Abstract
Motivation
The aim of this study is to assess the performance of RNA–RNA interaction prediction tools for all domains of life.
Results
Minimum free energy (MFE) and alignment methods constitute most of the current RNA interaction prediction algorithms. The MFE tools that include accessibility (i.e. RNAup, IntaRNA and RNAplex) to the final predicted binding energy have better true positive rates (TPRs) with a high positive predictive values (PPVs) in all datasets than other methods. They can also differentiate almost half of the native interactions from background. The algorithms that include effects of internal binding energies to their model and alignment methods seem to have high TPR but relatively low associated PPV compared to accessibility based methods.
Availability and Implementation
We shared our wrapper scripts and datasets at Github (github.com/UCanCompBio/RNA_Interactions_Benchmark). All parameters are documented for personal use.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
RNA biology has become more prominent after the discovery of non-coding RNAs (ncRNAs) and their versatile functions (Ambros, 2004; Barquist and Vogel, 2015; Kidner and Martienssen, 2005; Mattick, 2004; Mattick, 2009; Storz et al., 2011; Waters and Storz, 2009). The versatility of RNA molecules has led to the idea of an ‘RNA world’ where RNA formed the first primitive life forms (Gilbert, 1986). The importance of RNA biology is highlighted by the relatively small fraction of protein-coding regions of most eukaryotic genomes (Mattick, 2004, 2009). For example, 1.2% of the human genome contain protein coding genes, while 76% is transcribed into RNA (Pennisi, 2012). Likewise, prokaryotic cells contain various ncRNAs genes (Gottesman, 2004; Holmqvist and Vogel, 2013; Thébault et al., 2014; Vogel, 2009) and have also been shown to have transcriptional complexity like eukaryotes (Barquist and Vogel, 2015; Cohen et al., 2016; Güell et al., 2011, 2009; Lindgreen et al., 2014). ncRNA molecules often utilize RNA–RNA base pairing such as bacterial/archaeal small RNAs (sRNAs) (Prasse et al., 2013; Storz et al., 2011), small interfering RNAs (siRNAs) (Carthew and Sontheimer, 2009), microRNAs (miRNAs) (Carthew and Sontheimer, 2009; Cuperus et al., 2011), spliceosomal small nuclear RNAs (snRNAs) (Karijolich and Yu, 2010), small nucleolar RNAs (snoRNAs) (Brown et al., 2001; Gardner et al., 2010; Kiss, 2002; Omer et al., 2000), cajal-body specific small nuclear RNAs (scaRNAs) (Darzacq et al., 2002), clustered regularly-interspaced short palindromic repeats (CRISPR) RNA (Bhaya et al., 2011) and piwi-interacting RNAs (piRNAs) (Brennecke et al., 2007; Klattenhoff and Theurkauf, 2008). It seems some long-noncoding RNAs (lncRNAs) may also engage into RNA–RNA interactions (Kung et al., 2013), which are quite abundant in eukaryotes (Zhao et al., 2016).
In addition to endogenous ncRNAs genes, many experimental techniques take advantage of RNA–RNA interactions such as gene silencing (i.e. knock-out) by artificial siRNAs (Deleavey and Damha, 2012; Reynolds et al., 2004) and designing oligonucleotides for ribosomal RNA (rRNA) depletion in RNA-seq experiments (O’Neil et al., 2013).
Different clades of life utilize regulatory RNA–RNA interactions with different constraints: various mediator proteins (Carthew and Sontheimer, 2009; Vogel and Luisi, 2011), binding regions preference and distinct complementarity requirements (Ameres and Zamore, 2013; Millar and Waterhouse, 2005). Thus, many different tools have been developed to predict stable interactions. Some algorithms solve RNA–RNA interaction as an alignment problem using local alignment approaches (Hodas and Aalberts, 2004; Wenzel et al., 2012). Most of these use dynamic programming and minimum free energy methods (MFE) (Backofen and Hess, 2010; Dieterich and Stadler, 2012; Lorenz et al., 2011), which are also widely used methods for RNA secondary structure predictions (Markham and Zuker, 2008; McCaskill, 1990; Nussinov and Jacobson, 1980; Zuker, 2000; Zuker and Sankoff, 1984; Zuker and Stiegler, 1981). In bacteria, comparative methods are becoming popular (Kery et al., 2014; Pain et al., 2015; Wright et al., 2013), but they are restricted to conserved sRNAs, which are quite rare (Barquist and Vogel, 2015; Lindgreen et al., 2014).
RNA target detection is still a challenging task but it is vital to understand more about RNA–RNA interactions for functional annotation of unknown transcripts while making computationally feasible and biologically relevant prediction. In this study, we assessed the performance of available RNA–RNA interaction prediction tools on trusted, verified datasets from all domains of life. We evaluated their ability to recover established RNA–RNA pairs in eukaryotic, bacterial and archaeal systems. We also assessed how successfully they predict binding scores and reported the significance of these predictions.
2 Materials and methods
All RNA interaction prediction algorithms are freely available and cited in the manuscript. We used Python, R, Bash for the scripts and wrappers, which are shared in our Github repository (github.com/UCanCompBio/RNA_Interactions_Benchmark). A parser script (or a wrapper script) has been written for each of the tools benchmarked here. All the parameters and command line arguments are also accessible.
2.1 Benchmark datasets
We manually confirmed the correct interaction regions (which contain the binding base-pairs) for all dataset items and used entire target regions (i.e. UTRs, coding regions or target RNA) to make our benchmark as realistic as possible. We also manually confirmed that the true binding regions on target RNAs are consecutive with only few mismatches.
The eukaryotic benchmark dataset consisted of miRNAs from human, Arabidopsis, Caenorhabditis elegans (C. elegans) (Chou et al., 2015; Kozomara and Griffiths-Jones, 2013); C/D and H/ACA box snoRNAs from human, Arabidopsis, C. elegans, yeast (Brown et al., 2001; Lestrade and Weber, 2006; Piekna-Przybylska et al., 2007; Yoshihama et al., 2013); human and yeast U6/U2 snRNAs (Will and Lührmann, 2011); endogenous siRNAs from Arabidopsis (Addo-Quaye et al., 2008) and piRNAs from mouse (Gou et al., 2015). Experimentally verified miRNA/siRNA/piRNA-target mRNAs and snoRNA/snRNA-target RNAs were selected from different ncRNA families as much as possible (in total 88 pairs) (Supplementary Table S1).
We compiled a bacterial sRNA and target mRNA dataset from Salmonella, Escherichia coli (E. coli) and Listeria monocytogenes (L. monocytogenes) that consists of 60 verified sRNA-mRNA pairs (Cao et al., 2010; Lai and Meyer, 2015; Peer and Margalit, 2011). The target regions of bacterial sRNAs lie either in 5′UTR or downstream of start codon (Richter and Backofen, 2012; Storz et al., 2011). We selected regions 200 nucleotides (nts) upstream to 100 nts downstream of the start codons (i.e. 5′end mRNA) which contain verified binding regions. We extracted both sRNAs and target 5′end mRNAs from their associated genome sequences (Acces. AE006468.1, AL591824.1 and U00096.3) (Supplementary Table S1).
We gathered a set of archaeal C/D box snoRNAs consisting of 5 snoRNAs and their ribosomal RNA targets (Omer et al., 2000). We also added a member of less studied archaeal sRNA (from Methanosarcina mazei) (Jäger et al., 2012). Selected genes and targets were obtained from their associated archaeal genomes (AE008384.1) or Genbank (Supplementary Table S1).
2.2 Accuracy measures for binding site predictions
We calculated TPR (sensitivity) and PPV (precision) scores of each algorithm based on prediction of RNA–RNA binding regions for 154 manually curated interactions from the scientific literature. They include functionally characterized RNA–RNA interactions from Archaea, Bacteria and Eukaryotes. Verified binding regions between ncRNAs and target RNAs are annotated with published base-pairing interactions. These interactions can be used to assess overlaps between predicted and true binding regions on target RNAs. In this work, true positives (TPs) are the number of nucleotides on a correctly predicted binding region, false positive (FPs) are the number of nucleotides in a falsely predicted binding region (i.e. a predicted target that is not part of the curated set of interactions), and false negatives (FNs) are the number of nucleotides in a binding region where interactions are not predicted (Supplementary Fig. S1). True negatives (TNs) are generally not used for the treatment of RNA structure as the number of true negatives grows exponentially with sequence length while TP, FP and FN grow linearly (Wenzel et al., 2012). We can calculate an approximation to the Matthews correlation coefficient (MCC) (Matthews, 1975) by using the geometric mean of TPR and PPV (Gorodkin et al., 2001; Wenzel et al., 2012). These can be defined as:
| (1) |
| (2) |
| (3) |
2.3 A significance test for prediction scores
Besides these well-known accuracy measures, we also assessed the scores generated by the algorithms, which usually show the stability of interaction (e.g. a binding MFE). For each true and verified target (positive control), we created 200 dinucleotide shuffled sequences (negative controls) using the esl-shuffle tool (Eddy, 2011) to prevent possible biases caused by the nearest-neighbour energy model of structure prediction (Workman and Krogh, 1999). To determine the significance of native interactions, we fitted the binding energies shuffled interactions (as a background) into both normal and Gumbel distributions (using negative energies) (Gumbel, 1958), since MFE values mostly follow an extreme value distribution (Rehmsmeier et al., 2004; Tjaden, 2008). In short, we assessed significance of positive controls using a set of negative controls. A similar methodology showed an avoidance of crosstalk RNA–RNA interactions in prokaryotes which can be measured as a binding energy shift (Umu et al., 2016). We applied this approach only to bacterial dataset due to time constraints, and the uniform distribution of bacterial targets (i.e. identical 300 nucleotides long target mRNAs).
We selected the best scoring interaction as the native interaction if an algorithm produces more than one interaction, which is also true for all our analyses.
3 Results and discussion
3.1 RNA–RNA interaction prediction tools
The RNA–RNA interaction prediction methods are divided mainly into three groups: alignment like methods, MFE methods and comparative (homology) methods. We can also further divide the MFE methods into three different sub-classes based on whether their approach considers intramolecular base-pairs (internal structure), neglects intramolecular structure or measures the accessibility of the binding region. There are also other machine learning algorithms (Oğul et al., 2011; Yang et al., 2008), and probabilistic approaches like RactIP (Kato et al., 2010), which uses the CONTRAfold model (Do et al., 2006) for RNA interaction prediction.
RIsearch (Wenzel et al., 2012), Bindigo (Hodas and Aalberts, 2004) and Guugle (Gerlach and Giegerich, 2006) are examples of alignment-like methods. The RIsearch algorithm was mainly developed for rapidly searching genomes to detect RNA–RNA pairs from genome sequencing data by combining the Smith-Waterman-Gotoh algorithm with a nearest-neighbor energy model (Wenzel et al., 2012), while Bindigo adopts an optimized Smith-Waterman to find optimal oligonucleotide-RNA pairs (Hodas and Aalberts, 2004). Guugle uses suffix arrays to seek RNA targets based on RNA helix rules that allow G-U pairs (Gerlach and Giegerich, 2006).
Besides these alignment based methods, tools like BLAST (Altschul et al., 1990), Blat (Kent, 2002), ssearch (Pearson and Lipman, 1988) or other local alignment implementations can be used to rapidly collect long (reverse) complementary regions by including G-U pairs (C-U or G-A for the reverse complement) in the scoring matrix (Gerlach and Giegerich, 2006; Thébault et al., 2014; Wenzel et al., 2012).
MFE methods form the majority of the RNA–RNA interaction prediction tools (Backofen and Hess, 2010; Dieterich and Stadler, 2012; Lorenz et al., 2011). Many secondary structure prediction tools also utilize MFE methods (Lorenz et al., 2011; Markham and Zuker, 2008; Mathews and Turner, 2006; Zuker and Sankoff, 1984). Some MFE methods including RNAhybrid (Rehmsmeier et al., 2004), RNAduplex (Lorenz et al., 2011), DuplexFold (Reuter and Mathews, 2010) and TargetRNA (Tjaden, 2008) neglect intramolecular structures for the sake of algorithmic speed. Algorithms like Pairfold (Andronescu et al., 2005), RNAcofold (Bernhart et al., 2006) and bifold (Reuter and Mathews, 2010) take intramolecular base-pairing into account. RNAup (Mückstein et al., 2006), RNAplex (Tafer and Hofacker, 2008) and IntaRNA (Busch et al., 2008) compute the accessibility of binding regions to report the final MFE of the RNA duplex, which is considered more realistic biophysically (Richter and Backofen, 2012). AccessFold includes accessibility using a method defined as pseudo-energy minimization (DiChiacchio et al., 2015). BistaRNA also includes accessibility and can predict multiple binding sites (Poolsap et al., 2011). Lastly, tools like TargetRNA2 (Kery et al., 2014), CopraRNA (Wright et al., 2013), miRanda (John et al., 2004), TargetScan (Lewis et al., 2005), PETcofold (Seemann et al., 2011) and DIANA-microT (Kiriakidou et al., 2004) exploit homology and evolutionary conservation to predict interactions
Some RNA–RNA interaction prediction tools are developed to achieve a specific task or to predict very specific group of interactions. For example, PLEXY is designed for C/D snoRNAs (Kehr et al., 2011), RNAsnoop (Tafer et al., 2010) for H/ACA snoRNAs and TargetRNA (Tjaden, 2008) for bacterial sRNAs (E. coli and Salmonella). In this study, we tried to assess the versatility of prediction tools on different datasets as well as their prediction power where applicable. We excluded tools designed for specific RNA families such as specialized miRNA algorithms (reviewed in Witkos et al., 2011), specialized snoRNA target prediction algorithms and comparative bacterial sRNA prediction methods (reviewed in Backofen and Hess, 2010, Pain et al., 2015). We also excluded inteRNA (Alkan et al., 2006), IRIS (Pervouchine, 2004), piRNA (Chitsaz et al., 2009b) and biRNA (Chitsaz et al., 2009a), as they are either no longer supported or obsolete.
In summary, our final list of selected tools used for further analyses consisted of RIsearch (Wenzel et al., 2012), IntaRNA (Busch et al., 2008), RNAcofold (Bernhart et al., 2006), RNAhybrid (Rehmsmeier et al., 2004), RNAduplex (Lorenz et al., 2011), RNAplex (Tafer and Hofacker, 2008), RNAup (Mückstein et al., 2006), pairfold (Andronescu et al., 2005), bifold (Reuter and Mathews, 2010), DuplexFold (Reuter and Mathews, 2010), ssearch (Pearson, 1991), RactIP (Kato et al., 2010), bistaRNA (Poolsap et al., 2011), AccessFold (DiChiacchio et al., 2015) and NUPACK (Dirks et al., 2007) (Supplementary Table S2).
3.2 Overall prediction performances
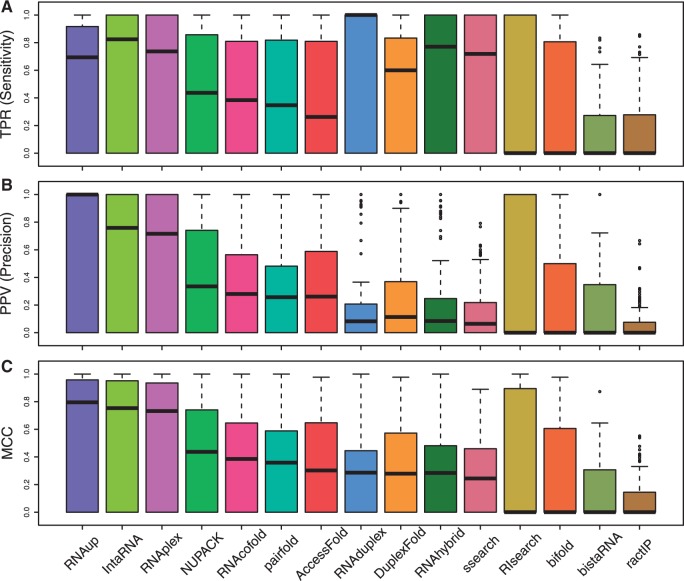
Our analyses of the overall performances of RNA interaction prediction algorithms show that three accessibility based algorithms (RNAup, IntaRNA and RNAplex) scored highest for sensitivity and precision. RNAup was highly precise compared to other tools (Fig. 1 and Table 1). IntaRNA was the second algorithm (almost identical to RNAup) with a reasonable running time. RNAplex was comparable to both algorithms. RNAduplex had the best overall TPR score, but it was not as precise as IntaRNA. Table 1 summarizes the ’cumulative’ TPR, PPV and MCC scores, while Figure 1 shows their distribution for all interactions (n = 154) on all domains of life.
Fig. 1.
The distribution of scores for RNA–RNA interaction prediction algorithms. (A) RNAduplex gave the highest median TPR (sensitivity) followed by IntaRNA. (B) RNAup was the most precise algorithm based on PPV score followed by the other accessibility based methods IntaRNA and RNAplex. (C) RNAup was the best prediction algorithm based on median MCC score, with IntaRNA and RNAplex giving similar scores. RactIP produced the worst overall MCC (Color version of this figure is available at Bioinformatics online.)
Table 1.
Total run time of algorithms, and the cumulative TPR, PPV and MCC scores
| Algorithm | Total run time (s) on | TPR | PPV | MCC |
|---|---|---|---|---|
| selected files (n = 50) | (Sensitivity) | (Precision) | ||
| AccessFold | 596.44 | 0.38 | 0.31 | 0.35 |
| bifold | 404.63 | 0.37 | 0.31 | 0.34 |
| bistaRNA | 102.29 | 0.15 | 0.16 | 0.15 |
| DuplexFold | 5.33 | 0.48 | 0.17 | 0.29 |
| IntaRNA | 24.44 | 0.59 | 0.56 | 0.58 |
| NUPACK | 794.2 | 0.42 | 0.42 | 0.42 |
| pairfold | 90.24 | 0.39 | 0.29 | 0.34 |
| ractIP | 87.62 | 0.16 | 0.06 | 0.1 |
| RIsearch | 4.16 | 0.36 | 0.45 | 0.40 |
| RNAcofold | 15.28 | 0.41 | 0.32 | 0.36 |
| RNAduplex | 6.45 | 0.66 | 0.12 | 0.27 |
| RNAhybrid | 32.84 | 0.56 | 0.12 | 0.26 |
| RNAplex | 17.19 | 0.55 | 0.57 | 0.56 |
| RNAup | 137.48 | 0.51 | 0.69 | 0.60 |
| ssearch | 4.69 | 0.56 | 0.1 | 0.23 |
The cumulative scores (i.e. TPR, PPV, MCC) are calculated by adding individual TP, FP and FN values for all predictions.
RIsearch and ssearch were the fastest methods, but they were not very sensitive or precise (Table 1). AccessFold and bifold had the longest run time, which appeared to increase for long RNA sequences like ribosomal RNAs or large target UTR regions. RIsearch and bifold gave inconsistent results, with combined MCCs of 0.33 and 0.40 respectively (Table 1). However, if we use a distribution of results as in Figure 1, the median MCCs appear to be zero for these algorithms. As bifold frequently returned no duplex structures for some RNA pairs (e.g. C. elegans miRNAs lin-4, lsy-6-3p, etc.), and RIsearch produced many unsuccessful predictions for bacterial sRNAs, which produced to zero MCC scores for both.
3.3 The significance test results of bacterial dataset
The MFE values produced by the algorithms are not very explicit, so it is common to use negative controls to determine the significance of predicted energy values (Rehmsmeier et al., 2004), especially for structure predictions (Workman and Krogh, 1999). As described in materials and methods, we created a set of negative controls for each native RNA–RNA interaction. Some algorithms were excluded from this assessment, because either they do not produce a score (i.e. RactIP, bistaRNA and ssearch) or are biased towards internal structures (i.e. pairfold, RNAcofold, bifold and NUPACK). Thus, the test of significance includes only 8 prediction algorithms (Table 2).
Table 2.
The test of significance results of selected algorithms on bacterial sRNAs.
| Algorithm | Total # of significant (P < 0.05) correct predictions for Gumbel dist. (n = 60) | Total # of significant (P < 0.05) correct predictions for normal dist. (n = 60) | Median rank of native interactions |
|---|---|---|---|
| AccessFold | 15 | 17 | 41.75 |
| DuplexFold | 2 | 8 | 63.5 |
| IntaRNA | 23 | 26 | 19 |
| RIsearch | 13 | 14 | 52.25 |
| RNAduplex | 8 | 11 | 54.25 |
| RNAhybrid | 5 | 6 | 76 |
| RNAplex | 23 | 30 | 10.5 |
| RNAup | 28 | 29 | 13.5 |
Higher is better for the second and third columns. Lower is better for the fourth column.
These results show that RNAplex and RNAup reported almost half of the native energies as significant if they are fitted to normal distributions. It seems the Gumbel fitting of scores is more conservative which likely decreases the risk of FP predictions on high-throughput predictions. RNAup results were almost identical for both distributions. IntaRNA performed slightly worse than these two algorithms. The last column of Table 2 shows the median rank of native interactions. If a prediction score of a native interaction has the highest score (e.g. lowest MFE), it is ranked 1 out of 201. Therefore, the median ranks in the last column can be interpreted as the expected number of FPs introduced by the algorithms before predicting the native interaction.
3.4 A summary of RNA–RNA interactions and algorithm performances for all domains of life
Eukaryotic RNA interactions mostly focus on RNA interference (RNAi) (i.e. miRNAs and siRNAs) (Ambros, 2004; Carthew and Sontheimer, 2009; Chen, 2008). In animal RNAi, miRNAs (∼20 nts long) prefer perfect complementarity in the seed region and have overall lower complementarity than plant counterparts (Ameres and Zamore, 2013; Axtell et al., 2011). In plants, high complementary target regions may lie in coding region as well as UTRs rather than only 3′UTRs (Ameres and Zamore, 2013; Axtell et al., 2011; Millar and Waterhouse, 2005). It is possible for a miRNA to target more than one region, especially in animals, which is known to increase efficiency of target gene downregulation (Millar and Waterhouse, 2005). However, in our benchmark we preferred to select miRNA targets containing a single designated binding region. Piwi associated piRNAs are also small endogenous RNAs (24–30 nts long) (Klattenhoff and Theurkauf, 2008; Zhang et al., 2015), some of which use antisense binding to regulate target RNAs (Gou et al., 2015) like miRNA and siRNA. H/ACA and C/D snoRNAs have roles in rRNA and snRNA maturation (Brown et al., 2001; Gardner et al., 2010; Kiss, 2002). These interactions differ in that C/D snoRNAs prefer a binding region on target RNA with consecutive nts around 7–20 bases long with a few mismatches (Gardner et al., 2010; Kehr et al., 2011), while H/ACA snoRNAs contain a stem loop within the binding region, which complicates target prediction (Gardner et al., 2010; Kiss et al., 2004; Tafer et al., 2010). Spliceosomal snRNAs form ribonucleoprotein (RNP) complexes with other snRNAs (Karijolich and Yu, 2010), and they are also targeted by snoRNAs (termed scaRNAs) (Darzacq et al., 2002). We included examples of both snRNA-snRNA and scaRNA-snRNA interactions to our dataset. It is also known that some lncRNAs use RNA–RNA interactions (Kung et al., 2013) but these were not included in our benchmark.
We found that in the eukaryotic dataset, accessibility based methods performed best based on the average MCC scores (except AccessFold and bistaRNA) (Fig. 2). IntaRNA (av. MCC: 0.51) slightly outperformed RNAup (av. MCC: 0.49) and produced a higher PPV than the other tools benchmarked. RNAplex (av. MCC: 0.48) and RIsearch (av. MCC: 0.48) (an alignment-like method) were also comparable with these two algorithms for eukaryotic datasets. Supplementary Table S3 explicitly shows the prediction scores for all 88 eukaryotic interactions.
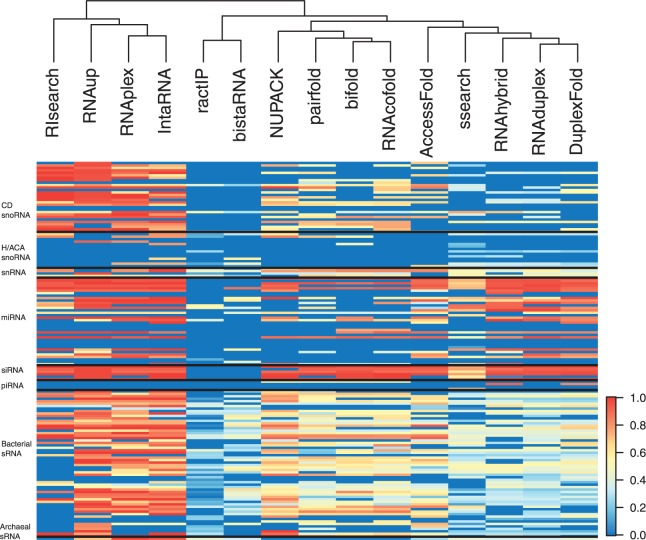
Fig. 2.
This heatmap shows MCC values of each tool for entire dataset. The red cells display a higher MCC value denoting a better prediction. Similar methods are mostly clustered together based on these predictions (dendrogram at top). Row labels show the type of interactions. Predictions for the single archaeal sRNA are on the last row. An in depth examination of these results show that the algorithms are poor at predicting human miRNA-mRNA interactions (av. MCC: 0.22), snoRNAs (weaker for H/ACA as expected, av. MCC: 0.09), mouse piRNAs (av. MCC: 0.07). Conversely, they perform best on Arabidopsis miRNAs (av. MCC: 0.72), siRNAs (av. MCC: 0.71) and bacterial sRNAs (av. MCC: 0.40), which is most likely an effect of high complementarity in binding regions for these
Bacterial small RNAs can be divided into three major types: antisense binding sRNAs, Hfq dependent sRNAs and csrA binding sRNAs (Storz et al., 2011; Vogel, 2009). However, in this study, bacterial sRNAs refer to either antisense or Hfq dependent sRNAs, which achieve their role via RNA–RNA base-pairing interactions. Bacterial sRNAs (50–200 nts long) prefer short binding regions relative to their size (Storz et al., 2011; Vogel, 2009). This was also true for our dataset, with an average binding region size of 23 nts, with the smallest just 7 nts long (Supplementary Table S1). Model bacterial organisms like E. coli or Salmonella contain hundreds of different sRNAs which points to a complex regulatory system in prokaryotic organisms (Waters and Storz, 2009). Moreover, increasing number of RNA-seq studies (Cohen et al., 2016; Sharma and Vogel, 2014; Sharma et al., 2010) reveal that there are novel regulatory ncRNAs are spanning in prokaryotes than previously anticipated (Barquist and Vogel, 2015; Chen et al., 2016; Lindgreen et al., 2014).
We found that in the bacterial dataset, accessibility based methods performed better than the others based on the average MCC scores, as with the eukaryotic dataset. RNAup (av. MCC: 0.68) slightly outperformed IntaRNA (av. MCC: 0.65) in bacterial sRNA interactions. RNAplex (av. MCC: 0.61) was comparable with the other two algorithms. In bacterial dataset, RIsearch (av. MCC: 0.31) did not perform as well as on the eukaryotic dataset, which decreased the overall performance (Fig. 2).
RNA interactions in archaea are not well characterized. Recent studies have shown that archaeal genomes contain a large number of ncRNA repositories similar to bacterial genomes (Lindgreen et al., 2014). Unfortunately, there are not many verified RNA interactions available in archaea, except archaeal snoRNAs. Archaeal genomes mostly contain C/D box snoRNAs; thus, we added 5 C/D box snoRNAs (Omer et al., 2000) and one archaeal sRNA (Jäger et al., 2012) as an archaeal benchmark dataset. The archaeal sRNA targets a bicistronic gene and trans-regulates expression of two protein coding genes concurrently (Jäger et al., 2012) (Figs 1 and 2 and Supplementary Table S3).
We found that in the archaeal dataset, RNAplex (av. 0.65) performed better than the other algorithms, followed by IntaRNA (av. MCC: 0.61). These two algorithms were followed by RNAup (av. MCC: 0.53) and RIsearch (av. MCC: 0.40). RIsearch was better on snoRNA predictions than the single archeal sRNA, which reduced the average overall performance. RNAplex recovered the binding region with a perfect MCC score, followed by IntaRNA.
3.5 Limitations of RNA–RNA interaction predictions algorithms
Unfortunately, 15 out of 154 RNA interaction pairs in our benchmark dataset could not be correctly predicted by any of the algorithms (i.e. an MCC score of 0 for all algorithms) (Fig. 2 and Supplementary Table S3) including 6 human miRNAs, and snoRNAs from yeast, human and archaea. The mouse piRNA results were also unsatisfactory, and one (piR-013474) could not be detected by any of the algorithms. The algorithms benchmarked performed best on Arabidopsis miRNAs, siRNAs and bacterial sRNAs (Fig. 2).
We applied the significance test to some of these failed eukaryotic interactions (e.g. mouse piRNAs, human miRNAs), aiming to see whether the predicted scores enabled the detection of true interactions (and separate scores for native interactions from background) rather than using correctly predicted binding regions. The comparison of two methods revealed consistent results as expected. For example, the native interaction of piR-013474 cannot be differentiated from background by any algorithm. This is also similar for other piRNAs and human miRNAs, where all algorithms consistently failed.
The lengths of target RNA regions (which include binding regions) seem to influence prediction quality (also discussed by Lai and Meyer, 2015). The average length of a eukaryotic target RNA is 1690 nts long in our dataset. However, this rises to around 2400 nts for those miRNAs which did not give prediction scores, and longer in piRNAs. As described in materials and methods, we did not truncate the targets (e.g. UTRs) that contained binding regions. We found a significant reverse correlation (Pearson’s r = -0.28, p < 0.05) between the lengths of target RNAs and average MCCs (i.e. overall performances). However, some of the algorithms (RNAup, RNAplex, RIsearch, RNAcofold and NUPACK) are less prone to this length bias (p > 0.05) (Supplementary Table S4), making them ideal for use on untruncated targets.
Another explanation for inadequate prediction may be the quality of the dataset. Not all experimental protocols are equally strong at detecting correct binding regions, functional characterization or identifying new targets (Chou et al., 2015; Kuhn et al., 2008; Thomson et al., 2011; Vogel and Wagner, 2007). However, the incorrectly predicted human miRNAs (hsa-miR-21-5p, hsa-miR-29b-3p, etc.) were validated by relatively strong evidence (Chou et al., 2015), which could rule out this explanation.
RNA structure prediction (and also RNA–RNA interaction prediction) algorithms are based on biophysical assumptions where the influence of tertiary interactions and other factors are neglected (Mathews, 2006; Mathews and Turner, 2006; Wuchty et al., 1999). RNA structures with the lowest free energy may not be the biologically active form, which may have multiple different conformations with different MFEs (Mathews, 2006; Mathews and Turner, 2006). Many algorithms ignore computationally expensive RNA structures (e.g. pseudoknots) (Do et al., 2006; Hofacker et al., 1994; Lorenz et al., 2011). MFE methods also become inaccurate with longer RNA sequences (Lai and Meyer, 2015; Lange et al., 2012; Mathews and Turner, 2006; Meyer, 2008). RNA interaction prediction algorithms generally do not consider multiple binding regions—only a few of which such as bistaRNA and ractIP, include multiple binding positions in their model (Kato et al., 2010; Poolsap et al., 2011). Cellular dynamics (i.e. interaction with other molecules, ion concentrations, etc.) can influence RNA structures (Onoa and Tinoco, 2004) and RNA interactions (Meyer, 2008; Mückstein et al., 2006), which is hard to factor into prediction models.
The ssearch tool uses the Smith-Waterman algorithm (Pearson and Lipman, 1988) and is the only pure alignment tool in our benchmark, although it is possible to use similar tools, such as BLAST or Blat, to extract complementary regions for high-throughput predictions. Once the gap penalty and scoring matrix parameters were tweaked to make it more suitable for RNA–RNA interaction prediction, ssearch was quite successful and even comparable with some MFE methods (e.g. RNAhybrid and DuplexFold) (Fig. 1).
Those MFE methods that include internal structures (e.g. pairfold, RNAcofold, bifold, NUPACK) are biased towards internal structures as many ncRNAs have stable internal structures (Clote et al., 2005). Therefore, using negative controls may lead to false significant predictions due to internal structures of interacting partners, giving misleading MFE scores. We also observed this effect in our predictions (data not shown), and so excluded those algorithms from the significance test. They also have relatively slow running times, and some have problems utilizing memory (e.g. bifold). NUPACK is the best among this type of prediction methods and RNAcofold is the fastest (Table 1).
It is apparent that the algorithms do not necessarily perform equally for all types of RNA–RNA interactions, and it is better to select algorithms appropriate to the input dataset. For example, RIsearch is fast and accurate for eukaryotic datasets, and would be suitable for high throughput predictions which can be combined statistical significance testing of the predicted scores. IntaRNA and RNAplex seem to be reliable and relatively fast for all datasets. RNAup is precise and less prone to length bias (Supplementary Table S4).
4 Conclusion
Here we present one of the most comprehensive benchmark of RNA–RNA interaction prediction methods that covers almost all RNA–RNA interactions in RNA biology. We extended the previous work (Lai and Meyer, 2015; Pain et al., 2015) by including all types of RNA–RNA interactions and the latest algorithms (DiChiacchio et al., 2015) in the RNA interaction prediction field. We have included a test to determine the statistical significance of the predicted scores by each algorithm. We have also reported that increasing length of target RNAs which contain binding regions also negatively influences overall prediction quality (Supplementary Table S4).
Three accessibility based algorithms, RNAup, IntaRNA and RNAplex, performed best for all types of interactions. We found that the accessibility based MFE methods could also differentiate almost half of the native interactions from background in our bacterial dataset (Table 2). Therefore, carefully designed negative controls (e.g. dinucleotide shuffling) allow for the use of predicted MFE values and separate scores for native interactions from the background. This makes the accessibility algorithms ideal tools for de novo predictions, especially those with smaller run-times such as IntaRNA and RNAplex, since candidate target RNAs can be thousands of nts long. RNAplex is also effective on detecting correct interaction regions buried in larger RNA targets (Results and Supplementary Table S4).
RNA interaction prediction is still an expanding field. Advances in sequencing technology has unveiled a vast number of novel uncharacterized ncRNA transcripts in different clades of life. These methods are also showing that many ncRNAs utilize RNA–RNA interactions (Kudla et al., 2011; Lu et al., 2016; Sharma et al., 2016) which makes RNA target prediction an important asset to determine functions of novel genes. Comparative methods are becoming popular (Lai and Meyer, 2015; Pain et al., 2015; Seemann et al., 2011; Wright et al., 2013), and may increase the prediction accuracy (Pain et al., 2015; Wright et al., 2013). However, some other results suggest that there is little to be gained from comparative approaches for predicting interactions (Lai and Meyer, 2015; Richter and Backofen, 2012) due to low conservation of many ncRNAs (Lindgreen et al., 2014). Unfortunately, most of the verified interactions in the RNA literature still belong to model species (human, C. elegans, Arabidopsis and E. coli, etc.) which also raises the risk of overfitting results to a modest numbers of known interactions. Weak prediction rates for piRNAs may suggest inadequacy of prediction methods for novel regulatory RNAs, but even well-known miRNA interaction predictions have failed to be detected by any of the algorithms benchmarked (Fig. 2). Archaeal regulatory systems are also not well studied, and only a handful of archaeal sRNAs have been identified. Therefore, non-comparative methods are still a robust way to produce ab initio interaction predictions. Our benchmark will help researchers to find an appropriate algorithm for functional annotation of unknown transcripts or a basis from which to improve or develop new methods. Our scripts and datasets are publicly available at Github (github.com/UCanCompBio/RNA_Interactions_Benchmark).
Funding
SUU is supported by a Biomolecular Interaction Centre and UC HPC (Bluefern) joint PhD Scholarship from the University of Canterbury. PPG is supported by Rutherford Discovery Fellowships, administered by the Royal Society of New Zealand.
Supplementary Material
Acknowledgments
We also thank Lars Barquist for valuable discussions and comments. We also thank Bethany Jose for extensive comments and proofreading.
Conflict of Interest: none declared.
References
- Addo-Quaye C. et al. (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol., 18, 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C. et al. (2006) RNA–RNA interaction prediction and antisense RNA target search. J. Comput. Biol., 13, 267–282. [DOI] [PubMed] [Google Scholar]
- Altschul S.F. et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ambros V. (2004) The functions of animal microRNAs. Nature, 431, 350–355. [DOI] [PubMed] [Google Scholar]
- Ameres S.L., Zamore P.D. (2013) Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol., 14, 475–488. [DOI] [PubMed] [Google Scholar]
- Andronescu M. et al. (2005) Secondary structure prediction of interacting RNA molecules. J. Mol. Biol., 345, 987–1001. [DOI] [PubMed] [Google Scholar]
- Axtell M.J. et al. (2011) Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol., 12, 221.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backofen R., Hess W.R. (2010) Computational prediction of sRNAs and their targets in bacteria. RNA Biol., 7, 33–42. [DOI] [PubMed] [Google Scholar]
- Barquist L., Vogel J. (2015) Accelerating discovery and functional analysis of small RNAs with new technologies. Annu. Rev. Genet, 49, 367–394. [DOI] [PubMed] [Google Scholar]
- Bernhart S.H. et al. (2006) Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol. Biol., 1, 3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D. et al. (2011) CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet., 45, 273–297. [DOI] [PubMed] [Google Scholar]
- Brennecke J. et al. (2007) Discrete small RNA-generating loci as master regulators of transposon activity in drosophila. Cell, 128, 1089–1103. [DOI] [PubMed] [Google Scholar]
- Brown J.W. et al. (2001) Multiple snoRNA gene clusters from Arabidopsis. RNA, 7, 1817–1832. [PMC free article] [PubMed] [Google Scholar]
- Busch A. et al. (2008) IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics, 24, 2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. et al. (2010) sRNATarBase: a comprehensive database of bacterial sRNA targets verified by experiments. RNA, 16, 2051–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. (2009) Origins and mechanisms of miRNAs and siRNAs. Cell, 136, 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.H. et al. (2016) Integration of multi-omics data of a genome-reduced bacterium: Prevalence of post-transcriptional regulation and its correlation with protein abundances. Nucleic Acids Res., 44, 1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2008) MicroRNA metabolism in plants. Curr. Top. Microbiol. Immunol., 320, 117–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitsaz H. et al. (2009a). biRNA: fast RNA–RNA binding sites prediction In: Salzberg S.L., Warnow T. (eds.) Algorithms in Bioinformatics, Lecture Notes in Computer Science, pages 25–36. Springer, Berlin, Heidelberg. [Google Scholar]
- Chitsaz H. et al. (2009b) A partition function algorithm for interacting nucleic acid strands. Bioinformatics, 25, i365–i373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.H. et al. (2015) miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res, 44, D239–D247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O. et al. (2016) Comparative transcriptomics across the prokaryotic tree of life. Nucleic Acids Res, 44, W46–W53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clote,P. et al. (2005) Structural RNA has lower folding energy than random RNA of the same dinucleotide frequency. RNA, 11, 578–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T. et al. (2011) Evolution and functional diversification of MIRNA genes. Plant Cell, 23, 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X. et al. (2002) Cajal body-specific small nuclear RNAs: a novel class of 2’-o-methylation and pseudouridylation guide RNAs. EMBO J., 21, 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleavey G.F., Damha M.J. (2012) Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol., 19, 937–954. [DOI] [PubMed] [Google Scholar]
- DiChiacchio L. et al. (2015) AccessFold: predicting RNA–RNA interactions with consideration for competing Self-Structure. Bioinformatics, 32, 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C., Stadler P.F. (2012) Computational Biology of RNA Interactions. Wiley Interdiscip. Rev. RNA, 4, 107–120. [DOI] [PubMed] [Google Scholar]
- Dirks R.M. et al. (2007) Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev., 49, 65–88. [Google Scholar]
- Do C.B. et al. (2006) CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics, 22, e90–e98. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. (2011) Accelerated profile HMM searches. PLoS Comput. Biol., 7, e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner P.P. et al. (2010) SnoPatrol: how many snoRNA genes are there?. J. Biol., 9, 4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach W., Giegerich R. (2006) GUUGle: a utility for fast exact matching under RNA complementary rules including G–U base pairing. Bioinformatics, 22, 762–764. [DOI] [PubMed] [Google Scholar]
- Gilbert W. (1986) Origin of life: The RNA world. Nature, 319, 618. [Google Scholar]
- Gorodkin J. et al. (2001) Discovering common stem–loop motifs in unaligned RNA sequences. Nucleic Acids Res., 29, 2135–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. (2004) The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu. Rev. Microbiol., 58, 303–328. [DOI] [PubMed] [Google Scholar]
- Gou L.T. et al. (2015) Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res., 25, 266.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güell M. et al. (2009) Transcriptome complexity in a genome-reduced bacterium. Science, 326, 1268–1271. [DOI] [PubMed] [Google Scholar]
- Güell M. et al. (2011) Bacterial transcriptomics: what is beyond the RNA horiz-ome?. Nat. Rev. Microbiol., 9, 658–669. [DOI] [PubMed] [Google Scholar]
- Gumbel E.J. (1958). Statistics of Extremes. 1958. Columbia Univ. Press, New York. [Google Scholar]
- Hodas N.O., Aalberts D.P. (2004) Efficient computation of optimal oligo-RNA binding. Nucleic Acids Res., 32, 6636–6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L. et al. (1994) Fast folding and comparison of RNA secondary structures. Monatsh. Chem., 125, 167–188. [Google Scholar]
- Holmqvist E., Vogel J. (2013) A small RNA serving both the hfq and CsrA regulons. Genes Dev., 27, 1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger D. et al. (2012) An archaeal sRNA targeting cis- and trans-encoded mRNAs via two distinct domains. Nucleic Acids Res., 40, 10964–10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B. et al. (2004) Human MicroRNA targets. PLoS Biol., 2, e363.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J., Yu Y.T. (2010) Spliceosomal snRNA modifications and their function. RNA Biol., 7, 192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y. et al. (2010) RactIP: fast and accurate prediction of RNA–RNA interaction using integer programming. Bioinformatics, 26, i460–i466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr S. et al. (2011) PLEXY: efficient target prediction for box C/D snoRNAs. Bioinformatics, 27, 279–280. [DOI] [PubMed] [Google Scholar]
- Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kery M.B. et al. (2014) TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res., 42, W124–W129., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C.A., Martienssen R.A. (2005) The developmental role of microRNA in plants. Curr. Opin. Plant Biol., 8, 38–44. [DOI] [PubMed] [Google Scholar]
- Kiriakidou M. et al. (2004) A combined computational-experimental approach predicts human microRNA targets. Genes Dev., 18, 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss A.M. et al. (2004) Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol., 24, 5797–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2002) Small nucleolar RNAs. Cell, 109, 145–148. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C., Theurkauf W. (2008) Biogenesis and germline functions of piRNAs. Development, 135, 3–9. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2013) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res., gkt1181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G. et al. (2011) Cross-linking, ligation, and sequencing of hybrids reveals RNA–RNA interactions in yeast. Proc. Natl. Acad. Sci. U. S. A., 108, 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn D.E. et al. (2008) Experimental validation of miRNA targets. Methods, 44, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung J.T.Y. et al. (2013) Long noncoding RNAs: past, present, and future. Genetics, 193, 651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D., Meyer I.M. (2015) A comprehensive comparison of general RNA–RNA interaction prediction methods. Nucleic Acids Res, 44, e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S.J. et al. (2012) Global or local? Predicting secondary structure and accessibility in mRNAs. Nucleic Acids Res., 40, 5215–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L., Weber M.J. (2006) snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res., 34, D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P. et al. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Lindgreen S. et al. (2014) Robust identification of noncoding RNA from transcriptomes requires phylogenetically-informed sampling. PLoS Comput. Biol., 10, e1003907.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R. et al. (2011) ViennaRNA package 2.0. Algorithms Mol. Biol., 6, 26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. et al. (2016) RNA duplex map in living cells reveals Higher-Order transcriptome structure. Cell, 165, 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham N.R., Zuker M. (2008) UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol., 453, 3–31. [DOI] [PubMed] [Google Scholar]
- Mathews D.H. (2006) Revolutions in RNA secondary structure prediction. J. Mol. Biol., 359, 526–532. [DOI] [PubMed] [Google Scholar]
- Mathews D.H., Turner D.H. (2006) Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol., 16, 270–278. [DOI] [PubMed] [Google Scholar]
- Matthews B.W. (1975) Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta, 405, 442–451. [DOI] [PubMed] [Google Scholar]
- Mattick J.S. (2004) RNA regulation: a new genetics?. Nat. Rev. Genet., 5, 316–323. [DOI] [PubMed] [Google Scholar]
- Mattick J.S. (2009) The genetic signatures of noncoding RNAs. PLoS Genet., 5, e1000459.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill J.S. (1990) The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers, 29, 1105–1119. [DOI] [PubMed] [Google Scholar]
- Meyer I.M. (2008) Predicting novel RNA–RNA interactions. Curr. Opin. Struct. Biol., 18, 387–393. [DOI] [PubMed] [Google Scholar]
- Millar A.A., Waterhouse P.M. (2005) Plant and animal microRNAs: similarities and differences. Funct. Integr. Genomics, 5, 129–135. [DOI] [PubMed] [Google Scholar]
- Mückstein U. et al. (2006) Thermodynamics of RNA–RNA binding. Bioinformatics, 22, 1177–1182. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Jacobson A.B. (1980) Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc. Natl. Acad. Sci. U. S. A., 77, 6309–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer A.D. et al. (2000) Homologs of small nucleolar RNAs in archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- O’Neil D. et al. (2013) Ribosomal RNA depletion for efficient use of RNA-Seq capacity. Curr. Protoc. Mol. Biol., DOI: 10.1002/0471142727.mb0419s103. [DOI] [PubMed] [Google Scholar]
- Onoa B., Tinoco I. Jr, (2004) RNA folding and unfolding. Curr. Opin. Struct. Biol., 14, 374–379. [DOI] [PubMed] [Google Scholar]
- Oğul H. et al. (2011) A probabilistic approach to microRNA-target binding. Biochem. Biophys. Res. Commun., 413, 111–115. [DOI] [PubMed] [Google Scholar]
- Pain A. et al. (2015) An assessment of bacterial small RNA target prediction programs. RNA Biol., 12, 509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W.R. (1991) Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics, 11, 635–650. [DOI] [PubMed] [Google Scholar]
- Pearson W.R., Lipman D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U. S. A., 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer A., Margalit H. (2011) Accessibility and evolutionary conservation mark bacterial small-rna target-binding regions. J. Bacteriol., 193, 1690–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. (2012) Genomics. ENCODE project writes eulogy for junk DNA. Science, 337, 1159, 1161.. [DOI] [PubMed] [Google Scholar]
- Pervouchine D.D. (2004) IRIS: intermolecular RNA interaction search. Genome Inform., 15, 92–101. [PubMed] [Google Scholar]
- Piekna-Przybylska D. et al. (2007) New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA, 13, 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolsap U. et al. (2011) Using binding profiles to predict binding sites of target RNAs. J. Bioinform. Comput. Biol., 9, 697–713. [DOI] [PubMed] [Google Scholar]
- Prasse D. et al. (2013) Regulatory RNAs in archaea: first target identification in methanoarchaea. Biochem. Soc. Trans., 41, 344–349. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M. et al. (2004) Fast and effective prediction of microRNA/target duplexes. RNA, 10, 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter J.S., Mathews D.H. (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinform., 11, 129.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. et al. (2004) Rational siRNA design for RNA interference. Nat. Biotechnol., 22, 326–330. [DOI] [PubMed] [Google Scholar]
- Richter A.S., Backofen R. (2012) Accessibility and conservation: General features of bacterial small RNA-mRNA interactions?. RNA Biol., 9, 954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann S.E. et al. (2011) PETcofold: predicting conserved interactions and structures of two multiple alignments of RNA sequences. Bioinformatics, 27, 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C.M., Vogel J. (2014) Differential RNA-seq: the approach behind and the biological insight gained. Curr. Opin. Microbiol., 19, 97–105. [DOI] [PubMed] [Google Scholar]
- Sharma C.M. et al. (2010) The primary transcriptome of the major human pathogen helicobacter pylori. Nature, 464, 250–255. [DOI] [PubMed] [Google Scholar]
- Sharma E. et al. (2016) Global mapping of human RNA–RNA interactions. Mol. Cell, 62, 618–626. [DOI] [PubMed] [Google Scholar]
- Storz G. et al. (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell, 43, 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafer H., Hofacker I.L. (2008) RNAplex: a fast tool for RNA–RNA interaction search. Bioinformatics, 24, 2657–2663. [DOI] [PubMed] [Google Scholar]
- Tafer H. et al. (2010) RNAsnoop: efficient target prediction for H/ACA snoRNAs. Bioinformatics, 26, 610–616. [DOI] [PubMed] [Google Scholar]
- Thébault P. et al. (2014) Advantages of mixing bioinformatics and visualization approaches for analyzing sRNA-mediated regulatory bacterial networks. Brief. Bioinform, 16, 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson D.W. et al. (2011) Experimental strategies for microRNA target identification. Nucleic Acids Res., 39, 6845–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B. (2008) TargetRNA: a tool for predicting targets of small RNA action in bacteria. Nucleic Acids Res., 36, W109–W113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umu S.U. et al. (2016) Avoidance of stochastic RNA interactions can be harnessed to control protein expression levels in bacteria and archaea. Elife. 5, e13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. (2009) A rough guide to the non-coding RNA world of salmonella. Mol. Microbiol., 71, 1–11. [DOI] [PubMed] [Google Scholar]
- Vogel J., Luisi B.F. (2011) Hfq and its constellation of RNA. Nat. Rev. Microbiol., 9, 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Wagner E.G.H. (2007) Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol., 10, 262–270. [DOI] [PubMed] [Google Scholar]
- Waters L.S., Storz G. (2009) Regulatory RNAs in bacteria. Cell, 136, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A. et al. (2012) RIsearch: fast RNA–RNA interaction search using a simplified nearest-neighbor energy model. Bioinformatics, 28, 2738–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. (2011) Spliceosome structure and function. Cold Spring Harb. Perspect. Biol., 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkos T.M. et al. (2011) Practical aspects of microRNA target prediction. Curr. Mol. Med., 11, 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C., Krogh A. (1999) No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res., 27, 4816–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P.R. et al. (2013) Comparative genomics boosts target prediction for bacterial small RNAs. Proc. Natl. Acad. Sci. U. S. A., 110, E3487–E3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchty S. et al. (1999) Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers, 49, 145–165. [DOI] [PubMed] [Google Scholar]
- Yang Y. et al. (2008) MiRTif: a support vector machine-based microRNA target interaction filter. BMC Bioinform., 9, S4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihama M. et al. (2013) snOPY: a small nucleolar RNA orthological gene database. BMC Res. Notes, 6, 426.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. et al. (2015) MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res., 25, 193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. (2016) NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res., 44, D203–D208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2000) Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol., 10, 303–310. [DOI] [PubMed] [Google Scholar]
- Zuker M., Sankoff D. (1984) RNA secondary structures and their prediction. Bltn. Mathcal. Biol., 46, 591–621. [Google Scholar]
- Zuker M., Stiegler P. (1981) Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res., 9, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.