Abstract
The genome and transcriptome are constantly modified by proteins in the cell. Recent advances in single-molecule techniques allow for high spatial and temporal observations of these interactions between proteins and nucleic acids. However, due to the difficulty of obtaining functional protein complexes, it remains challenging to study the interactions between macromolecular protein complexes and nucleic acids. Here, we combined single-molecule fluorescence with various protein complex pull-down techniques to determine the function and stoichiometry of ribonucleoprotein complexes. Through the use of three examples of protein complexes from eukaryotic cells (Drosha, Dicer, and TUT4 protein complexes), we provide step-by-step guidance for using novel single-molecule techniques. Our single-molecule methods provide sub-second and nanometer resolution and can be applied to other nucleoprotein complexes that are essential for cellular processes.
Keywords: Single-molecule fluorescence, Single-protein pull-down, Protein complex, RNA interference, Dicer, Drosha, TUT4
1. Introduction
Interactions between protein assemblies and nucleic acids are essential elements of cellular processes, such as transcription, translation, and chromatin remodeling. A well-known example of such a protein assembly is the spliceosome, a multi-megadalton ribonucleoprotein complex that uses numerous cofactors to catalyze the splicing of precursor messenger RNA [1,2]. The ribonucleoprotein complex called RISC (RNA-induced silencing complex) is a key player in RNA interference—a cellular process of translational repression [3]. The biogenesis and regulation of microRNA (non-coding RNA that mediates RNA interference) involves several protein complexes such as Drosha-DGCR8 [4,5], Dicer-TRBP [6,7], Dicer-Loqs [8,9] and TUTase-Trim25 [10].
A comprehensive analysis of nucleoprotein complexes is a stepping stone to understanding cellular processes. Recent advances in analytical and biochemical methods have led to numerous break-throughs in the characterization of multicomponent protein assemblies in complexes with nucleic acids. High-throughput approaches, including large-scale tandem affinity purification, the yeast two-hybrid system, and mass spectrometry analysis, have been used to identify thousands of new protein complexes in yeast [11–15], Drosophila melanogaster [16,17] and Caenorhabditis elegans [18]. In parallel, advanced computational methods have emerged during the past decade, which made it possible to predict the formation of protein complexes [19]. Major advances in sample preparation and detection techniques have also enabled crystallographers and electron microscopists to determine the structure of large protein complexes interacting with nucleic acid substrates at an atomic resolution [20,21].
Despite the wealth of information acquired from these analytical and biochemical methods, there is a need for complementary techniques that allow for real-time observations of the assembly and function of nucleoprotein complexes. Recently, we and other groups developed such single-molecule fluorescence methods. Hoskins et al. revealed the order of spliceosome assembly during pre-mRNA maturation in cell extract via single-molecule multi-color fluorescence [22,23]. Single-molecule pull-down FRET allowed Nils et al. to visualize in real time the splicing of pre-mRNA by the spliceosome [24,25]. Lee et al. used a single-molecule co-immunoprecipitation approach to investigate weak interactions between different proteins [26,27]. Jain et al. developed single-molecule pull-down techniques to determine the stoichiometry of protein complexes [28–33]. We developed a single-molecule pull-down method to gain insight into the molecular mechanism of large nucleoprotein complexes involved in micro-RNA uridylation [34].
Here, we describe various single-molecule pull-down approaches and provide protocols for the purification and immobilization of ribonucleoprotein complexes associated with their native cofactors. Our pull-down methods in combination with single-molecule fluorescence allow for real-time visualization of protein complexes and RNA interactions. We describe several different strategies used in our laboratory and list the challenges that we encountered during the development of these techniques. As a proof-of-concept, we show three examples of protein complexes involved in small RNA biogenesis (human Drosha-DGCR8, human Dicer-TRBP, Drosophila Dicer 2-Loqs-PD, and human TUT4 complex) and illustrate how we elucidate the molecular bases of their functions. With this protocol, single-molecule fluorescence can be widely used to study nucleoprotein complexes.
2. Materials and methods
2.1. Cell culture and transfection
2.1.1. HEK-293T cells
Human embryonic kidney cells (HEK-293T) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, 31885023, Gibco®) supplemented with 10% fetal bovine serum (FBS, heat-inactivated, Greiner Bio-One) at 37 °C and 5% CO2. Before transfection, cells were split into 10 cm cell culture dishes to a confluence of 25%. After 24 h of growth, plasmids of interest were transfected using a CaPO4 transfection method (Molecular Cloning: A Laboratory Manual, 3rd ed., Cold Spring Harbor Laboratory Press, 2001). For the in vivo biotinylation of human Dicer and Drosha proteins, an additional plasmid coding for the BirA enzyme was co-transfected. After 5 h, the medium was exchanged with fresh DMEM containing 1 μg/ml biotin (B4639, Sigma), and the transfected cells were incubated for another 48 h to enable protein expression and in vivo biotinylation.
2.1.2. SL2 cells
Schneider’s Drosophila Line 2 (SL2, CRL-1963™, ATCC®) was maintained in HyClone SFX-Insect Cell Culture medium (SH30278.LS, GE Healthcare HYCLONE) supplemented with 10% FBS (heat-inactivated, Greiner Bio-One) at 25 °C. When the culture reached a density of 0.5 × 106 cells/mL, the cells were transfected using the FuGENE® HD transfection method (E2311, Promega). After 24 h of incubation, 1 mM CuSO4 was added to the medium, and the cells were incubated for additional 48 h.
2.2. Cell harvest and lysis
Before the transfected cells were harvested with scrapers, DMEM was removed and the cells were washed with ice-cold Dulbecco’s Phosphate-Buffered Saline (DPBS, 14200 Gibco®). Subsequently, the cells were transferred to 15 mL tubes and centrifuged at 276×g and 4 °C for 5 min to form cell pellets. After the removal of the supernatant, the cell pellets were frozen and stored at −80 °C until further processing. Before lysis, the cells were thawed on ice for 30 min. Subsequently, HEK-293T and SL2 cells were resuspended in buffer D (20 mM Tris–HCl [pH 8.0], 200 mM KCl and 0.2 mM EDTA) and lysis buffer SL2 (30 mM HEPES-KOH [pH 7.4], 100 mM KOAc, 10% glycerol, 0.1% Triton X-100), respectively. Lysis was carried out by carefully passing the cells 10 times through a needle (30½ gauge, BD), while avoiding the formation of air bubbles. Afterwards, the lysate was centrifuged twice (16,100×g at 4 °C, for 20 min) to remove cell debris (pellet). The recovered cell extract (supernatant) was either directly used for single-molecule experiments (Drosha-DGCR8), or alternatively, tandem purification steps were carried out to obtain higher purity samples (dmDicer-2, hDicer and TUT4). To prevent disturbing the protein complexes, it is important to perform the cell lysis and immunoprecipitation in a gentle manner and in a physiologically relevant buffer. We do not recommend the use of sonication as a cell lysis method because this may cause protein complexes to disassemble and form aggregates [35].
2.3. Immunoprecipitation and elution
For immunoprecipitation of 1xFLAG-tagged proteins (dmDicer-2, hDicer, and TUT4), 1 mg of total protein in the cell extract was incubated with 2.5 μL of anti-FLAG antibody-conjugated agarose beads (50% slurry, anti-FLAG® M2 affinity gel, A2220, Sigma) under gentle agitation at 4 °C for 30–60 min. It is noted that a longer incubation time may increase the number of non-specific interactions and result in the pull-down of contaminant proteins. After incubation, the beads were gently washed five times with buffer D or buffer SL2 and resuspended in 10 μL of buffer D or buffer SL2, resulting in 100 μg/μL of total protein concentration. hDicer was eluted from the beads by site-specific cleavage using Tobacco Etch Virus TEV protease (0.05 U/μL) (V6101, Promega) at 30 °C for 90 min. Alternatively, the proteins of interest (dmDicer-2 and TUT4) were eluted from the beads using 2 mM 3xFLAG® peptide (F4799, Sigma). The eluted proteins were supplemented with glycerol to a final concentration of 10%, aliquoted and snap-frozen with liquid nitrogen for long-term storage at −80 °C. The immunoprecipitates (IPs) were tested for the enrichment of the proteins of interest using western blot analysis, while the catalytic activities of the IPs were tested with bulk assays (data not shown).
2.4. Single-molecule pull-down
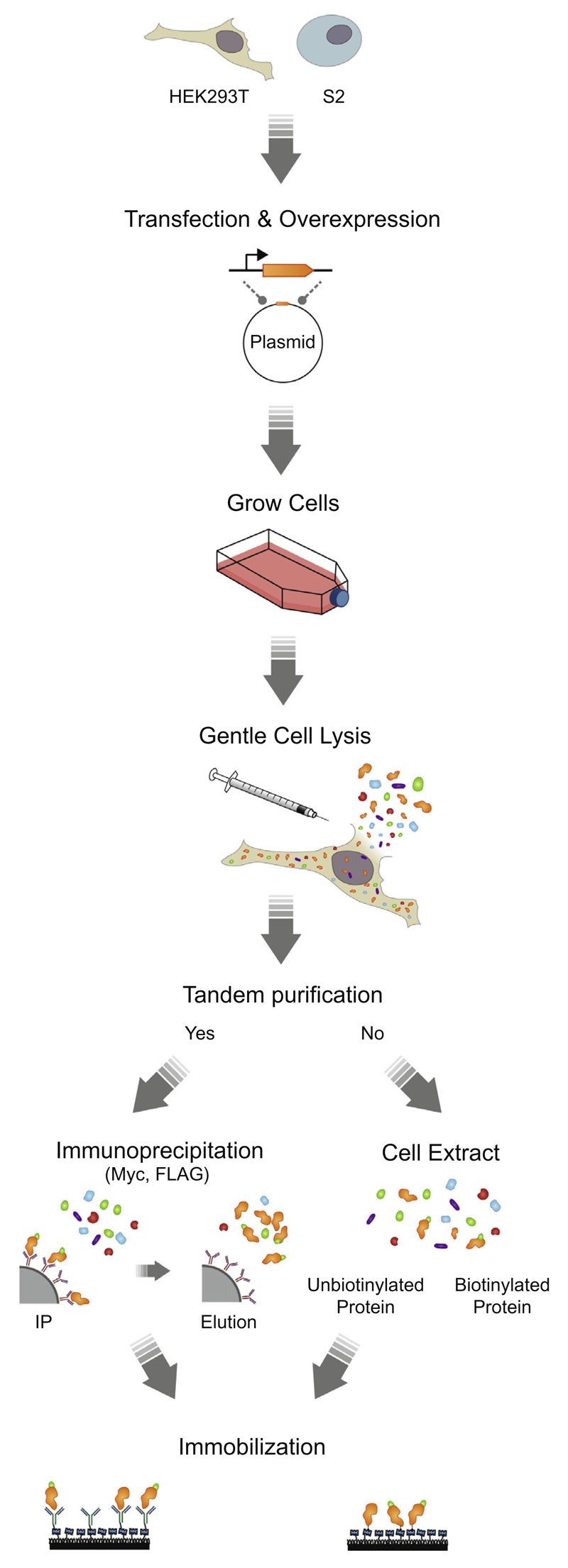
To increase purity of the IPs, an additional purification step was carried out directly on the surface of the imaging chamber using streptavidin or specific antibodies targeting the proteins of interest with nanomolar affinity range. This allowed for an efficient immobilization of the protein of interest, while discarding unwanted contaminant proteins (Fig. 1). Single-molecule pull-down procedures are described case by case in the Results and Discussion sections.
Fig. 1.
Overview of the single molecule pull-down techniques.
2.5. Nucleic acids preparation and labeling
2.5.1. Stem-loop RNA
All of the RNA constructs used in this study were synthesized by ST Pharm Co., Ltd., South Korea. Precursor-microRNA (pre-miRNA) molecules were constructed by ligating two synthetic RNAs. First, a single-stranded RNA containing a 5′ phosphate and a half of the terminal loop of pre-miRNA (100 pmol, strand J in Table 1) was mixed with the 5′ strand that contained the other half of the terminal loop (200 pmol, strand K in Table 1). The mixture (20 μL) in TE buffer supplemented with 100 mM NaCl was annealed by heating it to 80 °C, followed by a slow cooling down to 4 °C (−1 °C/4 min in a thermal cycler). The annealed substrate was ligated using 3 μL of T4 RNA ligase (5 U/μL, AM2140, Invitrogen), 3 μL of 0.1% BSA (AM2616, Ambion), 5 μL of the 10× ligation buffer provided, and 19 μL of H2O at 16 °C for 24 h. After acid phenol-chloroform extraction and ethanol precipitation, the RNA was purified with 12.5% urea polyacrylamide gel.
Table 1.
DNA and RNA sequences.
| Sequences (5′ → 3′) | |
|---|---|
| Pri-miRNA | A: GAU ACU AUA CUG AGA GCA UUC CGU UAU GUA GCA UUU CUU GGU UGU GAG GGG UUG UGC |
| Drosha-DGCR8 (Section 3.1) | B: AAG AAG AAU CUC ACG AUC AAG GAA UGC UAC AU |
| Strand C was Cy5-labeled | C: AAC GGA GuG UUU GAG CAG ACC CGC GAC U |
| dsRNA | D: AAG AAG AAU CUC ACG AUC AAG GAA UGC uAC AUA A |
| Drosophila Dicer-2 (Section 3.2.1) | E: pCGG AGU GUU UGA GCA GAC CCG CGA UCU UUC AUU GCC |
| Strand D was Cy3-labeled | F: CTC AAA CAC TCC GTT ATG TAG CAT TC |
| G: pGGC AAU GAA AGA UCG CGG GUC UGC UCA AAC ACU CCG UUA UGU AGC | |
| H: pAUU CCU UGA UCG UGA GAU UCU UCU U | |
| I: ACG ATC AAG GAA TGC TAC ATA ACG GA | |
| pre-let-7a-1 | J: UGA GGU AGU AGG UUG UAU AGU UUU AGG GUC ACA CC |
| Human Dicer (Section 3.2.2) | K: pCAC CAC UGG GAG AUA ACU AUA CAA UCU ACU GUC uUU CU |
| Strand K was Cy5-labeled | |
| Cy3-dA15 (section: 3.3) | X: Cy3-AAA AAA AAA AAA AAA |
p indicates phosphate; u represents dye-labeled nucleotide
The primary miRNA (pri-miRNA) substrate was constructed using the method described above. However, due to its length of 116 nucleotides (nt), pri-miRNA had to be ligated in two ligation steps. In the first ligation, a stem-loop structure was constructed (strands A and B in Table 1), followed by an additional ligation with a supplementary single-stranded RNA tail (strand C in Table 1) to obtain the full-length construct.
2.5.2. Double-stranded RNA
First, two 70-nt ssRNA strands were constructed by ligating two synthetic RNAs with a DNA splint following the method described in Section 2.5.1. The DNA splint was used to facilitate the ligation of the two RNA strands by T4 RNA ligase 2 (10 U/μL, M0239L, NEB). For the first strand, the ligation mixture contained a 34-nt Cy3-labeled RNA (200 pmol, strand D in Table 1), a 36-nt RNA containing a 5′ phosphate (200 pmol, strand E in Table 1) and a DNA splint (300 pmol, strand F in Table 1). For the second strand, the ligation mixture contained a 45-nt RNA with a 5′ phosphate (200 pmol, strand G in Table 1), a 25-nt RNA with a 5′ phosphate (200 pmol, strand H in Table 1) and a DNA splint (300 pmol, strand I in Table 1). After acid phenol-chloroform extraction and ethanol precipitation, both RNA strands were purified using a 10% urea polyacrylamide gel. Both RNA strands were annealed following the method described in Section 2.5.1.
2.5.3. DNA
The fluorescently labeled ssDNA (dA15, strand X in Table 1) was purchased from IDT DNA, USA.
2.5.4. RNA labeling
All RNA strands were labeled with the NHS-ester form of cyanine dyes, Cy3 or Cy5, (GE Healthcare), with an almost 100% efficiency, as described elsewhere [36]. The positions of the labeled bases are indicated in Table 1.
2.6. Single-molecule fluorescence microscopy
A prism-type total internal reflection microscope was used for the single-molecule experiments. eGFP molecules were excited with a 473-nm solid-state laser (OBIS LX 75 mW, Coherent), Cy3 molecules were excited with a 532-nm solid-state laser (Compass 215M-50, Coherent), and Cy5 molecules were excited with a 632-nm solid-state laser (25 LHP 928, CVI Melles Griot). To obtain the time traces, we excited eGFP, Cy3 and Cy5 molecules as weakly as possible to minimize their rapid photobleaching during the observation time. The fluorescence signals from single molecules were collected through a 60x water immersion objective (UPlanSApo, Olympus) with an inverted microscope (IX71, Olympus). To block 473 nm, 532 nm and 632 nm laser scattering, we used a 473-nm long-pass filter (Chroma), a 550-nm long-pass filter (Chroma) and a 633-nm notch filter (SemRock), respectively. Data were obtained in either single color or dual color mode. For dual color measurements, fluorescence signals were spatially split with a dichroic mirror (λcutoff = 645 nm, Chroma) and imaged onto two halves of an EMCCD camera (iXon 897, Andor Technology).
2.7. Microfluidic chamber preparation and immobilization schemes
To eliminate the nonspecific surface adsorption of proteins and nucleic acids to a quartz surface, piranha-etched slides (Finkenbeiner) were passivated with polyethylene glycol (PEG) over two rounds of PEGylation as described previously [37]. To further improve the surface quality for the experiments where crude cell extracts were used (Section 3.1), the assembled microfluidic flow chambers were incubated with 5% Tween-20 (v/v in T50 buffer: 10 mM Tris [pH 8.0], 50 mM NaCl) for 10 min, followed by a washing step with 100 μL of T50 buffer [38]. Afterwards, slides were incubated with 50 μL of streptavidin (0.1 mg/mL, S888, Invitrogen) for 2 min followed by a washing step with 100 μL of buffer of interest. Biotinylated proteins were specifically immobilized by incubating the chamber with 50 μL of immunoprecipitated protein or crude cell extract (500× diluted in buffer D) for 5 or 0.5 min, respectively. The remaining unbound proteins were washed away with 100 μL of the buffer of interest supplemented with an oxygen scavenging system (0.8% glucose (v/v), 0.1 mg/mL glucose oxidase (G2133, Sigma), 17 μg/μL catalase (Roche)). Oxygen scavenging system was used to reduce photobleaching and 1 mM Trolox (238813, Aldrich) was used to reduce photoblinking of the dyes [39]. The interaction between the biotinylated proteins and streptavidin was stable for several hours without any noticeable dissociation (data not shown).
Alternatively, when the proteins of interest were not biotinylated, commercially available biotinylated antibodies (see Table 2) were used for specific immobilization. In brief, after flushing the unbound streptavidin away, the chamber was incubated with 50 μL of biotinylated-antibody (66 nM) for 5 min. The remaining unbound antibodies were washed away with 100 μL of buffer of interest, and 20–50 μL of diluted immunoprecipitated protein or crude cell extract was introduced to the microfluidic chamber. After 5 min of incubation, the unbound proteins were washed away with the buffer of interest supplemented with an oxygen scavenging system and Trolox.
Table 2.
Antibodies used for immobilizing the proteins of interest.
| Antibodies | Final concentration | Reference |
|---|---|---|
| anti-FLAG antibody | 66 nM | F7425, Sigma |
| c-Myc antibody (A-14) | 66 nM | sc-789, Santa Cruz |
| Biotechnology | ||
| Biotin-SP (long spacer) AffinePure | 66 nM | 111-065-003, Jackson |
| Goat anti-Rabbit IgG (H + L) | ImmunoResearch | |
| anti-RFP antibody | 66 nM | ab65856, Abcam |
2.8. Single-molecule data acquisition and analysis
A series of CCD images were acquired with lab-made software written in Visual C++ with a time resolution of 0.03–1 s. Fluorescence images and time traces were extracted with programs written in IDL (ITT Visual Information Solutions) and analyzed with Matlab (MathWorks) and Origin (OriginLab Corporation). To systematically select single-molecule fluorescence signals of eGFP, Cy3 or Cy5 from the acquired images, we employed an algorithm written in IDL that searched for fluorescence spots with a defined Gaussian profile and with signals above a threshold. Apparent FRET (Förster resonance energy transfer) efficiency was defined as IA/(ID + IA), where ID and IA represent the donor (Cy3) and acceptor (Cy5) signals from two fluorescence spots from the same molecule, respectively.
3. Results and discussion
3.1. Stoichiometry determination: Drosha-DGCR8 protein complex
The Microprocessor complex, composed of Drosha and its cofactor DGCR8, plays an essential role in the initial stage of microRNA (miRNA) biogenesis. In the nucleus, the Microprocessor binds to and subsequently cleaves pri-miRNA transcripts, resulting in the production of hairpin-structured pre-miRNAs [40]. Drosha hosts catalytic sites that are required for cleavage, while its cofactor DGCR8 enhances binding to the substrate pri-miRNA [40]. Using a single-molecule pull-down method, we determined the stoichiometry of Drosha and its cofactor DGCR8 in naturally formed protein complexes.
We advanced a single-molecule pull-down method from Jain et al. [28] by introducing a biotin-streptavidin conjugation scheme (Fig. 1). We fused the N-terminus of Drosha with the acceptor peptide (AP) (Fig. 2A), which is covalently coupled with biotin by the Escherichia coli enzyme biotin ligase (BirA) [41,42]. In addition, we fused DGCR8 with a fluorescent protein (eGFP) to be able to observe interactions between Drosha and DGCR8.
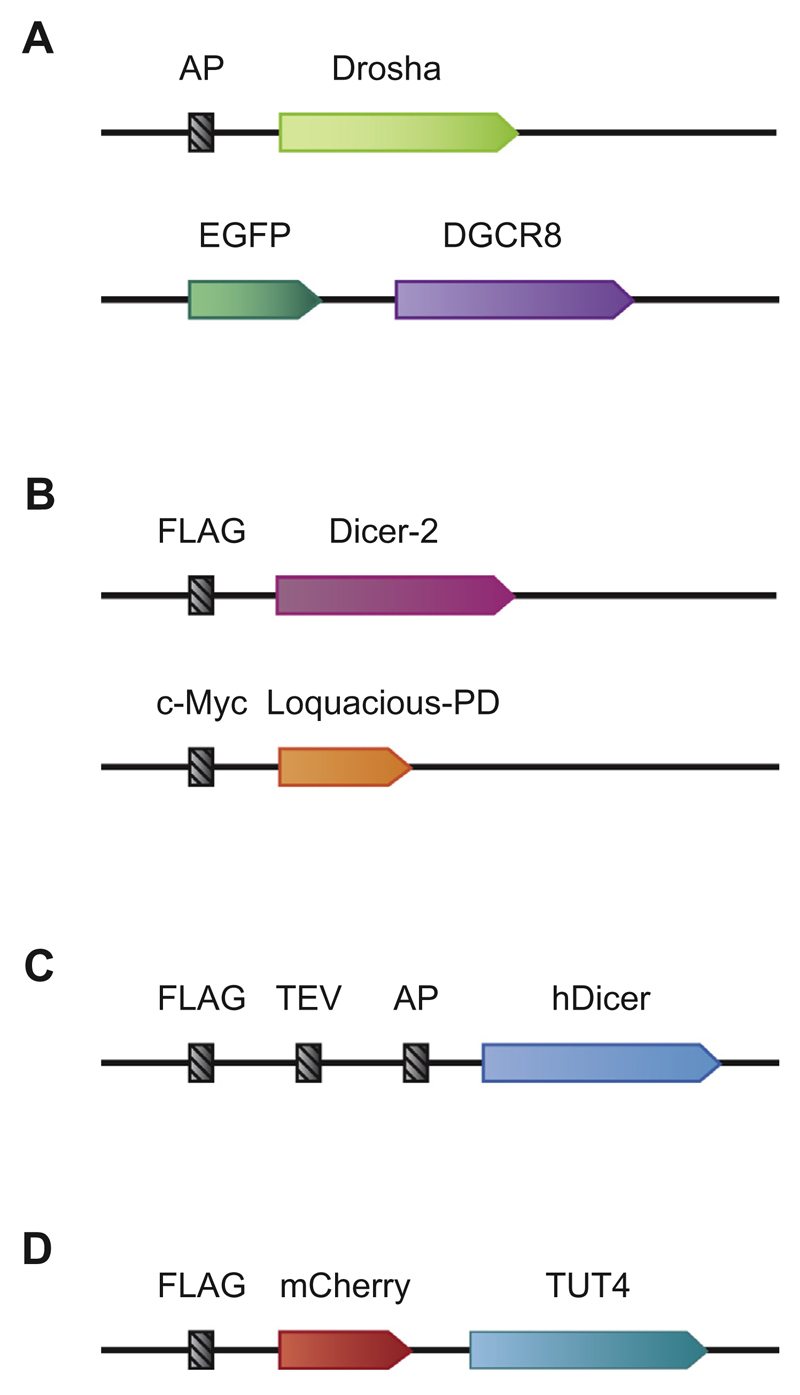
Fig. 2.
Plasmid constructs. A. Illustrated are the constructs used for Fig. 3. AP stands for the acceptor peptide that is recognized by BirA. DGCR8 is tagged with a fluorescent protein, eGFP. B. Illustrated are the constructs used for Fig. 4. Dicer-2 is tagged with the FLAG epitope. Loquacious-PD is tagged with the 1xc-Myc epitope. C. Illustrated is a construct used for Fig. 5. hDicer is tagged with 1xFLAG, TEV (Tobacco Etch Virus), and AP. D. Illustrated is a construct used for Fig. 6. TUT4 is tagged with FLAG epitope and a fluorescent protein (mCherry).
We immobilized the Drosha-DGCR8 complexes from the crude cell extract on the surface of the microfluidic chamber that was passivated with PEG with two rounds of PEGylation, as described by Chandradoss et al. in [37] and afterwards coated with streptavidin (Fig. 3A). However, we observed prominent non-specific adsorption of cellular proteins to the glass surface. Comparison with a control chamber that was not treated with streptavidin, which thus should not show immobilized proteins, revealed little difference in the number of detected molecules (data not shown). Tween-20 was recently reported to improve the surface passivation [38]. We observed that the additional treatment of the PEGylated surface with 5% Tween-20 reduced the number of non-specific adsorption of cellular proteins by a factor of two (data not shown). We note that it is not recommended to use BSA (bovine serum album) for surface passivation since BSA increases the degree of non-specific binding of proteins (unpublished observation).
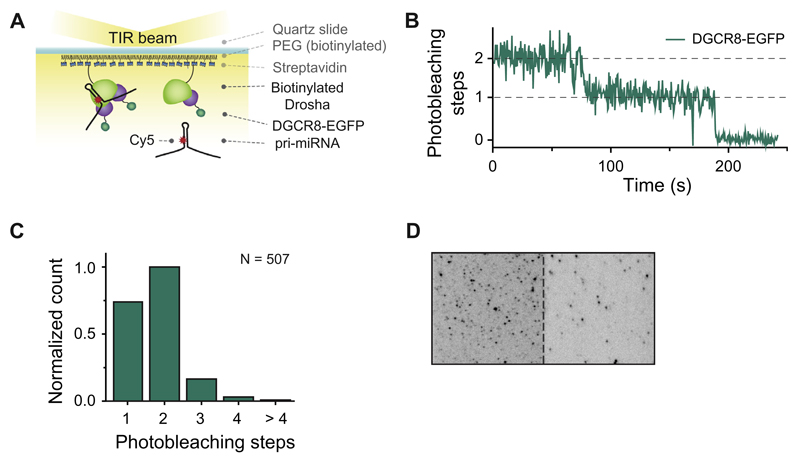
Fig. 3.
Single-molecule stoichiometry measurement. A. Schematic overview of a single-molecule stoichiometry measurement. Biotinylated Drosha-DGCR8/eGFP complexes were immobilized on a PEGylated surface using biotin-streptavidin conjugation. Cy5-labeled pri-miRNA molecules were introduced into the imaging chamber. B. Representative time trajectory of eGFP fluorescence. The stoichiometry of DGCR8 molecules associated with Drosha molecules was determined by counting the number of eGFP photobleaching steps (indicated with dashed lines). C. Bar plot showing the distribution of the Drosha/DGCR8 stoichiometry. D. Ability of Drosha-DGCR8 complexes to bind RNA molecules was confirmed using the co-localization of pri-miRNA and DGCR8. The figure shows a camera screenshot with eGFP molecules in the left channel and Cy5 molecules in the right channel. The red circles indicate co-localized DGCR8 (eGFP) and pri-miRNA (Cy5).
To determine the stoichiometry of the Drosha-DGCR8 complexes immobilized on the surface, we excited eGFP with a laser beam and recorded its fluorescence signal until all of the eGFP molecules were photobleached. The number of photobleaching steps, defined as a sudden decrease in eGFP fluorescence intensity (Fig. 3B), reflects the number of DGCR8 molecules associated with a single Drosha protein. Our photobleaching data (Fig. 3C) show that ~46% (236 among 513 analyzed molecules) of the Microprocessor complexes are composed of one Drosha and two DGCR8 proteins, in agreement with recently published work by Nguyen et al. [43].
To determine whether the immobilized Drosha-DGCR8 complexes were capable of binding RNA substrates, we introduced Cy5-labeled pri-miRNA, a known substrate of Microprocessor [44], into the microfluidic chamber (Fig. 3D). Simultaneous illumination of eGFP and Cy5 dyes allowed us to co-localize RNA bound to Drosha-DGCR8, suggesting that the immobilized Microprocessor complexes retained their RNA-binding activity.
We emphasize that when attempting to immobilize a protein of interest directly from the cell extract, it is important to pay attention to the high amount of proteins in the cell extract. It is crucial to maximize the quality of the glass surface and minimize the incubation time of the cell extract (we recommend an incubation for 30 s or less). As an alternative, one could employ an additional purification step to reduce the content of unwanted proteins before applying the protein sample to a microfluidic chamber. Examples of such tandem purification scheme are described in the following sections.
3.2. Binding affinity measurement: Dicer protein complexes
3.2.1. Drosophila Dicer-2 associated with Loquacious-PD
Drosophila melanogaster Dicer-2 (dmDicer-2) is an endoribonuclease that processes long double stranded RNA (dsRNA) molecules into 21-nt small interfering RNAs (siRNAs). For efficient cleavage of RNA substrates, dmDicer-2 requires a cofactor, Loquacious-PD [45]. Loquacious-PD also facilitates the loading of dsRNA substrates onto dmDicer-2 [46]. To visualize the binding of a dsRNA substrate by dmDicer-2 at the single-molecule level, we sought to develop a single-molecule pull-down assay advancing our previously reported SIMPlex technique (Single-molecule approach to Immunoprecipitated Protein complexes) [34]. For this purpose, we tested various immobilization schemes.
In the first attempt, we immobilized the dmDicer-2/Loquacious-PD complex from the crude cell extract using antibodies. We immobilized a primary biotinylated anti-rabbit IgG antibody, which allowed us to immobilize the secondary anti-c-Myc antibody that targets the 1xc-Myc tag fused to Loquacious-PD (Figs. 2B and 4A). Upon introduction of a 70-nt long Cy3-labeled dsRNA substrate, we observed a large number of binding events (dark spots on the CCD image, Fig. 4B, left panel). However, a control without antibodies also showed a substantial number of binding events (CCD image in the Fig. 4B, right panel). These results suggested that other RNA-binding proteins in the cell extract were non-specifically adsorbed on the surface and mediated RNA binding, which is consistent with our previous observation [34]. We note that the high concentration of proteins in the cell extract lead to a non-specific adsorption of many proteins on the surface of the imaging chamber, among which RNA binding proteins can interact with dye-labeled RNA molecules.
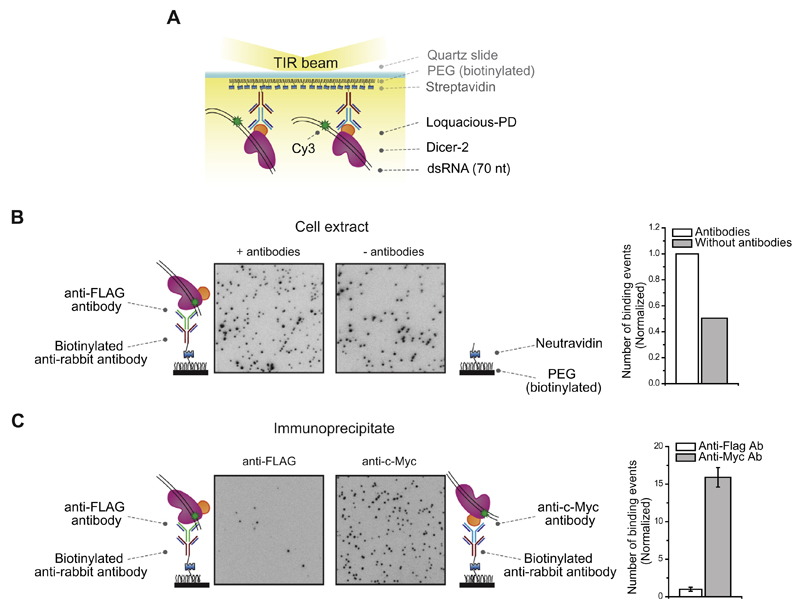
Fig. 4.
Single-molecule binding measurement. A. Schematic overview of a single-molecule pull-down assay for Dicer-RNA interactions. Immunoprecipitated dmDicer-2/Loquacious-PD is immobilized on a PEGylated surface using various antibodies. Binding of RNA was observed after injecting a 70-nt Cy3 labeled dsRNA. B. Crude cell extract is added on a PEGylated surface coated with a primary biotinylated anti-rabbit IgG antibody bound to a secondary anti-c-Myc antibody (left panel) or without a primary biotinylated anti-rabbit IgG antibody bound to a secondary anti-c-Myc antibody (right panel). Binding of RNA was observed after introducing a 70-nt Cy3 labeled dsRNA. The histogram on the right displays a normalized number of dsRNA molecules docked to the surface. C. Immunoprecipitated dmDicer-2/Loquacious-PD is immobilized on a PEGylated surface using biotinylated anti-rabbit IgG antibody bound to anti-FLAG antibody (left panel) or using biotinylated anti-rabbit IgG antibody bound to anti-c-Myc antibody (right panel). Binding of RNA was observed after introducing a 70-nt Cy3 labeled dsRNA. The histogram on the right displays a normalized number of dsRNA molecules docked to the surface.
To overcome non-specific adsorption, we prepared higher purity immunoprecipitates via two rounds of immunoprecipitation. In the first round of immunoprecipitation, the dmDicer-2/Loquacious-PD complex was pulled down using anti-c-Myc coated beads that target Loquacious-PD. After elution of the dmDicer-2/Loquacious-PD complex from the beads, a second round of immunoprecipitation was conducted on a single-molecule surface coated with a primary biotinylated anti-rabbit IgG antibody and a secondary anti-FLAG antibody that targeted FLAG-tagged dmDicer-2. Upon introduction of the Cy3-labeled dsRNA, we observed that this immobilization scheme generated little fluorescence (34.3 ± 9.0 binding events per field of view; CCD image in Fig. 4C, left panel). This observation suggests that two rounds of immunoprecipitation improved the purity of the IP, resulting in a reduced background of non-specific interactions. However, the lack of Cy3 fluorescence signals indicated that there were few dmDicer-2/Loquacious-PD immobilized on the surface. This could be due to the overrepresentation of Loquacious-PD compared to dmDicer-2 in the crude cell extract. Pull-down with anti-c-Myc coated beads might have resulted in a large quantity of Loquacious-PD that was not associated with dmDicer-2. Alternatively, even if dmDicer-2/Loquacious-PD complex was immobilized, the FLAG antibody might have affected the ability of dmDicer-2 to bind to RNA substrates.
To enrich the immunoprecipitate with dmDicer-2, we changed the order of the purification scheme. We pulled down dmDicer-2 in the first round of immunoprecipitation using an anti-FLAG antibody. For the second round of immunoprecipitation on the single-molecule surface, we used the anti-c-Myc antibody that targets Loquacious-PD. With this scheme, we observed a large amount of fluorescence signal upon introduction of Cy3-labeled dsRNA, suggesting that the immobilized complexes are potent for RNA binding (546.0 ± 44.7 binding events per field of view; CCD image in Fig. 4C, right panel). In addition, a control surface without antibodies (data not shown) showed hardly any fluorescence signal. These results suggest that a tandem purification scheme can be used to purify protein complexes with high purity. In summary, it is essential to determine how many rounds of pull-down are required to reach purity that is suitable for single-molecule observation, to optimize the order of pull-down to obtain protein complexes in a high yield, and to empirically select the position of tags that allows for reliable immobilization.
3.2.2. Human Dicer associated with TRBP
In humans, the endoribonuclease Dicer processes pre-miRNAs into mature miRNAs [3]. Dicer is associated with dsRNA-binding protein TRBP. A static picture of pre-miRNA maturation by the Dicer complex was provided by structural and biochemical studies, but a more dynamic view of this process remains to be established.
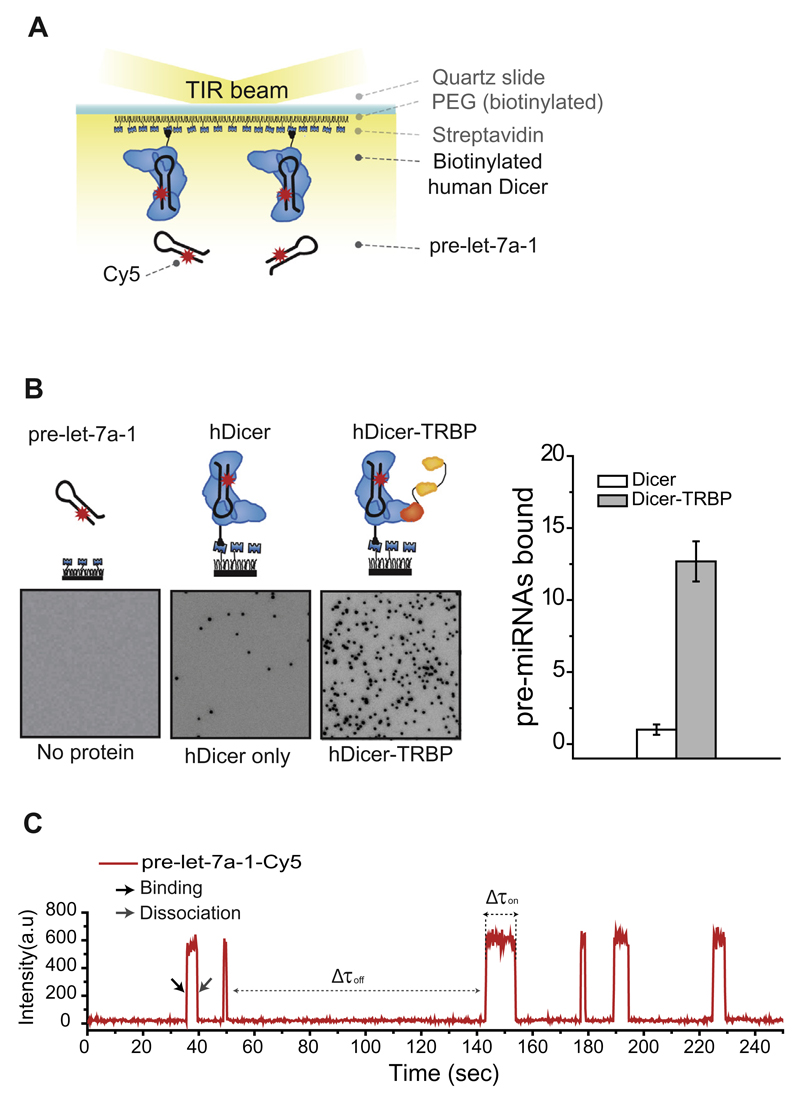
To visualize miRNA processing by the Dicer-TRBP complex at the single-molecule level, we employed a tandem purification method that allows the pull-down and surface immobilization of protein complexes. To pull down and immobilize TRBP-associated Dicer on a surface, we cloned FLAG, TEV (tobacco etch virus) and AP tags upstream of the human Dicer coding sequence (Fareh et al., in preparation). The three tags were used for immunoprecipitation, elution and in vivo biotinylation, respectively (Figs. 1 and 2C). BirA enzyme was co-expressed for in vivo biotinylation of the AP tag. After the purification process (in vivo biotinylation, FLAG immunoprecipitation and TEV elution), the IPs were immobilized on the surface of the imaging chamber via biotinstreptavidin conjugation (Figs. 1 and 5A).
Fig. 5.
Single-molecule kinetics measurement. A. Schematic overview of a single-molecule pull-down assay for Dicer-RNA interactions. Immunoprecipitated human Dicer-TRBP complexes were immobilized on a PEGylated surface using biotin-streptavidin interaction. Cy5-labeled pre-let-7a-1 was introduced into the imaging chamber by flow. Interactions between surface-immobilized Dicer-TRBP and Cy5-labeled pre-let-7a-1 were recorded in real time. B. CCD images display the RNA binding activity of Dicer (middle) and the Dicer-TRBP complex (right). Passivated surface without Dicer immobilized was used as negative control (left). The histogram on the right displays a normalized number of pre-let-7a-1 stably bound to Dicer (white) or to Dicer-TRBP (gray). C. Representative time trajectory (obtained with a time resolution 300 ms) displays six binding events of Cy5-labeled pre-let-7a-1 to a single Dicer-TRBP complex. The black arrow indicates the binding and the gray arrow indicates the dissociation of pre-let-7a-1.
We introduced a dye-labeled pre-miRNA into the imaging chamber. After 5 min of incubation, we washed away the unbound pre-miRNA and quantified the RNA binding activity of each protein complex by taking snapshots of different fields of view (Fig. 5B). The dark spots on the CCD image represent single Cy5-labeled pre-miRNAs that are stably bound to single Dicer proteins. Compared to Dicer alone, Dicer-TRBP showed an increase in the RNA binding activity of one order of magnitude (Fig. 5B). A passivated surface without immobilized Dicer did not show any adsorption of RNA, ruling out any nonspecific interactions between RNA and the quartz surface (Fig. 5B). This single-molecule approach can be used to quantify the previously reported enhancement of RNA binding activity mediated by TRBP [47].
To visualize the interaction between the Dicer-TRBP complex and pre-miRNA in real time, we introduced dye-labeled RNA into the imaging chamber while recording the binding events. The representative time trace (Fig. 5C) shows a sudden increase of the fluorescence, which reflects the interaction of surface-immobilized Dicer-TRBP with the RNA molecule, followed by RNA dissociation that is reflected by the loss of the fluorescence signal. The time trace can be further analyzed to determine kinetic parameters, including the binding and dissociation rates, which allows to draw the energy landscape of substrate recognition and processing by Dicer complexes. The average binding dwell-time (<Δτon>) is obtained from a distribution of the interaction time between Dicer complexes and pre-miRNA (Δτon). The dissociation rate (koff) is the inverse of <Δτon>. The association rate (kon) is the inverse of the average time interval between two successive binding events (Δτoff).
3.3. Single-molecule FRET measurements: TUT4 protein complexes
Terminal uridylyl transferases (TUTs) function as integral regulators of miRNA biogenesis. Recent studies have shown that TUT4 (ZCCHC11), TUT7 (ZCCHC6) and TUT2 (GLD2/PAPD4) enhance the maturation of pre-miRNAs through distributive mono-uridylation [48,49]. In contrast, in embryonic stem cells and cancer cells, where Lin28 is enriched, TUT4 and TUT7 inhibit pre-miRNA maturation through oligo-uridylation [50–52]. The oligo U-tail promotes degradation by the exonuclease DIS3L2 [34,50,53,54]. Although the general mechanism of oligo-uridylation has been well established, the underlying molecular mechanism remains poorly understood.
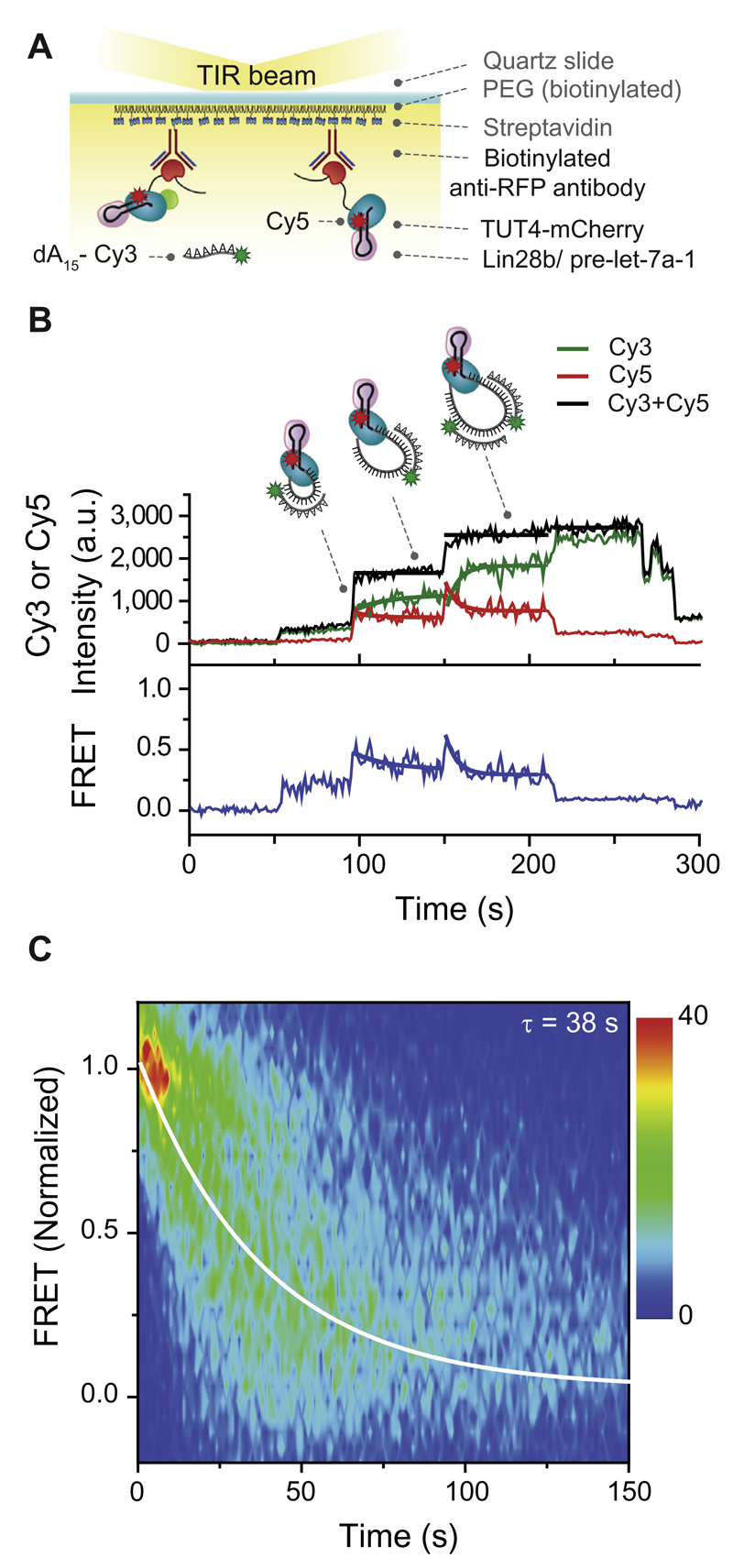
This limited understanding is mainly due to the lack of full-length recombinant TUT4. Therefore, we employed our single-molecule pull-down method that makes use of tandem purification to obtain high purity full-length TUT4 [34]. In brief, TUT4-FLAG-mCherry proteins were pulled down from a crude cell extract using beads coated with FLAG antibodies, followed by a second round of immunoprecipitation directly in the single-molecule chamber (Fig. 1). This scheme resulted in the immobilization of TUT4 through mCherry anti-RFP conjugation (Fig. 6A). Next, we reconstituted the ternary complex required for oligo-uridylation by introducing a Cy5-labeled pre-miRNA substrate that was pre-incubated with the processivity factor Lin28b. After equilibration, the Lin28b-bound pre-miRNA complex docked to the immobilized TUT4 proteins (Fig. 6A) [34].
Fig. 6.
Single-molecule FRET measurement. A. Schematic overview of a single-molecule FRET assay. Immunoprecipitated TUT4-Flag-mCherry proteins were immobilized on a PEGylated surface using anti-RFP antibodies (RFP, red fluorescent protein). Next, Lin28b/pre-let-7a-1 complexes were immobilized to the surface in a TUT4 specific manner. Oligo uridylation was tracked in real-time by simultaneously injecting UTP with Cy3-labeled oligo dA15. B. Representative time trajectory of the donor (Cy3, green), acceptor (Cy5, red), total fluorescence intensity (black), and the corresponding FRET values are in blue. Thick lines in green (Cy3), red (Cy5), and blue represent single-exponential fits of each oligo dA15 hybridization event. Thick lines in black represent the mean value of the total intensity during the hybridization of an oligo dA15. C. Contour plot of the evolution of FRET over time, measured with 749 single-molecule TUT4 complexes.
To initiate oligo-uridylation by the immobilized ternary TUT4/Lin28b/pre-miRNA complex, we injected a solution containing 100 μM UTP into the microfluidic chamber. To track the molecular dynamics of oligo-uridylation in real time, we included 10 nM Cy3-labeled oligonucleotide dA15 (oligo-dA15). Upon elongation of the U-tail, oligo-dA15 hybridized with the U-tail, resulting in a step-wise increase of the total fluorescence intensity (Fig. 6B, black line). Intriguingly, we obtained a signal from the Cy5-labeled pre-miRNA while exciting the Cy3-labeled oligo-dA15, suggesting that Förster resonance energy transfer (FRET) occurred between these two dyes (Fig. 6B, green and red line). Apparent FRET efficiency is the ratio between IA (acceptor signal) and (ID + IA) (total signals summing donor and acceptor signals). Upon hybridization of the first oligo-dA15, we observed an increase in FRET (Fig. 6B, blue line) that gradually decreased during the elongation of the U-tail. When the U-tail had reached a sufficient length, a second oligo-dA15 hybridized, leading to another increase in FRET that again gradually decreased over time (Fig. 6B and C). This suggests that TUT4 and Lin28 maintain a tightly associated complex, which captures the 3′ end of pre-miRNA and brings it to its own catalytic domain. This mechanism hints at the formation of a unique closed loop of the U-tail during oligo-uridylation by TUT4 (Fig. 6B).
To obtain a kinetic understanding of the oligo-uridylation process, we analyzed the gradually decreasing FRET events (Fig. 6C). Each oligo-dA15 hybridization event was selected using home-written Matlab software. The starting time and the FRET values of selected events were normalized such that all the events start with a FRET efficiency value 1. To visualize the distribution among the decaying FRET traces, a contour plot was made of the normalized FRET data. The kinetic rate was extracted by fitting the data from the contour plot with a single-exponential decay, which resulted in an average dwell time of 38 s per oligo-dA15 hybridization event.
Our oligo-uridylation data suggest that we were able to obtain functional full-length TUT4 molecules with high purity through the single-molecule pull-down method. In addition, by using FRET, we uncovered that TUT4 and Lin28 remain in tight contact while making use of a unique closed loop formation during oligo-uridylation. Recent crystal structures of the yeast homolog Cid1 showed that the surface of the C-terminal domain of Cid1 is mostly positively charged [55], which might facilitate loop formation by wrapping the U-tail around the protein. This suggests that the loop formation may be a general feature of TUTs.
4. Conclusion
We have shown that when integrated with protein complex pull-down methods, single-molecule fluorescence techniques become pertinent tools to obtain mechanistic insights into ribonucleoprotein complexes. These techniques can be applied to study the function and stoichiometry of any nucleoprotein that is difficult to obtain using traditional biochemical methods. However, special consideration must be given to protein complex purification and surface immobilization to attain single-molecule observation of functionally active proteins that are free from surface artifacts.
Acknowledgments
C.J. was funded by European Research Council under the European Union’s Seventh Framework Programme [FP7/2007-2013]/ERC grant agreement n° [309509]. We thank V. Narry Kim (Seoul National University), Mikiko Siomi (Keio University), and John Strouboulis (B.S.R.C. Alexander Fleming) for sharing plasmids. We thank Joo lab members for their help.
References
- [1].Hang J, Wan R, Yan C, Shi Y. Structural basis of pre-mRNA splicing. Science. 2015;349:1191–1198. doi: 10.1126/science.aac8159. [DOI] [PubMed] [Google Scholar]
- [2].Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- [3].Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- [4].Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- [5].Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Choudhury NR, Nowak JS, Zuo J, Rappsilber J, Spoel SH, Michlewski G. Trim25 is an RNA-specific activator of Lin28a/TuT4-mediated uridylation. Cell Rep. 2014;9:1265–1272. doi: 10.1016/j.celrep.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- [12].Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- [13].Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, et al. Superti-Furga, Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- [14].Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, et al. Greenblatt, Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- [15].Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- [16].Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- [17].Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, McKillip E, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Srihari S, Yong CH, Patil A, Wong L. Methods for protein complex prediction and their contributions towards understanding the organisation, function and dynamics of complexes. FEBS Lett. 2015;589:2590–2602. doi: 10.1016/j.febslet.2015.04.026. [DOI] [PubMed] [Google Scholar]
- [20].Maskell DP, Renault L, Serrao E, Lesbats P, Matadeen R, Hare S, Lindemann D, Engelman AN, Costa A, Cherepanov P. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rodgers ML, Paulson J, Hoskins AA. Rapid isolation and single-molecule analysis of ribonucleoproteins from cell lysate by SNAP-SiMPull. RNA. 2015;21:1031–1041. doi: 10.1261/rna.047845.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Krishnan R, Blanco MR, Kahlscheuer ML, Abelson J, Guthrie C, Walter NG. Biased Brownian ratcheting leads to pre-mRNA remodeling and capture prior to first-step splicing. Nat Struct Mol Biol. 2013;20:1450–1457. doi: 10.1038/nsmb.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kahlscheuer ML, Widom J, Walter NG. Single-molecule pull-down FRET to dissect the mechanisms of biomolecular machines. Methods Enzymol. 2015;558:539–570. doi: 10.1016/bs.mie.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lee HW, Ryu JY, Yoo J, Choi B, Kim K, Yoon TY. Real-time single-molecule coimmunoprecipitation of weak protein-protein interactions. Nat Protoc. 2013;8:2045–2060. doi: 10.1038/nprot.2013.116. [DOI] [PubMed] [Google Scholar]
- [27].Lee HW, Kyung T, Yoo J, Kim T, Chung C, Ryu JY, Lee H, Park K, Lee S, Jones WD, Lim DS, et al. Real-time single-molecule co-immunoprecipitation analyses reveal cancer-specific Ras signalling dynamics. Nat Commun. 2013;4:1505. doi: 10.1038/ncomms2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang Y, Ha T. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jain A, Arauz E, Aggarwal V, Ikon N, Chen J, Ha T. Stoichiometry and assembly of mTOR complexes revealed by single-molecule pulldown. Proc Natl Acad Sci USA. 2014;111:17833–17838. doi: 10.1073/pnas.1419425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Panter MS, Jain A, Leonhardt RM, Ha T, Cresswell P. Dynamics of major histocompatibility complex class I association with the human peptide-loading complex. J Biol Chem. 2012;287:31172–31184. doi: 10.1074/jbc.M112.387704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shen Z, Chakraborty A, Jain A, Giri S, Ha T, Prasanth KV, Prasanth SG. Dynamic association of ORCA with prereplicative complex components regulates DNA replication initiation. Mol Cell Biol. 2012;32:3107–3120. doi: 10.1128/MCB.00362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jain A, Liu R, Xiang YK, Ha T. Single-molecule pull-down for studying protein interactions. Nat Protoc. 2012;7:445–452. doi: 10.1038/nprot.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Means CK, Lygren B, Langeberg LK, Jain A, Dixon RE, Vega AL, Gold MG, Petrosyan S, Taylor SS, Murphy AN, Ha T, et al. An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc Natl Acad Sci USA. 2011;108:E1227–E1235. doi: 10.1073/pnas.1107182108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yeom KH, Heo I, Lee J, Hohng S, Kim VN, Joo C. Single-molecule approach to immunoprecipitated protein complexes: insights into miRNA uridylation. EMBO Rep. 2011;12:690–696. doi: 10.1038/embor.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stathopulos PB, Scholz GA, Hwang YM, Rumfeldt JA, Lepock JR, Meiering EM. Sonication of proteins causes formation of aggregates that resemble amyloid. Protein Sci. 2004;13:3017–3027. doi: 10.1110/ps.04831804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Selvin PR, Ha T. Single-molecule techniques: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. [Google Scholar]
- [37].Chandradoss SD, Haagsma AC, Lee YK, Hwang JH, Nam JM, Joo C. Surface passivation for single-molecule protein studies. J Vis Exp. 2014 doi: 10.3791/50549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pan H, Xia Y, Qin M, Cao Y, Wang W. A simple procedure to improve the surface passivation for single molecule fluorescence studies. Phys Biol. 2015;12:045006. doi: 10.1088/1478-3975/12/4/045006. [DOI] [PubMed] [Google Scholar]
- [39].Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- [40].Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- [41].Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, Hohng S, Kim VN, Woo JS. Functional anatomy of the human microprocessor. Cell. 2015;161:1374–1387. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- [44].Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- [45].Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–546. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sinha NK, Trettin KD, Aruscavage PJ, Bass BL. Drosophila dicer-2 cleavage is mediated by helicase- and dsRNA termini-dependent states that are modulated by Loquacious-PD. Mol Cell. 2015;58:406–417. doi: 10.1016/j.molcel.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- [49].Kim B, Ha M, Loeff L, Chang H, Simanshu DK, Li S, Fareh M, Patel DJ, Joo C, Kim VN. TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J. 2015;34:1801–1815. doi: 10.15252/embj.201590931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- [51].Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- [52].Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013;19:1632–1638. doi: 10.1261/rna.040055.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Munoz-Tello P, Gabus C, Thore S. Functional implications from the Cid1 poly (U) polymerase crystal structure. Structure. 2012;20:977–986. doi: 10.1016/j.str.2012.04.006. [DOI] [PubMed] [Google Scholar]