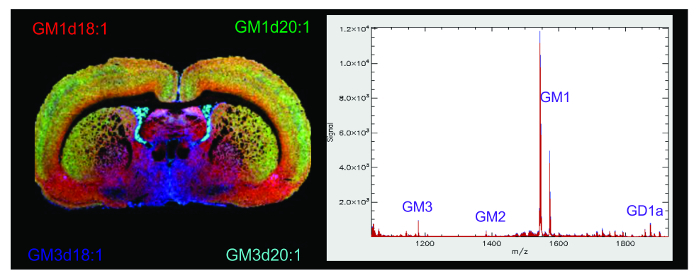 Figure 4. MALDI IMS of Gangliosides in the Rat Brain. Representative MALDI IMS molecular image and mass spectrum of gangliosides in the rat brain after sublimation with DAN matrix. The molecular image is a composite of multiple ganglioside species in the spectrum overlaid and pseudocolored using Image J software to show the unique anatomical distribution of ganglioside species throughout the brain. Species GM1d18:1 (red, mass ~1,547 Da), GM1d20:1 (green, mass ~1,573 Da), GM3d18:1 (blue, mass ~1,178 Da), and GM3d20:1 (teal, mass ~1,207 Da) are represented. The mass spectrum displays ionic abundance of analytes within a 1,000 - 2,000 mass range. The major A-series gangliosides are labelled along the spectrum. Please click here to view a larger version of this figure.
Figure 4. MALDI IMS of Gangliosides in the Rat Brain. Representative MALDI IMS molecular image and mass spectrum of gangliosides in the rat brain after sublimation with DAN matrix. The molecular image is a composite of multiple ganglioside species in the spectrum overlaid and pseudocolored using Image J software to show the unique anatomical distribution of ganglioside species throughout the brain. Species GM1d18:1 (red, mass ~1,547 Da), GM1d20:1 (green, mass ~1,573 Da), GM3d18:1 (blue, mass ~1,178 Da), and GM3d20:1 (teal, mass ~1,207 Da) are represented. The mass spectrum displays ionic abundance of analytes within a 1,000 - 2,000 mass range. The major A-series gangliosides are labelled along the spectrum. Please click here to view a larger version of this figure.
Discussion
This work details a standardized protocol of matrix sublimation on tissue for the detection of negatively charged lipids, such as gangliosides, in MALDI IMS experiments. Sample preparation for MALDI IMS is highly variable and must be customized to suit the unique properties of the analyte of interest. The application of matrix onto the tissue sample surface is a crucial aspect of the sample preparation process with regards to the quality of results in IMS. Particular care should be taken when selecting matrices, ensuring that the matrix is compatible with the analyte of interest and is free of matrix-derived artifacts in the spectrum. Sublimation is a dry matrix deposition technique which provides a high degree of matrix crystal homogeneity with little delocalization of lipids on tissue. DAN matrix in particular shows a high degree of sensitivity for the desorption of a group of negatively charged membrane lipids called gangliosides, as demonstrated in rat brain sections, and is stable under vacuum for extended periods of time, allowing for compatibility with the majority of imaging applications. DAN matrix has the additional benefit of also having a high affinity for positively charged lipid species, thus increasing the versatility of this matrix for MALDI IMS applications 2. It should also be noted that multiple lipid species, including phospholipids, can be detected simultaneously when using this IMS protocol.
A thorough examination of the properties of DAN matrix as compared to 9 other commonly used matrices has been previously published 2. In this article, the authors highlight the increased sensitivity of DAN matrix in negative ion mode as compared to the other matrices examined as well as the capability for high resolution imaging using DAN. DAN matrix was found to have the highest overall performance for negative polarity ions. Interestingly, it performed equally well for positive polarity ions. Other common matrices which have demonstrated high affinity for negative polarity ions include DHA, DHB, and 9-AA. The efficiency of DHA as a matrix is limited by its low stability under vacuum which makes it impractical for most imaging applications 2. 9-AA has the benefit of producing low background noise and demonstrated enrichment of sulfatide signals in the 750 - 950 mass range in negative mode compared to DHB matrix 13. Although DHB has been shown to be very effective in positive mode, it yields lower negative polarity ion signal as compared to DAN matrix 2. DAN matrix also demonstrated increased sensitivity in negative ion mode in the lower mass range (650 - 950) compared to DHB 2. Additionally, it has been reported that DHB can form significant matrix clusters in negative ion mode up to 750 Da 2.
Another method of dry matrix deposition has been described by Puolitaival et al., in which ground matrix is passed through a fine sieve and deposited onto the sample surface. The authors compared this dry method of matrix deposition to a wet matrix manual spray technique and found that they gave comparable signal for phospholipids in the 450 - 1,200 mass range 14. One study used both sublimation and dry-coating with a sieve to examine phospholipid expression, however, the authors did not comment on how the two techniques compared in terms of image resolution or ion signal yield 15.
In order to ensure the widest possible range of applicability, the protocol presented was a basic workflow that will allow both new and more specialized users of MALDI IMS to achieve highly reproducible results for a wide variety of lipid imaging applications. However, sublimation protocols can be modified to meet the unique circumstances of the experiment. For example, the fine matrix crystal deposition provided by sublimation allows for the option to reverse the order of sample preparation by pre-coating slides/plates with matrix prior to mounting the tissue. The matrix can be sublimated onto the conductive surface in advance and stored in a dark environment for later use thus reducing sample preparation time post-sectioning while still maintaining high sensitivity for lipid detection 16,17. Another modification that can be made to this protocol to enhance signal for the detection of lipids is a 2 min wash with ammonium formate 8.
Although sublimation can circumvent many of the drawbacks of most wet deposition techniques, such as smaller, more homogenous matrix crystal deposition 1, decreased delocalization of analytes, and low cost as compared to automated sprayers, some variability in the sample preparation process is unavoidable. In the case of sublimation, there are several factors which may cause minute differences from one experiment to another and can include the following which have been observed in our lab. There can be differences in the position of the sublimation apparatus on the sand. When running several sublimation experiments back-to-back, the sand in the sand bath may begin to shift its position which can affect the dispersion of heat throughout the sand bath. Flattening the sand before each round of sublimation can also change the dispersion of heat as sand that has been displaced may take time to return to a uniform temperature. Sublimation time may have to be adjusted slightly between each round to compensate for shifting sand in the sand bath. Alternatively, use of an oil bath may lead to more homogenous heat dispersion.
Secondly, the amount of matrix at the bottom of the sublimator can vary. DAN matrix has a clumpy grey appearance and has a tendency to form large clusters. These clusters can be manually broken up and arranged in the bottom of the sublimator but cannot be completely eliminated. Large clusters of matrix will take longer to heat and sublimate than smaller pieces which can affect the end product of the sublimation.
The vacuum output on the sublimation apparatus is another critical factor. Most sublimators are made with a single vacuum output onto which tubing is attached to connect it to the vacuum pump during sublimation. Because the pressure is being regulated on that side of the apparatus, there may be a slight unevenness of the matrix deposition, with more being deposited on the side with the vacuum output. This can be somewhat compensated for by shifting either the position of the apparatus in the sand, the position of the matrix in the bottom of the sublimator, or by changing the position of the slide/plate on the condenser. These factors are the main drawbacks of the sublimation technique and for this reason it is crucial to run a test slide through the sublimation process before running experimental sample so that time can be adjusted to compensate for these variables.
Another reported drawback of sublimation is that the detection of higher mass analytes is limited by insufficient analyte extraction and/or mixing with matrix. A rehydration step after sublimation can help pull out these higher mass analytes to circumvent this issue. This is typically achieved using a humidity chamber 18. However, in this protocol, a freezing step is added after sublimation which achieves the same purpose. The freezing and subsequent thawing (in a desiccator) of the samples before insertion in the MALDI instrument causes condensation to form on the surface of the sample which temporarily rehydrates the sample to allow for the extraction of higher mass lipids while preserving the tissue for analysis 12,17.
Overall, sublimation is a highly sensitive and cost effective method of matrix application for the detection of lipids in MALDI IMS experiments. Indeed, our group has noticed significant improvements to IMS results when we shifted from using an air-spray method 3 to a sublimation method 4 for matrix deposition. The present protocol is appropriate for a number of lipid imaging applications but can be modified to suit the needs of the experimenter.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We would like to acknowledge the technical assistance of Kristina Jurcic in the University of Western Ontario MALDI MS facility, as well as the National Sciences and Engineering Council (NSERC) for funding this work. The authors would also like to acknowledge the Caprioli group (Vanderbilt University, TN) and Chaurand group (Université de Montreal, QC) for their advice in optimizing the sublimation technique presented in this manuscript.
References
- Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 2007;18:1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Charbonneau JL, Fournaise E, Chaurand P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal. Chem. 2012;84:2048–2054. doi: 10.1021/ac2033547. [DOI] [PubMed] [Google Scholar]
- Caughlin S, et al. Increased Expression of Simple Ganglioside Species GM2 and GM3 Detected by MALDI Imaging Mass Spectrometry in a Combined Rat Model of Aβ Toxicity and Stroke. PLoS ONE. 2015;10:0130364. doi: 10.1371/journal.pone.0130364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weishaupt N, Caughlin S, Yeung K, Whitehead S. Differential Anatomical Expression of Ganglioside GM1 Species Containing d18:1 or d20:1 Sphingosine Detected by MALDI Imaging Mass Spectrometry in Mature Rat Brain. Front Neuroanatomy. 2015;9(155) doi: 10.3389/fnana.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead S, et al. Imaging Mass Spectrometry Detection of Gangliosides Species in the Mouse Brain following Transient Focal Cerebral Ischemia and Long-Term Recovery. PLoS ONE. 2011;6(6):20808. doi: 10.1371/journal.pone.0020808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B, Süß R, Schiller J. An update of MALDI-TOF mass spectrometry in lipid research. Progress in Lipid Research. 2010;49:450–475. doi: 10.1016/j.plipres.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Barceló-Coblijn G, Fernández JA. Mass spectrometry coupled to imaging techniques: the better the view the greater the challenge. Front Physiol. 2015;6(3):1–5. doi: 10.3389/fphys.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel PM, Spraggins JM, Baldwin HS, Caprioli R. Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry. Anal. Chem. 2012;84:1557–1564. doi: 10.1021/ac202383m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RC, Hankin JA, Barkley RM, Zemski Berry KA. MALDI imaging of lipids after matrix sublimation/deposition. Biochim. Biophys. Acta. 2011;1811:970–975. doi: 10.1016/j.bbalip.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskolla TW, Karas M, Roth U, Steinert K. Comparison between vacuum sublimed matrices and conventional dried droplet preparation in MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2009;20:1104–1115. doi: 10.1016/j.jasms.2009.02.010. [DOI] [PubMed] [Google Scholar]
- O'Rourke MB, Raymond BB, Djordjevic SP, Padula MP. A versatile cost-effective method for the analysis of fresh frozen tissue sections via matrix-assisted laser desorption/ionisation imaging mass spectrometry. Rapid Commun. Mass Spectrom. 2015;29:637–644. doi: 10.1002/rcm.7138. [DOI] [PubMed] [Google Scholar]
- Patterson NH, Thomas A, Chaurand P. Monitoring time-dependent degradation of phospholipids in sectioned tissues by MALDI imaging mass spectrometry. J Mass Spectrom. 2014;49:622–627. doi: 10.1002/jms.3382. [DOI] [PubMed] [Google Scholar]
- Cheng H, Sun G, Yang K, Gross RW, Han X. Selective desorption/ionization of sulfatides by MALDI-MS facilitated using 9-aminoacridine as matrix. J. Lipid Res. 2010;51:1599–1609. doi: 10.1194/jlr.D004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J. Am. Soc. Mass Spectrom. 2008;19:882–886. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurand P, Cornett D, Angel P, Caprioli R. From Whole-body Sections Down to Cellular Level, Multiscale Imaging of Phospholipids by MALDI Mass Spectrometry. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.O110.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove KJ, Frappier SL, Caprioli RM. Matrix pre-coated MALDI MS targets for small molecule imaging in tissues. J. Am. Soc. Mass Spectrom. 2011;22:192–195. doi: 10.1007/s13361-010-0013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Caprioli RM. Matrix precoated targets for direct lipid analysis and imaging of tissue. Anal. Chem. 2013;85:2907–2912. doi: 10.1021/ac303554e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemperline E, Rawson S, Li L. Optimization and comparison of multiple MALDI matrix application methods for small molecule mass spectrometric imaging. Anal. Chem. 2014;86:10030–10035. doi: 10.1021/ac5028534. [DOI] [PMC free article] [PubMed] [Google Scholar]


