Abstract
Background: Breast cancer patients with residual disease after neoadjuvant chemotherapy (NAC) have increased recurrence risk. Molecular characterization, knowledge of NAC response, and simultaneous generation of patient-derived xenografts (PDXs) may accelerate drug development. However, the feasibility of this approach is unknown.
Methods: We conducted a prospective study of 140 breast cancer patients treated with NAC and performed tumor and germline sequencing and generated patient-derived xenografts (PDXs) using core needle biopsies. Chemotherapy response was assessed at surgery.
Results: Recurrent “targetable” alterations were not enriched in patients without pathologic complete response (pCR); however, upregulation of steroid receptor signaling and lower pCR rates (16.7%, 1/6) were observed in triple-negative breast cancer (TNBC) patients with luminal androgen receptor (LAR) vs basal subtypes (60.0%, 21/35). Within TNBC, TP53 mutation frequency (75.6%, 31/41) did not differ comparing basal (74.3%, 26/35) and LAR (83.3%, 5/6); however, TP53 stop-gain mutations were more common in basal (22.9%, 8/35) vs LAR (0.0%, 0/6), which was confirmed in The Cancer Genome Atlas and British Columbia data sets. In luminal B tumors, Ki-67 responses were observed in tumors that harbored mutations conferring endocrine resistance (p53, AKT, and IKBKE). PDX take rate (27.4%, 31/113) varied according to tumor subtype, and in a patient with progression on NAC, sequencing data informed drug selection (olaparib) with in vivo antitumor activity observed in the primary and resistant (postchemotherapy) PDXs.
Conclusions: In this study, we demonstrate the feasibility of tumor sequencing and PDX generation in the NAC setting. “Targetable” alterations were not enriched in chemotherapy-resistant tumors; however, prioritization of drug testing based on sequence data may accelerate drug development.
Breast cancer patients who receive neoadjuvant chemotherapy (NAC) and achieve pathologic complete response (pCR; elimination of invasive tumor in breast and axillary lymph nodes) have excellent long-term survival; conversely, patients with residual disease are at increased risk of disease recurrence and early death (1). Multiple studies have evaluated the association of clinical (2) and molecular subtypes (eg, intrinsic subtypes [3]) with pCR. While more recent reports have detailed the molecular landscape of breast cancer (4–6), the implications of these alterations in terms of chemotherapy response have not been well studied.
Patient-derived xenografts (PDXs) are increasingly used as a tool to study the impact of new drugs (7,8). However, the clinical information from many of these models is unknown. Therefore, in the setting of NAC, a detailed molecular characterization of breast cancer along with the collection of PDX models would allow the opportunity to prioritize the development of new agents.
We developed a prospective neoadjuvant clinical study (the Breast Cancer Genome Guided Therapy Study) in women with stage I–III breast cancer. The goals of this study were to 1) perform comprehensive sequencing of germline (9) and tumor tissues (exome and RNA sequencing), 2) identify whether common genomic alterations and perturbed pathways were associated with response/resistance to standard chemotherapy, and 3) develop PDXs from percutaneous needle biopsies for testing of therapeutic regimens chosen on the basis of alterations identified by genetic sequencing.
Methods
The Breast Cancer Genome Guided Therapy Study (BEAUTY) is a prospective institutional review board–approved NAC clinical study (NCT02022202) enrolling patients age 18 years or older with stage I–III breast cancer 1.5 cm in size or larger being recommended treatment with NAC by their treating physicians from March 5, 2012, to May 1, 2014, across Mayo Clinic Rochester, Mayo Clinic Florida, and Mayo Clinic Arizona (Supplementary Figure 1, available online). All patients provided written informed consent and were to receive 12 weeks of weekly paclitaxel (with trastuzumab for human epidermal growth factor receptor 2–positive [HER2+] malignancies), followed by four cycles of an anthracycline-based regimen (see Supplementary Table 1, available online). Pertuzumab was allowed along with trastuzumab for HER2+ disease after September 2012. Carboplatin was allowed after June 2013 for triple-negative breast cancer (TNBC).
Ultrasound-guided percutaneous tumor biopsies and blood specimens were collected prior to NAC, and tumor tissue was collected from surgery. Baseline and surgical samples were injected into mice for xenografts and fresh-frozen for sequencing (Supplementary Figure 2, available online)
Surgery was performed following completion of NAC, and residual cancer burden (RCB) scores were calculated (10). Pathologic complete response (pCR) was defined as absence of invasive tumor in the breast and axillary lymph nodes (ypT0/Tis, ypN0) (11). Clinical approximated subtypes were defined using the St. Gallen Criteria (12): luminal A (estrogen receptor [ER] > 10% + grade 1 [13] or ER > 10% + grade 2 + Ki-67 < 15%); luminal B (ER > 10% + grade 2 + Ki67 ≥ 15% or ER > 10% + grade 3); ER+/human epidermal growth factor receptor 2 positive [HER2+] (ER > 10% + HER2+ [3+] by immunohistochemistry [IHC] or amplified by fluorescence in situ hybridization [FISH]); ER-/HER2+ (ER ≤ 10% + HER2+ [3+] by IHC or amplified by FISH); and TNBC (ER ≤ 10% + progesterone receptor–[PR] ≤ 10% HER2-).
Sample Size Justification
With 200 subjects, we would be able to detect 80% of the variants in a region (assuming 20 variants in the region) with minor allele frequency of 0.005 with probability 0.86. We anticipated a minimum detected odds ratio for pCR assuming that 40% of the 200 subjects enrolled would have a pCR. The odds ratios are presented in the supplementary methods, available online for a variety of minor allele frequencies/variable frequencies (5%, 10%, and 20%) using a test of association with statistical significance level α (0.00001, 0.0001, and 0.001). The statistical significance levels used are to account for the testing of multiple hypotheses. Power was computed using the software package Quanto for a binary end point and assumed a dominant genetic mode. Given a slower accrual rate than expected, the study was closed early after accrual of 140 patients over 2.5 years.
Xenograft Generation
All procedures of animal studies were performed according to the National Institutes of Health guideline with approval obtained from the Mayo Clinic Institutional Animal Care and Use Committee (IACUC) and Biosafety Committee. Female nonobese diabetic (NOD)-SCID (NOD.CB17-Prkdcscid/J) mice or NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (six to eight weeks) were purchased from Jackson Laboratories (Bar Harbor, ME) and fed water containing 0.16 µg/mL 17β-estradiol (purchased from sigma Aldrich, St. Louis, MO) for one to two weeks prior to xenografting. Percutaneous breast cancer biopsies obtained prior to NAC in all patients were used for attempted xenograft establishment. Percutaneous needle biopsies (one to two cores from 14 gauge needles) were implanted with Matrigel (BD Biosciences, Heidelberg, Germany) within an hour of sample collection in the flanks of immunodeficient NOD-SCID or NSG mice. Low-dose estradiol was maintained in the drinking water and freshly made every week. Mice were killed by CO2 inhalation once the tumor size met the IACUC guideline. Tumors were formalin fixed and paraffin embedded, and the histology of the PDX was confirmed by hematoxylin and eosin and immunohistochemical stains. Take rate was defined as the number of unique subjects with histologically verified tumor growth with passage to second-generation mice.
In Vivo Drug Testing
A PDX (both pretreatment biopsy and residual surgical specimen) that was derived from a patient who progressed on chemotherapy and rapidly developed brain metastases and death following surgery was selected for in vivo drug testing. Once PDX tumors grew to 1 cm in diameter, tumors were resected and dissected into 8 mm3 pieces and re-injected subcutaneously into the rear flank of NOD-SCID mice. Fourteen mice were implanted with the pretreatment biopsy specimen, and following tumor growth to 200–250 mm3, mice were randomized 1:1 to placebo or olaparib (15 mg/kg, once daily, i.p.) using stratified sampling randomization based on tumor volume (StudyDirector software, StudyLog, San Francisco, CA) The process was repeated for 14 mice implanted with the PDX that grew from the residual surgical specimen. Tumor size and body weight were measured on days 0, 2, 5, 9, and 12. Mice were killed after nine days (those bearing the pretreatment PDX) or 12 days (mice bearing residual surgical specimen PDX).
Statistical Analysis
We analyzed the somatic mutation profiles at the gene level for all samples in the study as well as the breast cancer subtypes using the program MutSigCV (version 1.4) (14) to identify somatically mutated genes that were more frequently observed than expected by chance as determined by adjusting for covariates and background mutation processes. We filtered out somatic variants that were found in the 1000 genomes database or Exome Variant Server at greater than 1.0%. The somatic alteration profiles of the 30 most frequently mutated genes as identified by MutSigCV are presented in the results. The P values were determined using a one-sided bivariate beta-binomial test where the gene’s observed mutation rates and mutational functional categories across all samples are compared with the corresponding gene-specific background mutation rate and functional category distribution. For each gene in the top 30 of each subtype and across all subtypes, the P values were less than .025. The q value is calculated using the Benjamini and Hochberg (15) method and is a statistic to measure false discovery rate and adjust for multiple testing. Only a portion of the genes in the top 30 are statistically significant with respect to this q value. Genes with statistically significant q values are listed in the figure captions. In addition, the gene expression analysis for individual breast cancer subtypes was conducted using EdgeR (16). The details of the gene expression, pathways, and single patient analyses are provided in the Supplementary Methods (available online).
Relative tumor volume was defined as the tumor volume at the time of death divided by the tumor volume prior to start of treatment. Wilcoxon rank-sum tests were used to assess whether the relative tumor volume at time of death differed with respect to treatment.
All statistical tests were two-sided, and a P value of less than .05 was considered statistically significant. SAS 9.3 was used to carry out statistical analyses.
Results
Patient Characteristics
Of the 140 women enrolled, eight withdrew consent prior to NAC (Supplementary Figure 3, available online, CONSORT diagram). Of the remaining 132, 65 (49.2%) had clinical cT3/4 tumors, and 76 (57.6%) had node-positive disease (Table 1). The clinical molecular subtypes were: TN: 33.3%; ER-/HER2+: 15.2%; ER+/HER2+: 12.1%; luminal B: 29.5%; luminal A: 8.3%; and luminal unknown: 1.5%.
Table 1.
Baseline characteristics of patients in the BEAUTY study
| Patient and disease features | Total (n = 132) No. (%) |
|---|---|
| Age, y | |
| <30 | 2 (1.5) |
| 30–39 | 21 (15.9) |
| 40–49 | 36 (27.3) |
| 50–59 | 40 (30.3) |
| 60–69 | 24 (18.2) |
| 70+ | 9 (6.8) |
| Ethnicity | |
| Hispanic or Latino | 1 (0.8) |
| Not Hispanic or Latino | 129 (97.7) |
| Not Reported | 2 (1.5) |
| Clinical molecular subtype | |
| Triple-negative* | 44 (33.3) |
| Basal-like 1 | 6 (14.6) |
| Basal-like 2 | 5 (12.2) |
| Immunomodulatory | 10 (24.3) |
| Mesenchymal | 6 (14.6) |
| Mesenchymal stem–like | 6 (14.6) |
| Luminal androgen receptor | 4 (9.7) |
| Unstable or ER-positive | 4 (9.7) |
| ER-/HER2+ | 20 (15.2) |
| ER+/HER2+ | 16 (12.1) |
| Luminal B | 39 (29.5) |
| Luminal A | 11 (8.3) |
| Luminal unknown | 2 (1.5) |
| Clinical T-stage | |
| T1 | 12 (9.1) |
| T2 | 55 (41.7) |
| T3 | 60 (45.5) |
| T4 | 5 (3.8) |
| Clinical N-stage | |
| N0 | 56 (42.4) |
| N1 | 69 (52.3) |
| N2 | 4 (3.0) |
| N3 | 3 (2.3) |
| Nottingham Grade | |
| 1 | 8 (6.1) |
| 2 | 51 (38.6) |
| 3 | 73 (55.3) |
| Histology | |
| Infiltrating ductal | 114 (86.4) |
| Infiltrating lobular | 5 (3.8) |
| Mixed lobular ductal | 10 (7.6) |
| Other | 3 (2.3) |
| Pre-NAC Ki67 | |
| ≥15% | 113 (89.0) |
| <15% | 14 (11.0) |
| Missing | 5 |
Lehman Classification derived based on RNA seq for 41/44 TNBC. ER = estrogen receptor; HER2 = human epidermal growth factor receptor 2.
Treatment Course and Outcome
Eight patients failed to complete NAC due to disease progression (three patients) and patient refusal (five patients). The pCR rate varied by clinical molecular subtype (Table 2) and by PAM50 (Supplementary Table 2, available online). Of the 132 who began NAC, pCR rates were: TN: 54.5%; ER-/HER2+: 60.0%; ER+/HER2+: 25.0%; luminal B: 10.3%; and luminal A/unknown: 0%.
Table 2.
Residual disease by clinical molecular subtype
| Pathological extent of residual disease | Clinical molecular subtype |
Total (n = 132) No. (%) | |||||
|---|---|---|---|---|---|---|---|
| Triple-negative (n = 44) No. (%) | ER-/HER2+ (n = 20) No. (%) | ER+/HER2+ (n = 16) No. (%) | Luminal B (n = 39) No. (%) | Luminal A (n = 11) No. (%) | Luminal unknown (n = 2) No. (%) | ||
| Residual cancer burden class | |||||||
| RCB-0 | 24 (54.5) | 12 (60.0) | 4 (25.0) | 4 (10.3) | 0 (0.0) | 0 (0.0) | 44 (33.3) |
| RCB-I | 6 (13.6) | 3 (15.0) | 2 (12.5) | 3 (7.7) | 0 (0.0) | 0 (0.0) | 14 (10.6) |
| RCB-II | 7 (15.9) | 2 (10.0) | 6 (37.5) | 14 (35.9) | 7 (63.6) | 0 (0.0) | 36 (27.3) |
| RCB-III | 5 (11.4) | 3 (15.0) | 4 (25.0) | 16 (41.0) | 4 (36.4) | 2 (100.0) | 34 (25.8) |
| Not evaluable because of progression* | 2 (4.5) | 0 (0.0) | 0 (0.0) | 2 (5.1) | 0 (0.0) | 0 (0.0) | 4 (3.0) |
| pCR | |||||||
| Yes | 24 (54.5) | 12 (60.0) | 4 (25.0) | 4 (10.3) | 0 (0.0) | 0 (0.0) | 44 (33.3) |
| No | 20 (45.5) | 8 (40.0) | 12 (75.0) | 35 (89.7) | 11 (100.0) | 2 (100.0) | 88 (66.7) |
* These patients did not have surgery because of progression. ER = estrogen receptor; HER2 = human epidermal growth factor receptor 2; RCB = residual cancer burden; pCR = pathologic complete response.
Sequencing Analysis
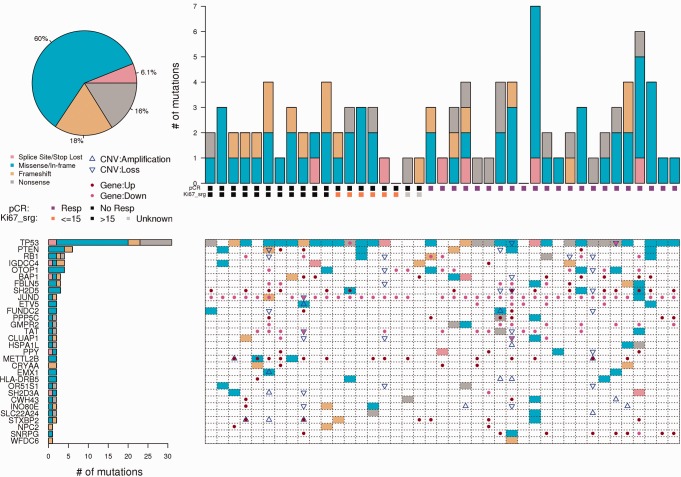
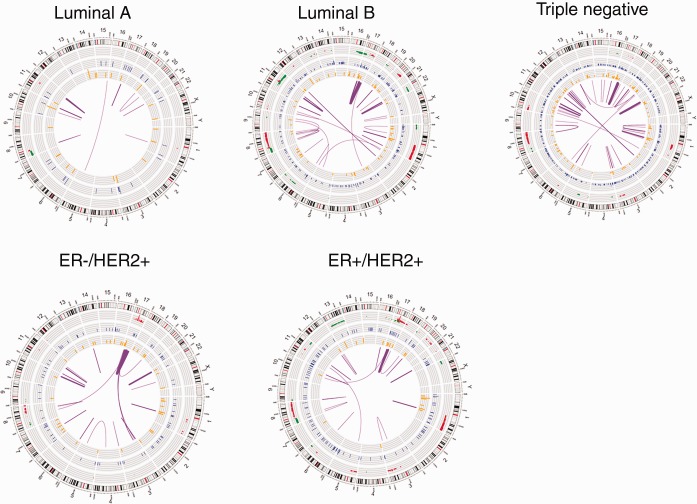
Pre-NAC tumor and germline sequencing data were generated for 122 patients. RNA and exome sequencing results are in Supplementary Tables 3–12 (available online) and Supplementary Figure 4, A and B (available online). The profiles of the most frequently mutated breast cancer genes, somatic copy number alteration (CNA), and gene expression modulation are provided by clinical molecular subtype in Figure 1 and Supplementary Figures 5–8 (available online). Somatic mutations, CNAs, and fusion transcripts according to breast cancer subtype are illustrated in Figure 2 . Detailed CNAs by clinical molecular subtype are provided in Supplementary Tables 7–11 (available online).
Figure 1.
Somatically mutated genes in patients with triple-negative breast cancer (TNBC) clinical molecular subtype. The mutation profile of the 30 most frequently mutated genes (P < .006) for triple-negative subtype, as determined by the MutSigCV method. After adjustment for multiple testing (q < .05), three genes (TP53, PTEN, and OTOP1) remained statistically significant. The upper left panel shows the distribution of the mutation type across all patients; the upper right panel shows the distribution of mutations by chemotherapy response for each patient. The lower left panel shows the number and type of mutations seen per gene for the 30 mutated genes. We order the genes in the figures by the number of patients with mutations. For genes with equal numbers of patients with somatic SNV/indel alterations, the presentation order is based on estimated cancer gene statistical significance. The lower right panel gives the mutation, the copy number gain and loss, and modulation of expression (described in the “Methods” section) by patient and by gene. CNV = copy number variants; INDEL = insertion and deletion; pCR = pathologic complete response; SNV = single-nucleotide variant.
Figure 2.
Circos plots of mutations in clinical molecular breast cancer subtypes. This plot depicts the human chromosomes arranged in a circular pattern, with individual chromosomes represented as sections. From outside to inner side: The outermost track represents the copy number alteration (CNA), amplification in red, and deletion in green); the second track presents somatic single-nucleotide variants (SNVs) identified in whole-exome sequencing data (blue); the third track is RNA nonsynonymous tumor-specific expressed SNV(eSNVs; orange). The radius height in the outer three tracks represents the CNA ratio across the samples of this subtype. The innermost arches (purple) are the fusions; the two ends of an arch indicate the location of the two fused genes, and the thickness of the arch is proportional to the frequency of the fusion.
We analyzed the frequency of somatic mutation profiles across genes using the program MutSigCV (version 1.4) (Supplementary Methods, available online). At the genome-wide level, a frequent somatically mutated gene observed across all tumor subtypes was PIK3CA (TN: 2/41; ER-/HER2+: 4/20; ER+/HER2+: 3/15; luminal B: 8/33; and luminal A: 4/11). Additionally, TP53 was statistically significantly mutated in TN (75.6%, 31/41) and HER2+ tumors, regardless of ER status (62.9%, 22/35), but less frequent in luminal B (9.1%, 3/33) and luminal A tumors (0/11). Other frequently mutated genes included OTOP1 (9.0%, 11/122), MAP3K1 (6.6%, 8/122), PTEN (6.6%, 8/122), ARID1A (6.6%, 8/122), GATA3 (5.7%, 7/122), AKT1 (4.9%, 6/122), KDM6A (4.9%, 6/122), IGDCC4 (4.9%, 6/122), and CDH1 (4.1%, 5/122) (Supplementary Tables 5, a–o, available online). The somatic mutations from exome sequencing were confirmed using tumor RNA-Seq (Supplementary Table 5, a–o, available online); a statistically significantly greater number of somatic expressed single-nucleotide variants (eSNVs) were present in TNBC compared with other subtypes (P = .001) (Figure 2).
Recurrent Gene Mutations and Chemotherapy Response
Triple-Negative Subtype
By PAM50, 38 of 41 (92.7%) were “basal” (Supplementary Table 2, available online), and responses to chemotherapy (pCR following NAC) were seen in both the basal and nonbasal groups. The most frequently mutated genes by MutSigCV analysis were TP53 (31/41), PTEN (6/41), and OTOP1 (4/41) (Figure 1), but their frequency did not differ by chemotherapy response. Regarding CNAs, we observed amplifications in chr8q12.1-.3 (5/22), chr8q13.1-.3 (7/22), and chr8q21.11-.12 (7/22) only in tumors that achieved pCR. These CNA regions in tumors that achieved pCR were associated with expression of 28 genes on chromosome 8 (Supplementary Table 13, available online). Evaluating tumors without pCR (n = 20), we observed frequent amplifications in chr15q26.3 (4/20) (Supplementary Table 7, available online). Evaluation of mutation and CNA data identified variations in HYDIN and RP1 more often in chemosensitive tumors (Supplementary Table 5 and 6, available online).
Evaluating differential gene expression in TN tumors, we identified 419 genes (with at least a twofold change and P value of less than .05; additional details of gene expression analysis are included in the Supplementary Materials, available online), and multiple statistically significant pathways were associated with pCR, including glucocorticoid receptor (GR) regulatory network; FOXA1, FOXA2, and FOXA3 networks; integrins; SMAD2/3; and androgen receptor signaling (Supplementary Table 13, available online). We observed overexpression of AR, ERBB4, FOXA1, ESR1, PGR, EGF, MAPK10, KIT, and FGFR2 in non-pCR patients compared with patients with pCR.
As androgen and other hormone-related genes/signaling pathways were upregulated in the overall TN cohort with chemotherapy resistance, we applied a shrunken centroid model (Supplementary Material, available online) to TN RNA-Seq data and demonstrated that the luminal androgen receptor (LAR) subtype comprised 14.6% (6/41) of the TN group, while the remaining 35 tumors were basal, with the basal subtype exhibiting fewer hormone-related pathways associated with pCR (Supplementary Table 14, available online). Lower pCR rates were seen in the LAR subtype (16.6%, 1/6) compared with the basal subtype (60.0%, 21/35).
Within clinical TNBC, the TP53 mutation frequency was 75.66% (31/41), which compared similarly with the TCGA (4) and British Columbia (BC) cohorts (Supplementary Table 22, available online) (17) and did not differ comparing basal (74.3%, 26/35) and LAR (83.3%, 5/6) subtypes. Given that TP53 mutations are known to be associated with pCR (18), we evaluated whether TP53 mutation type differed between the basal and LAR groups. Indeed, the frequency of TP53 stop-gain mutations was higher within the basal (22.9, 8/35) vs LAR (0%; 0/6) subtypes, a finding confirmed within TCGA (4) and BC (17) data sets where TP53 stop-gain alterations were observed in 15.0% (16/107) and 13.2% (7/53) of basal tumors, respectively, compared with 8.3%, (2/24) and 8.3% (1/12) of the LAR tumors, respectively. Given different frequencies of TP53 mutations within two distinct TNBC subgroups that additionally exhibit differential response to chemotherapy, we evaluated the pCR rate according to type of TP53 mutation. Of the 31 TNBCs with TP53 mutations, the overall pCR was 17 of 31 (54.8%) but higher, 75.0% (6/8), in patients with TP53 stop-gain alteration vs 47.8% (11/23) for all other TP53 mutations (frame-shift, nonsynonymous coding and splice site mutations).
ER-/HER2+ Subtype
PAM50 analysis demonstrated that 13 of 20 were HER2 enriched (Supplementary Table 2, available online) and eight of 13 and four of seven achieved pCR in the HER2-enriched and HER2-nonenriched subsets, respectively. The most commonly mutated genes were TP53 (12/20) and PIK3CA (4/20) (Supplementary Figure 5, available online). All four patients with PIKC3A mutations had pCR whereas the frequency of TP53 mutations did not differ according to chemotherapy response.
CNA amplifications in chr3q23 (3/12), chr17q24.3 (3/12), and chr20q13.33 (3/12) were only observed in patients achieving pCR. In patients without pCR (n = 8), we observed amplifications in chr1q21.1-.3 (3/8), chr1q23.3 (3/8), and chr11q13.3 (3/8) and deletions in chr8p12 (3/8), 8p22 (3/8), 9p21.2 (3/8), and 9p22.3 (3/8) (Supplementary Table 8, available online), but not in patients with pCR (0/12).
We identified 445 differentially expressed genes (analysis details discussed in Supplementary Material, available online) and pathways comparing non-pCR (n = 8) and pCR patients (n = 12) including FGFR and EGFR1, androgen and ER signaling, angiopoietin receptor Tie2-mediated signaling, and signaling events regulated by RET (Supplementary Table 16, available online). Network analysis of 445 genes demonstrated TNNI3, COL11A2, AGT, GRIN2B, and ACTN2 genes to be upregulated and highly connected, whereas ESR1, KIT, IRF4, IL12B, and IGF1 genes were demonstrated to be highly connected and downregulated in patients without pCR compared with patients with pCR.
ER+/HER2+ Subtype
By PAM50, 10 of 15 were HER2 enriched (Supplementary Table 2, available online). The most frequently mutated genes included TP53 (10/15), ARID1A (4/15), PIK3CA (3/15), and KDM6A (3/15) (Supplementary Figure 6, available online). The mutation frequency did not differ by chemotherapy response except for KDM6A, where mutations were observed in three of 11 tumors with residual disease compared with zero of four with pCR.
With regard to CNA, amplifications in chr17q22 (4/11), chr17q23.1-.2 (4/11), chr17q24.2-.3 (3/11), chr17q25.1 (3/11), and chr20q13.32 (3/11) and deletions in chr16q22.2 (4/11) were observed only in patients without pCR (Supplementary Table 9, available online). In contrast, patients with pCR exhibited amplifications in chr11q13.2‐13.3 (2/11), chr5p13.1‐13.3 (2/11), chr5p14.1,14.3 (2/11), chr5p15.1‐15.2 (2/11), and chr5p15.31‐15.33 (2/11) (Supplementary Table 9, available online).
Differential gene expression analysis for tumors with (n = 4) and without pCR (n = 11) identified 354 differentially expressed genes (analysis details discussed in the Supplementary Material, available online) and pathways including Ret, ERbB2/ErB3, glucocorticoid, nongenomic androgen receptor signaling, and angiopoietin receptor Tie2-mediated signaling (Supplementary Table 17, available online). Network analysis of 354 genes demonstrated the following highly connected upregulated genes including; ERBB4, GATA2, ERBB3, MYB, GFRA1, KCNJ3, and ZBTB16; and downregulated genes: VEGFA, APOA1, CAMK2B, ASCL1, FGF19, FGB, FGA, NKX2‐5, HAP1, and COL19A1 in patients without and with pCR.
Clinical Luminal B Subtype
Within the clinical luminal B tumors (n = 33), PAM50 demonstrated: luminal B (13/33), luminal A (7/33), basal (7/33), HER2 (3/33), missing (1/33), and normal (2/33) (Supplementary Table 2, available online). The most commonly observed mutations included PIK3CA (8), AKT1 (4), MAP3K1 (4), TP53 (3), CDH1 (3), and GATA3 (3) (Supplementary Figure 7, available online). Because pCR was rarely observed (n = 2) and because postchemotherapy (surgical) Ki-67 values are prognostic in luminal breast cancers (19), luminal B tumors were defined as “chemosensitive” (pCR or surgical Ki-67 ≤ 15%, n = 23), “chemoresistant” (surgical Ki-67 > 15%, n = 5), or not classified (surgical Ki-67 not available, n = 5).
Of the eight patients with PIK3CA mutations, seven were evaluable for response including chemosensitive (6/23) and chemoresistant (1/5) tumors (Supplementary Figure 7, available online). Ki-67 responses were observed in tumors that harbored genetic alterations that confer endocrine resistance (p53, AKT, and IKBKE).
Regarding CNA, amplifications in chr14q21.3 and chr14q22.1 (6/23) and deletions in chr17p11.2 (6/23) and chr17p13.1‐3 (6/23) were only observed in chemosensitive tumors. In contrast, there were no chromosomal bands that were clearly specific to chemoresistant tumors in this group (Supplementary Table 11, available online).
Differential gene expression analysis comparing chemoresistant (n = 5) and chemosensitive (n = 23) tumors identified 863 genes (analysis details discussed in Supplementary Material, available online). Pathways statistically significantly associated with chemosensitivity (FDR < 0.00001) included GR signaling, GMCSF-mediated signaling, pathways involved in chronic myeloid leukemia, Pertussis, and IL2-mediated signaling events (Supplementary Table 18, available online). Genes such as GFRA1, PGR, ESR1, ERBB4, RET, IGF1R, BCL2, and GATA3 were key upregulated nodes, whereas MMP7, PSMA8, PRAME, WT1, CFTR, CRYAB, and EPHB6 were downregulated (highly connected) in the network analysis of 863 genes.
Clinical Luminal A Subtype
Evaluation by PAM50 demonstrated the following: luminal A (6/11), luminal B (2/11), basal (1/11), HER2 (1/11), and normal (1/11) (Supplementary Table 2, available online). While the clinical luminal A tumors were characterized by a lack of genomic instability compared with other subtypes (Figure 2), the most frequently mutated genes by MutSigCV analysis included PIK3CA (4/11), CBFB (4/11), and GATA3 (3/11) (Supplementary Figure 8, available online). The most common CNAs included chr8p11.22 (2/11) and chr8p11.23 (2/11).
Patient-Derived Xenografts
In 120 patients, tissue was obtained from pre-NAC percutaneous biopsy samples and implanted subcutaneously into 412 mice. PDX from seven patients had some tumor growth but without pathological confirmation. Therefore, 113 patients were evaluable for assessment of PDX take rate. The overall take rate was 27.4% (31/113) (Table 3). TNBC yielded the highest take rate (51.3%, 20/39), followed by HER2+ (26.5%, 9/34) and luminal B (5.0%, 2/40). No PDXs were derived from luminal A tumors. From 35 patients with residual tumor after NAC, we established six PDX models (17.1%, 6/38), and five out of six were TNBC.
Table 3.
Breast cancer PDX take rate by clinical molecular subtype*
| Clinical molecular subtype | Tumor tissue implanted into xenograft | Breast cancer PDX generated | PDX take rate, % |
|---|---|---|---|
| Triple-negative | 39 | 20 | 51.3 |
| ER-/HER2+ | 20 | 5 | 25.0 |
| ER+/HER2+ | 14 | 4 | 28.6 |
| Luminal B | 30 | 2 | 6.7 |
| Luminal A | 9 | 0 | 0 |
| Luminal unknown | 1 | 0 | 0 |
| Total | 113 | 31 | 27.4 |
* ER = estrogen receptor; HER2 = human epidermal growth factor receptor 2; PDX = patient-derived xenograft.
Potential Usefulness of Xenografts for Validating Drug Targets
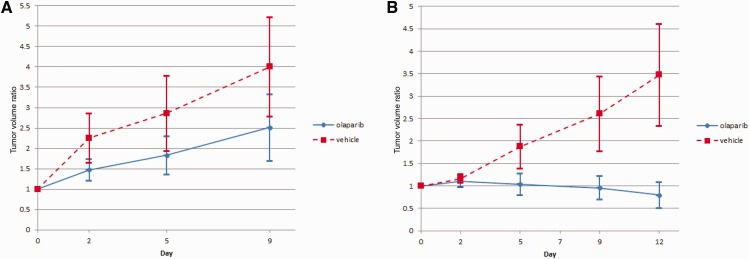
PDXs were established from two of six patients with progression during chemotherapy, including one with rapid development of metastases to brain and death. In this patient, based on baseline tumor sequencing demonstrating deleterious somatic alterations in PTEN, PALB2, BRCA1, CHEK1, DICER1, and NF1, as well as IRF4 and MYC amplification, olaparib was chosen for PDX testing. Fourteen mice were implanted with PDXs generated from pretreatment biopsy specimen and another 14 with PDXs generated from residual disease found at surgery. Seven mice from each group were randomized to either olaparib or placebo. Relative tumor volumes at day 9 were statistically significantly smaller among the mice treated with olaparib than mice treated with placebo in the PDXs generated from pretreatment specimen (P = .04). Similarly, relative tumor volumes at day 12 were statistically significantly smaller among the mice treated with olaparib than mice treated with placebo in the PDXs generated from the residual tumor (P < .001) (Figure 3 ).
Figure 3.
Tumor growth in patient-derived xenografts (PDXs) generated from patient specimens. A) PDXs were generated from a pre–neoadjuvant chemotherapy (NAC) specimen treated with olaparib or placebo and B) PDXs generated from residual tumor after NAC treated with either olaparib or placebo. Error bars represent the standard deviation. NAC = neoadjuvant chemotherapy; PDX = patient-derived xenograft.
Discussion
This is the first prospective study to our knowledge to demonstrate the feasibility of using percutaneous core needle tumor biopsies for the dual purpose of generating massive parallel gene sequencing and PDXs in the NAC setting. Our a priori hypothesis was that alterations in oncogenic drivers and tumor suppressors previously reported as “targetable” would be commonly observed in patients with chemotherapy resistance. However, results from this study indicated that common “targetable” alterations were not enriched in chemotherapy-resistant tumors.
Transcriptome sequencing revealed that a substantial proportion of the resistant TNBC had upregulation of key nodes and pathways involved in steroid hormone receptor (HR) signaling including AR, FOXA1, FOXA2, and FOXA3, as well as genes/nodes involved in HR cross talk including ERBB4, EGF, MAPK10, KIT, and FGFR2. Given prior reports that these genes/pathways are enriched in the LAR subset, we hypothesized that resistant TNBC would be enriched with LAR subtype. Indeed, the LAR subtype exhibited limited response to chemotherapy compared with non-LAR tumors, confirming a recent report (20). Although TP53 mutations did not differ comparing the basal and LAR subtypes, we observed a higher frequency of TP53 stop-gain alterations comparing the basal (22.9%) and LAR (0%) subtypes, which we confirmed with the TCGA and BC data sets. Given our preliminary observation that higher rates of pCR were observed within tumors with the stop-gain TP53 mutations (75.0%) vs other TP53 mutations (47.8%), further studies in larger cohorts are needed to confirm whether TP53 mutation type as well as the presence/absence of hormonal pathway activation impacts chemotherapy response within the TNBC subgroup.
In luminal B, Ki-67 responses (Ki-67 ≤ 15%) were observed in tumors that harbored mutations that confer endocrine resistance (p53, AKT, and IKBKE). These data provide further evidence for the importance of chemotherapy in tumors with mutations that drive de novo endocrine resistance.
A major limitation to drug development is access to tumor models that faithfully replicate the human situation. PDXs mirror the biology of human tumors (21) and more accurately replicate the “host” factors critical for evaluating drug response phenotypes. Petrillo et al. (22) established xenografts from five of 20 breast tumors, of which four out of five were TNBC. Li et al. (23) established PDXs from 20 of 152 (13.1%), and some (seven PDX) were obtained from primary tumors. Notably, in this cohort using needle biopsies, our take rate of 27.4% compares favorably with prior studies (13%–27%) (24) that mainly utilized surgical samples. Like Petrillo, we observed that ER expression was a major determinant of take rate. The potential power of this approach is illustrated in the patient with progression on chemotherapy in which tumor sequencing identified potential drug targets and PDX testing of the primary and resistant tumors demonstrated antitumor activity with olaparib. However, this information was not available to this patient in a timely manner given the rapid development of distant metastases and death.
A limitation to this study is accrual was lower than anticipated and limited statistical power to assess whether the frequency of rare genomic alterations differed comparing responders with nonresponders. Because of this, larger cohorts are required to assess association with chemotherapy response. Additionally, work is ongoing within this cohort to define the immune transcriptome and its association with chemotherapy response.
In summary, this is the first study to demonstrate the feasibility of obtaining tumor sequencing data and PDX in the NAC setting. While “targetable” alterations were not enriched in chemotherapy-resistant tumors, the simultaneous generation of PDX and sequence data will be an invaluable tool for drug development, especially in chemotherapy-resistant tumors. Larger studies are necessary to evaluate whether targeting infrequent tumor alterations in patients with chemoresistance will improve clinical outcomes.
Funding
This work was supported by the Mayo Clinic Center for Individualized Medicine; Nadia’s Gift Foundation; John P. Guider; The Eveleigh Family; the Pharmacogenomics Research Network (grant number U10GM 61388‐15 to RW, LW, MPG); the National Institutes of Health (grant number RO1 GM28157 to RW); the Mayo Clinic Cancer Center (grant number CA15083‐40A2 to MPG, JNI); George M. Eisenberg Foundation for Charities; the Mayo Clinic Breast SPORE (grant number P50CA 116201‐9 to MPG, JNI, DWV, VJS, KRK); the Prospect Creek Foundation, and the Randy Shaver Cancer Research and Community Fund.
Notes
The study sponsors had no involvement in the design, collection, analysis, data interpretation, report writing, or decision to submit the paper for publication.
A subset of the xenografts described here are available through WuXi AppTec, and Mayo receives financial compensation for their commercial use.
We have made our data available through database of Genotypes and Phenotypes (dbGap) (25) developed by the National Center for Biotechnology Information. The manuscript reports results from 122 patients; however, patient consent to release data are available for 118 patients. Thereore, the individual sequencing and phenotypic data for 118 samples (118*3 = 354 bam files) are available through the dbGap ID phs001050.v1.p1 NIGMS/BreastCancerGenGuidedTherapy and the Supplementary Materials (available online).
The authors would like to thank Judith A. Gilbert for her assistance with manuscript writing, Vicki Shea for manuscript preparation, and the following people for their assistance with data handling and the clinical trial: Cloann Schultz, Janet Lensing, Sharon Mercill, Jean Stahl, Amy Headlee, Michelle Roos, Tom Stock, Roxann Neumann, Tabitha Hanson, Karla Kopp, Jeanne Hansen, Daniel O’ Brien, Aditya Bhagwate, and Matthew Bockol from Mayo Rochester; Jacqueline Joy from Mayo Florida; and Jennifer Roedig and Shelly Perez from Mayo Arizona.
Authors: Matthew P. Goetz, Krishna R. Kalari, Vera J. Suman, Ann M. Moyer, Jia Yu, Daniel W. Visscher, Travis J. Dockter, Peter T. Vedell, Jason P. Sinnwell, Xiaojia Tang, Kevin J. Thompson, Poulami Barman, Douglas W. Mahoney, Erin Carlson, Steven N. Hart, Ping Yin, Bo Qin, Sara J. Felten, Sarah A. McLaughlin, Alvaro Moreno-Aspitia, John A. Copland, Donald W. Northfelt, Richard J. Gray, Katie Hunt, Amy Conners, Hugues Sicotte, Jeanette E. Eckel-Passow, Jean-Pierre Kocher, James N. Ingle, Marissa S. Ellingson, Michelle McDonough, Eric D. Wieben, Richard Weinshilboum, Liewei Wang, Judy C. Boughey.
Affiliations of authors: Medical Oncology (MPG, JNI), Department of Molecular Pharmacology and Experimental Therapeutics (MPG, JY, PY, BQ, RW, LW), Department of Health Sciences Research (KRK, VJS, TJD, PTV, JPS, XT, KJT, PB, DWM, EC, SNH, SJF, HS, JEEP, JPK), Department of Laboratory Medicine and Pathology (AMM, DWV), Department of Radiology (KH), Center for Individualized Medicine (AC, RW), Biochemistry (MM), and Department of Surgery (JCB), Mayo Clinic, Rochester, MN; Department of Surgery (SAM), Hematology/Oncology (AMA), Department of Cancer Biology (JAC), and Department of Radiology (MSE, EDW), Mayo Clinic, Jacksonville, FL; Hematology/Oncology (DWN) and Department of Surgery (RJG), Mayo Clinic, Scottsdale, AZ.
Supplementary Material
References
- 1.van der Hage JH, van de Velde CCJH, Mieog SJSD. Preoperative chemotherapy for women with operable breast cancer (review). The Cochrane Library. 2012;5:1–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172. [DOI] [PubMed] [Google Scholar]
- 3.Prat A, Fan C, Fernandez A, et al. Response and survival of breast cancer intrinsic subtypes following multi-agent neoadjuvant chemotherapy. BMC Med. 2015;13:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis MJ, Ding L, Shen D, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao H, Korn JM, Ferretti S, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21(11):1318–1325. [DOI] [PubMed] [Google Scholar]
- 8.Ter Brugge P, Kristel P, van der Burg E, et al. Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1-deficient breast cancer. J Natl Cancer Inst. 2016;108(11):djw148. [DOI] [PubMed] [Google Scholar]
- 9.Ellingson MS, Hart SN, Kalari KR, et al. Exome sequencing reveals frequent deleterious germline variants in cancer susceptibility genes in women with invasive breast cancer undergoing neoadjuvant chemotherapy. Breast Cancer Res Treat. 2015;153(2):435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Symmans WF, Peintinger F, Hatzis C, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25(28):4414–4422. [DOI] [PubMed] [Google Scholar]
- 11.Edge SB, Byrd DR, Compton CC, et al. AJCC Cancer Staging Manual. 7th ed New York: Springer; 2010. [Google Scholar]
- 12.Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes—dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22(8):1736–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. [DOI] [PubMed] [Google Scholar]
- 14.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes, Nature. 2013;499:214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Statistical Society Series B. 1995;57:289. [Google Scholar]
- 16.Nikolayeva O, Robinson MD. edgeR for differential RNA-seq and ChIP-seq analysis: An application to stem cell biology. Methods Mol Biol. 2014;1150:45–79. [DOI] [PubMed] [Google Scholar]
- 17.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen MB, Zhu YQ, Xu JY, et al. Value of TP53 status for predicting response to neoadjuvant chemotherapy in breast cancer: A meta-analysis. PLoS One. 2012;7(6):e39655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Minckwitz G, Schmitt WD, Loibl S, et al. Ki67 measured after neoadjuvant chemotherapy for primary breast cancer. Clin Cancer Res. 2013;19(16):4521–4531. [DOI] [PubMed] [Google Scholar]
- 20.Masuda H, Baggerly KA, Wang Y, et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res. 2013;19(19):5533–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodhia KA, Hadley A, Haluska P, et al. Prioritizing therapeutic targets using patient-derived xenograft models. Biochim Biophys Acta. 2015;1855(2):223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrillo LA, Wolf DM, Kapoun AM, et al. Xenografts faithfully recapitulate breast cancer-specific gene expression patterns of parent primary breast tumors. Breast Cancer Res Treat. 2012;135(3):913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Shen D, Shao J, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4(6):1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whittle JR, Lewis MT, Lindeman GJ, et al. Patient-derived xenograft models of breast cancer and their predictive power. Breast Cancer Res. 2015;17:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39(10):1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.