Abstract
Eukaryotic translation initiation factor eIF5A promotes protein synthesis by resolving polyproline-induced ribosomal stalling. Here, we report a 3.25-Å resolution crystal structure of eIF5A bound to the yeast 80S ribosome. The structure reveals a previously unseen conformation of an eIF5A–ribosome complex and highlights a possible functional link between conformational changes of the ribosome during protein synthesis and the eIF5A–ribosome association.
Keywords: structure, crystallography, ribosome, eIF5A, hypusine
Introduction
The universally conserved translation factor eIF5A in eukaryotes and its bacterial homolog EF-P help ribosomes produce proteins containing three or more consecutive proline residues by resolving polyproline-induced stalling [1–3]. In eukaryotes, eIF5A activity depends on its unusual post-translationally modified residue hypusine, which is thought to augment the rate of peptide bond formation in the ribosomal catalytic center [4–6]. Structural studies of EF-P bound to the bacterial 70S ribosome from Thermus thermophilus and of eIF5A bound to the eukaryotic 80S ribosome from yeast showed that both EF-P and eIF5A bind the ribosome in the vicinity of the ribosomal catalytic center [1,7,8], positioning the critical hypusine residue (in the eIF5A structure) to contact the acceptor stem of the P-site tRNA [8]. Although these studies provided mechanistic insights into EF-P/eIF5A activity, in both structural studies the ribosome was in the same classical state conformation. Therefore, these studies left unexplored how eIF5A activity is related to ribosome dynamics during protein synthesis. In fact, during translation, the ribosomal subunits make rotation-like motions, which serve to coordinate tRNA and mRNA translocation between the active sites of the ribosome [9,10]. It is unknown whether changes in the ribosome conformation alter the structure the eIF5A-binding site and possibly facilitate eIF5A recruitment or release from the ribosome.
Structure of the eukaryotic ribosome bound to hypusinated eIF5A
Our interest in eIF5A arose from crystallographic studies of the eukaryotic ribosome. Curiously, we observed that 80S ribosomes purified from glucose-starved yeast cells carry two additional proteins—-stress-related factor Stm1 and eIF5A (Table S1) [11]. While Stm1 appeared in the electron density map, eIF5A was absent, possibly due to its dissociation during extensive post-crystallization treatments of the crystals (see Materials and Methods in Sup. Data). In this study, we modified the post-crystallization treatments and supplemented the soaking solutions with recombinant eIF5A. These changes have enabled us to determine the structure of hypusinated eIF5A from Saccharomyces cerevisiae bound to the yeast 80S ribosome at 3.25 Å resolution (I/σ = 1, Table 1; Fig. 1a).
Table 1.
Data collection and refinement statistics
| 80S/eIF5A–Hyp51 | 80S/eIF5A–Lys51 | |
|---|---|---|
| Data collection | ||
| No. of crystals | 3 | 2 |
| Space group | P21 | P21 |
| Unit cell | ||
| a, b, c (Å) | 435.63/286.45/303.41 | 438.23/289.33/305.47 |
| α = β = 90°, γ(°) | 98.95 | 98.92 |
| Resolution | 189.16–3.15 (3.25–3.15) | 190.48–3.25 (3.35–3.25) |
| Rmeas | 36.0 (212.0) | 43.4 (235.6) |
| I/σI | 6.54 (1.03) | 6.38 (0.98) |
| CC1/2 | 99.4 (39.6) | 99.5 (35.5) |
| Resolution at CC1/2 = 0.5 (Å) | 3.25 | 3.35 |
| Completeness (%) | 99.9 (99.7) | 99.9 (99.9) |
| Redundancy | 9.1 | 8.2 |
| Refinement | ||
| Resolution | 189.16–3.15 | 190.48–3.25 |
| No. reflections | 1,261,726 | 1,176,370 |
| Rwork/Rfree | 25.56/26.37 | 25.43/29.54 |
| No. of atoms | 407,909 | 407,903 |
| Protein | 184,678 | 185,006 |
| RNA and ions | 223,231 | 220,897 |
| B-factors | ||
| Protein | 85.5 | 103.9 |
| RNA and ions | 73.5 | 98.2 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.006 |
| Bond angles (Å) | 1.039 | 1.016 |
| eIF5A-occupancy refinement (%) | 97 | 88 |
| PDB ID | 5dat | 5dc3 |
Values in parenthesis show statistics of the highest resolution shell.
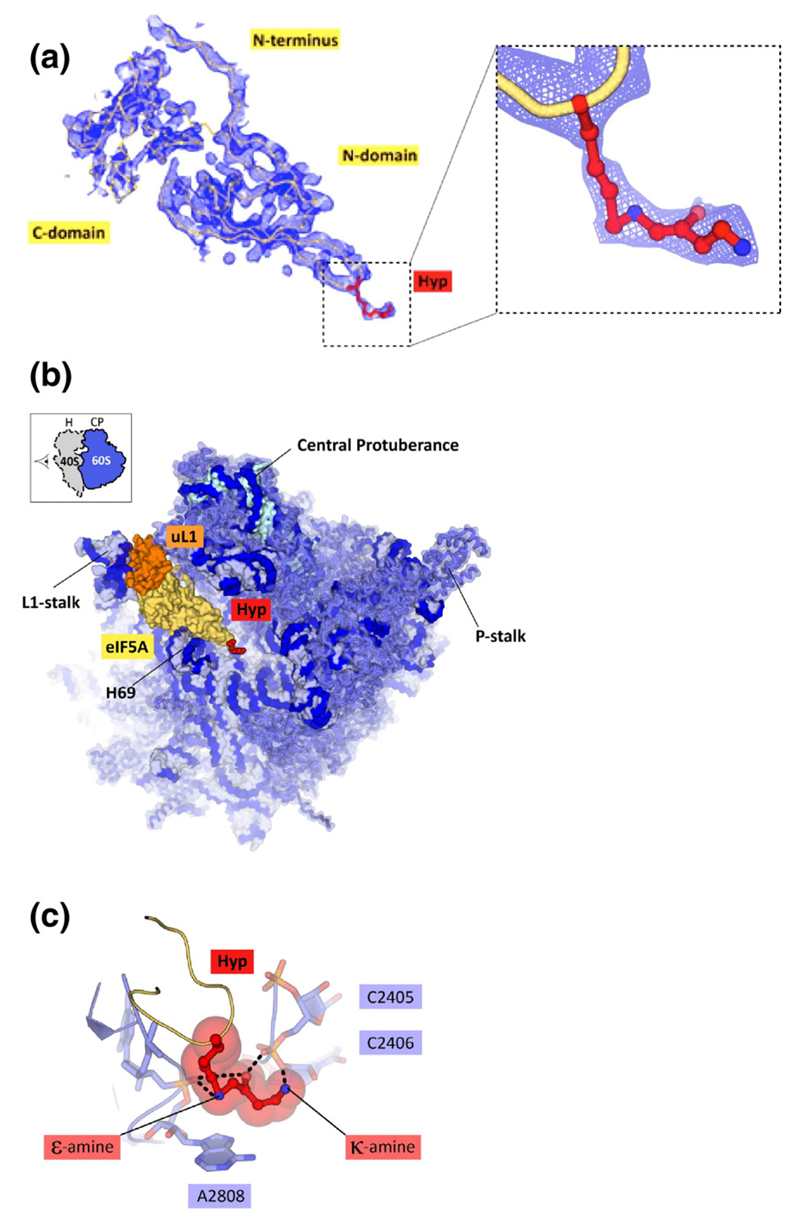
Fig. 1. Electron density map reveals ribosome-bound eIF5A. Views of the 80S ribosome/eIF5A structure.
(a) Unbiased Fo-Fc electron density map, contoured at 3σ, reveals the hypusine conformation and allows nearly complete modeling of ribosome-bound eIF5A. Yellow cartoon traces the Cα atoms, red sticks show hypusine. (b) View of the 60S ribosomal subunit shows the eIF5A-binding site and eIF5A interactions with two dynamic ribosome features—the L1-stalk and helix H69 of the 25S rRNA. The inset shows the view orientation. (c), The hypusine-binding site. The hypusine conformation is stabilized by extensive contacts with the 25S rRNA.
The electron density map revealed hypusinated eIF5A bound to one of two 80S ribosomes in the asymmetric unit of the crystal (Fig. 1a). Although our crystal structure lacks a P-site tRNA, we found that eIF5A occupies the same position as was observed in the 3.9 Å cryo-EM structure of the 80S ribosome complex with A- and P-site tRNAs [8] (Fig. 2a and S1). In this position, eIF5A binds adjacent to the P site, where it interacts with the intersubunit interface and two dynamic features of the large ribosomal subunit—the L1-stalk and helix H69 of the 25S rRNA (Fig. 1b). In addition, eIF5A forms several eukaryote-specific contacts with the large ribosomal subunit (Fig. S1). As in the cryo-EM structure of the 80S ribosome/eIF5A complex [8], the hypusine residue is located in direct vicinity of the ribosomal P site. The hypusine residue adopts a sharply bent conformation in which its positively charged κ-amino group is directed toward the CCA-end of the P-site tRNA, pointing between the 2′- and 3′-OH groups on the C75 ribose (Fig. S2). This conformation of hypusine is similar to the hypusine conformation in the cryo-EM structure of the 80S ribosome/eIF5A complex [8], although the κ-amino group is slightly shifted toward residue A2808 of the 25S rRNA in the cryo-EM structure (Fig. 1c and S2). This subtle difference might possibly result from the presence of the tRNA in the P site or might partially reflect the coordinate error in both structures. Although it remains unclear how hypusine stimulates the rate of protein synthesis, our structure illustrates that the hypusine position in the P site of the ribosome is determined predominantly by its interaction with the ribosome, rather than by interactions with the tRNA bound to the ribosomal P site.
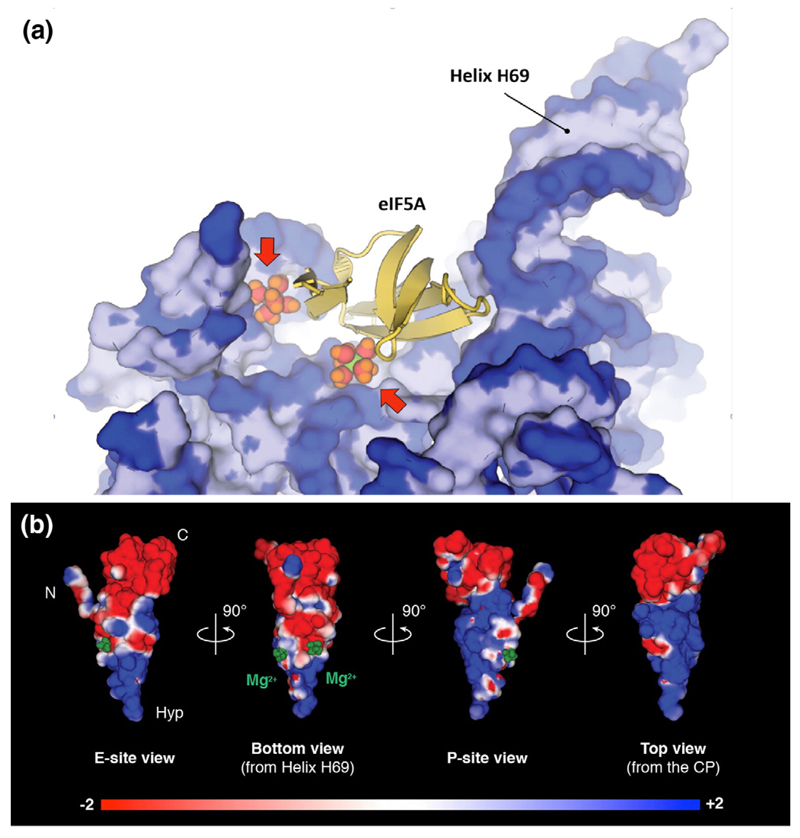
Fig. 2. Binding of eIF5A to the ribosome is mediated by magnesium ions.
(a) View of the eIF5A/25S rRNA interface shows two magnesium ions that stabilize the eIF5A N-terminal domain at the ribosomal interface. Arrows point to the magnesium ions. (b) The eIF5A electrostatic surface highlights positions of the magnesium ions in negatively charged cavities of the eIF5A molecule.
In a parallel experiment, we determined the structure of unmodified eIF5A from S. cerevisiae bound to the yeast 80S ribosome at 3.35 Å resolution (I/σ = 1, Table 1). The eIF5A lacking hypusine occupied the same position as hypusinated eIF5A in the eIF5A/80S ribosome complex; however, the unmodified eIF5A showed only partial occupancy in the binding site (Table 1). This partial occupancy is consistent with previous observations showing that, in addition to its essential role in promoting protein synthesis [1,6,12], hypusine stabilizes eIF5A association with the ribosome [13,14].
Binding of eIF5A to the ribosome is stabilized by magnesium ions
Importantly, we observed that interactions between eIF5A and the ribosome are not limited to direct eIF5A–ribosome contacts but include two indirect contacts mediated by magnesium ions (Fig. 2a). These ions—coordinated by eIF5A residues Gln22, Asp72, Ser74, and Asn79; and 25S rRNA residues G2418, A2419, and C2792 in the ribosome—enable binding between the negatively charged rRNA and the negatively charged interface of the N-terminal domain of eIF5A (Fig. 2b). Along with previously described tRNA-like properties of eIF5A and EF-P including their tRNA-like shape, position in the ribosome, and binding to protein uL1 [7,15], the Mg2+-mediated contacts between eIF5A and the rRNA are reminiscent of Mg2+-mediated contacts between the tRNA and the ribosome [16] and illustrate another aspect of structural similarity between eIF5A and tRNA.
Ribosome rotation alters the eIF5A-binding site
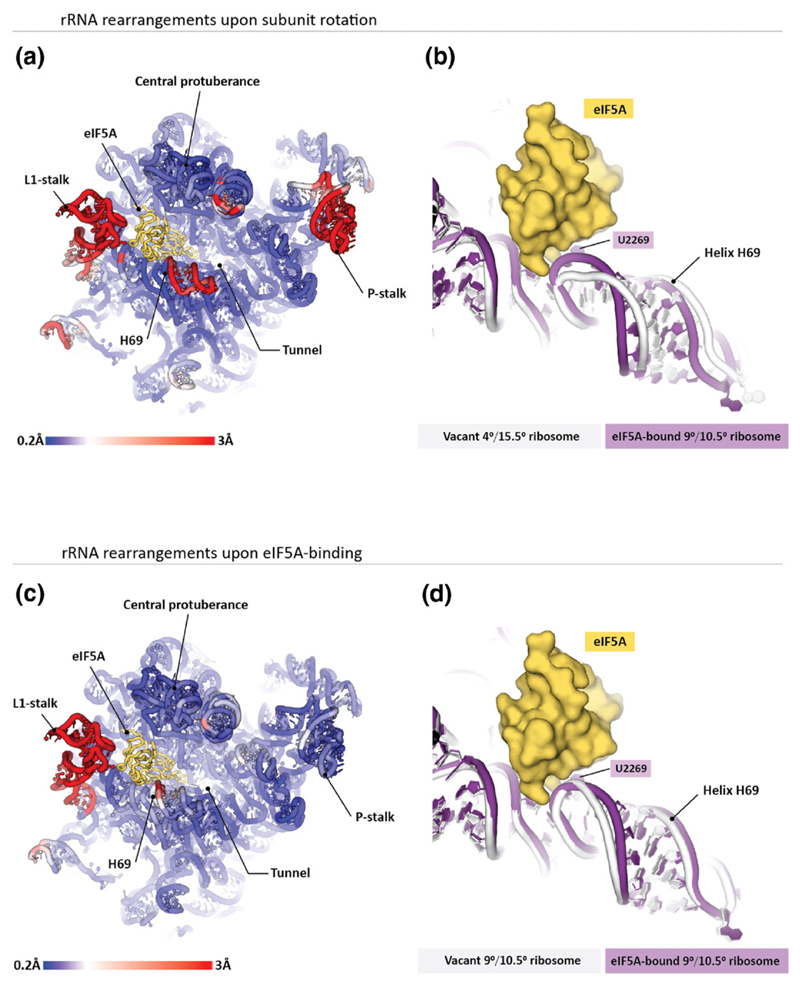
As noted above, eIF5A binds to one of two 80S ribosomes in the asymmetric unit of the crystal. In the eIF5A-bound ribosome, the 40S subunit is rotated ~9° relative to the 60S, the 40S head domain is swiveled ~10.5° (9°/10.5°), and the N-terminal domain of eIF5A interacts with dynamic helix H69 of the 25S rRNA (Fig. 3a). In the second ribosome of the asymmetric unit, which lacks eIF5A, the 40S subunit is rotated ~4°, the head swivel is ~15.5° (4°/15.5°), and helix H69 moves ~6 Å away from the eIF5A docking site (Fig. 3b). To discriminate whether the structural changes including the helix H69 movements in the eIF5A-bound 80S ribosome are induced by rotation of the ribosomal subunits or caused by eIF5A binding to the ribosome, we compared the eIF5A-binding pocket in our eIF5A/80S complex with the corresponding site in a previously described [11] vacant 80S ribosome with the identical rotation state (9°/10.5°) (Fig. 3c). Interestingly, only local and relatively minor changes in the helix H69 structure were observed when comparing the eIF5A-bound and unbound ribosomes with the same rotation state (Fig. 3d). Collectively, these observations indicate that the altered position of helix H69 in the eIF5A-binding site on the ribosome is triggered not by eIF5A binding but rather by the transition of the ribosome between different rotation states. It is noteworthy that the head swiveling and 40S body rotation observed in our eIF5A/80S complex are similar to conformations adopted in different translocation intermediates of the ribosome [17,18], suggesting that ribosome dynamics during the elongation cycle may modulate the structure of the eIF5A-binding pocket.
Fig. 3. Structure of the eIF5A-binding site depends on the ribosome rotation state. Panels (a and b) illustrate that ribosome transitions between the rotated states cause marked changes in the eIF5A-binding site.
(a) The 25S rRNA from the S. cerevisiae 80S ribosome is colored by displacement of the corresponding nucleotides between the 9°/10.5° (40S rotation/40S head swivel; eIF5A-bound) and 4°/15.5° (no eIF5A) rotated states of the two ribosomes in the asymmetric unit. The most prominent difference in the eIF5A-binding site is observed in helix H69. (b) Zoom on helix H69 shows that in the eIF5A-bound ribosome, helix H69 directly contacts eIF5A, whereas in the vacant ribosome, helix H69 moves ~6 Å away when eIF5A is docked in the equivalent position. Panels (c and d) illustrate structural changes in the 25S rRNA upon eIF5A binding to the ribosome. Two crystal structures of the yeast 80S ribosome are compared: the vacant 4°/15.5°-rotated ribosome (pdb 4v7r) and the eIF5A-bound 4°/15.5°-rotated ribosome (this study). (c), The 25S rRNA from the S. cerevisiae 80S ribosome is colored as in (a). (d) Zoom on helix H69 illustrates that eIF5A binding induces relatively subtle changes in the helix H69 structure.
Discussion
In summary, our structure describes eIF5A interactions with the rotated state of the 80S ribosome. Moreover, we show that the relative rotation of the ribosomal subunits provokes changes in the eIF5A-binding site by altering the conformation of helix H69 in the 25S rRNA. As it is not clear when, how, and in which conformation ribosomes recruit eIF5A during protein synthesis, our crystal structure, along with prior structural studies, may prove valuable in designing biochemical experiments to shed light on this unexplored aspect of eIF5A function. The observation that eIF5A interacts with two dynamic features of the large ribosomal subunit, the L1-stalk and helix H69, raises the possibility that eIF5A affinity to the ribosome may depend on the ribosome conformation. Accordingly, the resemblance between eIF5A and tRNA molecules might extend beyond their similar binding sites to include common mechanisms of recruitment or release that are coupled to the dynamic motions in the ribosome during translation elongation.
Accession Codes
The structures were deposited to the protein data bank with accession codes 5dat and 5dc3.
Appendix A. Supplementary Data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2016.05.011.
Acknowledgments
We are grateful to the staff of PROXIMA 1 beamline at the synchrotron SOLEIL (France) and, in particular, to Andrew Thompson and Pierre Legrand for providing rapid access and assisting with data collection, Corwin Miller (Yale University) for critical reading of the manuscript, and to the members of R.M., T.D., and M.Y. teams for insightful and inspiring discussions! This work was supported by Foundation pour la Recherche Medicale en France SPF20111223404 (to S.M.), the Austrian Science Fund FWF (P21641 and I1040 to R.M.), the French National Research Agency ANR-11-BSV8-006 01 (to G.Y.), the European Research Council advanced grant 294312 and the Human Frontier Science Program grant RGP0062/2012 (both to M.Y.), the Russian Government Program of Competitive Growth of Kazan Federal University (both to M.Y. and G.Y.), and in part by the Intramural Research Program of the National Institutes of Health, NICHD (T.D.).
References
- [1].Gutierrez E, et al. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51(1):35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Doerfel LK, et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339(6115):85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- [3].Ude S, et al. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339(6115):82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- [4].Cooper HL, Park MH, Folk JE, Safer B, Braverman R. Identification of the hypusine-containing protein hy + as translation initiation factor eIF-4D. Proc Natl Acad Sci U S A. 1983;80(7):1854–1857. doi: 10.1073/pnas.80.7.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Park MH, Abbruzzese A, Folk JE. Post-translational formation of hypusine: biogenesis of translation initiation factor eIF-4D. Adv Exp Med Biol. 1988;231:633–640. doi: 10.1007/978-1-4684-9042-8_53. [DOI] [PubMed] [Google Scholar]
- [6].Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459(7243):118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325(5943):966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schmidt C, et al. Structure of the hypusinylated eukaryotic translation factor eIF-5 A bound to the ribosome. Nucleic Acids Res. 2016;44(4):1944–1951. doi: 10.1093/nar/gkv1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dunkle JA, Cate JH. Ribosome structure and dynamics during translocation and termination. Annu Rev Biophys. 2010;39:227–244. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- [10].Yamamoto H, et al. EF-G and EF4: translocation and back-translocation on the bacterial ribosome. Nat Rev Microbiol. 2014;12(2):89–100. doi: 10.1038/nrmicro3176. [DOI] [PubMed] [Google Scholar]
- [11].Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- [12].Dever TE, Gutierrez E, Shin BS. The hypusine-containing translation factor eIF5A. Crit Rev Biochem Mol Biol. 2014;49(5):413–425. doi: 10.3109/10409238.2014.939608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jao DL, Chen KY. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5 A to the translating 80S ribosomal complex. J Cell Biochem. 2006;97(3):583–598. doi: 10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- [14].Zanelli CF, et al. eIF5A binds to translational machinery components and affects translation in yeast. Biochem Biophys Res Commun. 2006;348(4):1358–1366. doi: 10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]
- [15].Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S. A paralog of lysyl–tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat Struct Mol Biol. 2010;17(9):1136–1143. doi: 10.1038/nsmb.1889. [DOI] [PubMed] [Google Scholar]
- [16].Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- [17].Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468(7324):713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.