Abstract
Histone deacetylase (HDAC) inhibitors are efficacious epigenetic-based therapies for some cancers and neurological disorders; however, each of these drugs inhibits multiple HDACs and has detrimental effects on the skeleton. To better understand how HDAC inhibitors affect endochondral bone formation, we conditionally deleted one of their targets, Hdac3, pre- and postnatally in type II collagen α1 (Col2α1)–expressing chondrocytes. Embryonic deletion was lethal, but postnatal deletion of Hdac3 delayed secondary ossification center formation, altered maturation of growth plate chondrocytes, and increased osteoclast activity in the primary spongiosa. HDAC3-deficient chondrocytes exhibited increased expression of cytokine and matrix-degrading genes (Il-6, Mmp3, Mmp13, and Saa3) and a reduced abundance of genes related to extracellular matrix production, bone development, and ossification (Acan, Col2a1, Ihh, and Col10a1). Histone acetylation increased at and near genes that had increased expression. The acetylation and activation of nuclear factor κB (NF-κB) were also increased in HDAC3-deficient chondrocytes. Increased cytokine signaling promoted autocrine activation of Janus kinase (JAK)–signal transducer and activator of transcription (STAT) and NF-κB pathways to suppress chondrocyte maturation, as well as paracrine activation of osteoclasts and bone resorption. Blockade of interleukin-6 (IL-6)–JAK–STAT signaling, NF-κB signaling, and bromodomain extraterminal proteins, which recognize acetylated lysines and promote transcriptional elongation, significantly reduced Il-6 and Mmp13 expression in HDAC3-deficient chondrocytes and secondary activation in osteoclasts. The JAK inhibitor ruxolitinib also reduced osteoclast activity in Hdac3 conditional knockout mice. Thus, HDAC3 controls the temporal and spatial expression of tissue-remodeling genes and inflammatory responses in chondrocytes to ensure proper endochondral ossification during development.
INTRODUCTION
Endochondral ossification is a dynamic developmental process wherein mesenchymal progenitor cells differentiate into chondrocytes and form a cartilaginous template for bone mineralization. During periods of skeletal growth, chondrocytes produce a matrix rich in type II collagen and proteoglycans such as aggrecan. Growth plate chondrocytes eventually become hypertrophic and stimulate vasculogenesis to recruit the osteoclasts and osteoblasts that remodel and ossify bone (1, 2). Chronic inflammation can alter endochondral bone development and interrupt skeletal growth (3–6). Despite the negative structural consequences of chronic inflammation, growth plate chondrocytes produce inflammatory cytokines and matrix-degrading enzymes during development (3, 7, 8). Thus, controlled local expression of these proteins may have a role in skeletal formation and maintenance. Understanding the complex molecular regulatory pathways controlling each step of endochondral ossification and production of cytokines by chondrocytes is needed to improve our understanding of skeletal development, as well as that of regeneration, because tissue repair processes mediated by inflammatory cytokines and endochondral bone formation are also essential and sequential steps of bone fracture repair (9–11).
Histone deacetylases (HDACs) affect various cellular processes but are best known as transcriptional corepressors that epigenetically control gene transcription by removing acetyl groups from lysine side chains of histone tails. The removal of these posttranslational modifications from histones prevents the recruitment of readers, such as bromodomain- or YEATS domain–containing proteins, thereby promoting chromatin compaction and repression of RNA polymerase II–dependent gene expression (12–14). Humans and mice have just 18 HDACs, which are divided into four classes on the basis of their structure and function. Class I HDACs (HDAC1, HDAC2, HDAC3, and HDAC8) predominately localize to the nucleus, although HDAC3 has also been detected at plasma membranes (15). Class I HDACs are ubiquitously expressed and have high enzymatic activity toward histone substrates and thus serve as the enzymatic subunits of multiprotein repressive complexes. Class II HDACs (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10) vary from class I HDACs in that they shuttle between the nucleus and cytoplasm and have more temporal and spatial gene expression patterns. Class II HDACs have low intrinsic enzymatic activity and therefore often recruit class I HDACs for most of their enzymatic activity. Class III HDACs [sirtuins (SIRTs)] are substantially different from the other HDACs in that they require nicotinamide adenine dinucleotide (NAD+) instead of zinc (Zn2+) for their catalytic activity and thus are inhibited by different small molecules. Class IV consists of only HDAC11, which shares characteristics of both class I and class II HDACs.
HDACs can deacetylate proteins other than histones, including transcription factors [nuclear factor κB (NF-κB), RUNX2, p53, and signal transducer and activator of transcription 3 (STAT3)], to posttranslationally influence their stability and activity (16–23). HDACs are particularly important during development when gene expression programs change quickly as cell fate and function are determined. Small molecules with inhibitory activity for class I and II HDACs have been used to treat many cancers and mood disorders, and are in clinical trials for the treatment of neurological disorders and arthritis (24–26). However, many of these inhibitors, such as suberanilohydroxamic acid (SAHA), which is also known as vorinostat, are nonspecific and target multiple HDACs. As a result, off-target effects are common, particularly in the context of skeletal development and repair. For example, in utero exposure to HDAC inhibitors causes birth defects (27–31), and long-term exposure increases fracture risk in children and adults and reduces bone density in mice (32–36). Therefore, it is important to define the biological roles of individual HDACs in the skeleton.
HDAC3, HDAC4, HDAC5, and HDAC7 are crucial for endochondral ossification (24, 37). HDAC3 is highly expressed by osteoblasts and chondrocytes and acts as a corepressor for RUNX2, ZFP521, class II HDACs (HDAC4, HDAC5, and HDAC7), and other transcriptional regulators (38–40). Germline deletion of Hdac3 causes embryonic lethality during midgestation [embryonic day 9.5 (E9.5)] several days before skeletogenesis begins (41). Tissue-specific ablation of Hdac3 in osteoblasts [using osteocalcin (OCN)–Cre], in osteoprogenitors [using osterix (OSX1)–Cre], and in neural crest cells (using WNT1-Cre or PAX3-Cre) has demonstrated its importance in long bone and craniofacial bone development (42–46); however, the specific role of HDAC3 in chondrocyte maturation remains to be investigated.
Here, we examined the role of HDAC3 in hyaline cartilage development by genetically deleting it in vitro and in vivo. Prenatal deletion of Hdac3 in chondrocytes caused embryonic lethality, but postnatal deletion using an inducible conditional knockout (CKO) system produced animals with several skeletal abnormalities, including delayed secondary ossification center (SOC) formation, delayed maturation of epiphyseal plate cartilage, and increased osteoclastogenesis. Transcriptomic and chromatin analyses of HDAC3-deficient immature murine chondrocytes (IMCs) revealed increased expression of cytokine and matrix-degrading genes and reduced expression of genes related to extracellular matrix composition and deposition, bone development, and ossification. The altered secretome of HDAC3-deficient chondrocytes had autocrine and paracrine effects on bone formation and matrix remodeling, suggesting an important role of HDAC3 in controlling the coupling of chondrocyte maturation and bone modeling. Inhibition of cytokine-controlled pathways [namely, Janus kinase (JAK)–STAT or NF-κB] or of bromodomain extraterminal (BET) proteins that recognize acetylated proteins rescued many of the defects observed in Hdac3-depleted chondrocytes. Furthermore, inhibition of JAK signaling suppressed osteoclast recruitment in Hdac3-CKO mice. Together, these results demonstrate that HDAC3 plays an important role in endochondral bone formation by maintaining temporal and spatial regulation of gene expression and downstream signaling factors that are crucial in chondrocyte maturation, proper long bone growth, and ossification. Furthermore, our study provides genetic evidence that HDAC3 normally attenuates cytokine expression during skeletal development to control the JAK-STAT and NF-κB pathways, and thereby supports cartilage extracellular matrix formation and remodeling.
RESULTS
HDAC3 is essential for chondrocyte maturation
Previous experiments showed that HDAC3 is abundant in proliferating and hypertrophic growth plate chondrocytes of 1-month-old mice (44). Further analysis revealed that HDAC3 is detectable in these zones as early as E14.5 (fig. S1). To determine the physiological role of HDAC3 in chondrocytes during endochondral ossification, Hdac3 was first conditionally deleted in cells expressing Cre recombinase under the control of the Col2a1 promoter (Hdac3-CKOCol2a1 mice). Col2a1 is expressed in embryonic cartilaginous structures as early as E10.5 and in developing skeletal structures at E13.5 (47–49). Hdac3 conditional heterozygous mice (Hdac3fl/+; Col2a1-Cre+, hereafter called HET mice) were born at expected Mendelian ratios (fig. S2A) and were normal in size (fig. S2B). However, no Hdac3-CKOCol2a1 animals were born in nine litters (fig. S2A). Hdac3-CKOCol2a1 and HET mice were detected at E10.5, but only HET mice were present at E16.5. Thus, Hdac3 expression in chondrocytes is necessary during these early stages of skeletal formation between E10.5 and E16.5.
Postnatal ablation of Hdac3 in chondrocytes delays endochondral ossification
To further understand the role of HDAC3 in endochondral ossification, we next used a model that would allow for postnatal deletion of Hdac3 and bypass the embryonic lethality of Hdac3-CKOCol2a1 mice. Hdac3fl/fl mice were crossed to mice carrying the tamoxifen-inducible Col2a1-Cre transgene to generate Hdac3-CKOCol2ERT pups. These animals were given a single subcutaneous injection of tamoxifen or vehicle (control) on post-natal day 5 (P5). HDAC3 protein abundance was significantly lower in the growth plates of 4-week-old Hdac3-CKOCol2ERT animals (Fig. 1A), and Hdac3 mRNA expression in the xiphoid process was on average 60% below normal at 8 weeks of age (Fig. 1B). Hdac3-CKOCol2ERT animals weighed about 50% less at 4 weeks of age than did vehicle-injected mice. Weight deficiencies were resolved by 8 weeks, but femur length was not corrected (Fig. 1, C and D).
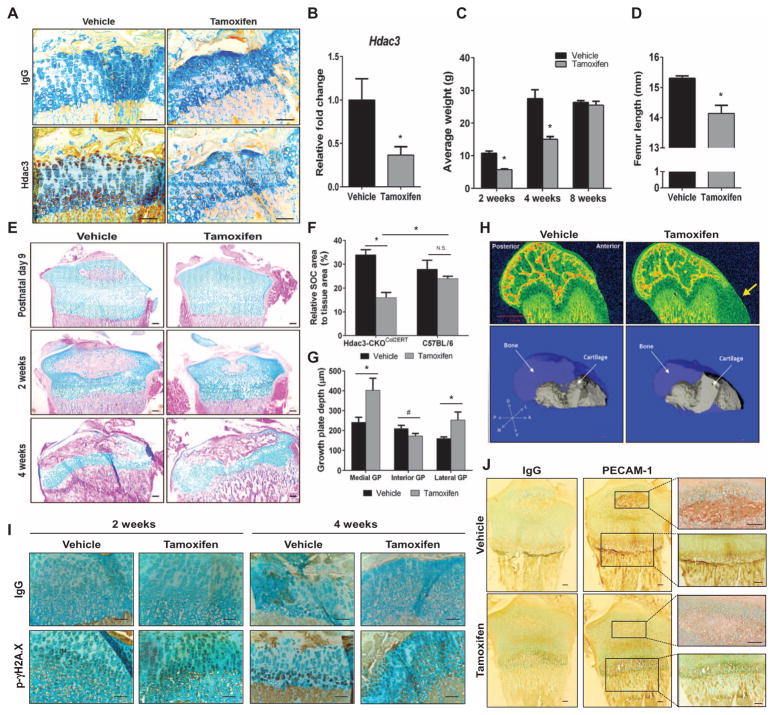
Fig. 1. Hdac3 ablation in chondrocytes during postnatal growth delays skeletal development.
(A) Immunohistochemistry for HDAC3 on tibial growth plates of 4-week-old Hdac3-CKOCol2ERT mice that had been injected with tamoxifen or vehicle at P5. Scale bars, 200 μm. (B) qPCR for Hdac3 in the xiphoid processes of 8-week-old mice described in (A) (n = 3 mice per group). (C) Weights of male mice at indicated ages (n = 5 mice per group). (D) Femur lengths of 8-week-old male animals described in (A) were measured in micro-CT (computed tomography) scans (n = 5 mice per group). (E) Alcian blue/eosin-stained tibial sections isolated at the indicated ages from mice described in (A). Scale bars, 100 μm. (F) Quantification of SOC area to total tissue area of epiphyses from 2-week-old Hdac3-CKOCol2ERT or C57BL/6 mice injected at P5 with tamoxifen or vehicle (n = 5 mice per group). N.S., not significant. (G) Quantification of total growth plate (GP) depth across the indicated regions in 4-week-old animals described in (A) (n = 5 mice per group). (H) Equilibrium partitioning of an ionic contrast agent (EPIC)–CT scans of femoral condyles from 4-week-old mice described in (A) (top). The yellow arrow points to residual cartilage along the growth plate in the Hdac3-CKOCol2ERT mice. Three-dimensional reconstructions of the femoral condyles show the growth plate (solid gray) within the bone architecture (semitransparent blue). A, anterior; D, dorsal; L, lateral; M, medial; P, posterior; V, ventral axes. (I) Immunohistochemistry for phosphorylated γH2A.X on tibiae of 2- and 4-week-old mice described in (A). Scale bars, 100 μm. (J) Immunohistochemistry for PECAM-1 on tibiae of P9 pups described in (A). Scale bars, 100 μm. *P = 0.05, #P = 0.075, Student’s t test.
Severe delays in SOC formation were first observed in tibiae of 9-day-old Hdac3-CKOCol2ERT animals, 4 days after administration of tamoxifen, and were more pronounced at 14 days of age (Fig. 1, E and F). By 4 weeks of age, the SOCs were fully formed in both control and Hdac3-CKOCol2ERT animals, but defects in the growth plates of the Hdac3-CKOCol2ERT animals were evident. The growth plates of Hdac3-CKOCol2ERT animals were 18% thinner than those in control animals in the interior region of the growth plate, but were 67% thicker on the medial side and 57% thicker on the lateral region (Fig. 1, E, G, and H). Tamoxifen did not affect the growth plates of wild-type C57BL/6 mice (fig. S3). We previously showed that Hdac3 deletion in osteoblasts increases phosphorylated γH2A.X, which is a marker of DNA double-strand breaks and a possible indicator for both senescence and apoptosis (43). Immunohistochemistry revealed an increased abundance and earlier presence of phosphorylated γH2A.X in the growth plates of 2- and 4-week-old Hdac3-CKOCol2ERT mice (Fig. 1I). In contrast, abundance of the angiogenic marker platelet endothelial cell adhesion molecule–1 (PECAM-1) was reduced in the epiphyses of postnatal Hdac3-CKOCol2ERT animals, both in the SOC and at the transition of hypertrophic chondrocytes of the growth plate into the primary spongiosa (Fig. 1J), indicating delayed vascularization in Hdac3-CKOCol2ERT. Together, these data demonstrate that HDAC3 is required for postnatal long bone development and indicate that functions of HDAC3 in chondrocytes include protecting DNA integrity, promoting vasculogenesis, and coupling the final stages of chondrocyte maturation to ossification.
HDAC3 regulates the chondrocyte transcriptome and chromatin landscape
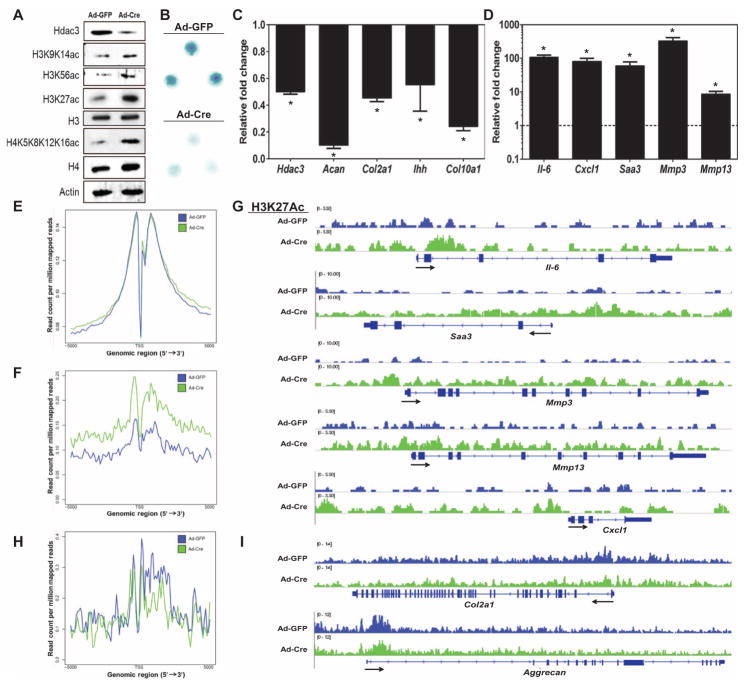
To understand molecular and epigenomic mechanisms by which HDAC3 controls chondrocyte maturation and endochondral ossification, IMCs were isolated from 7-day-old Hdac3f l/f l mice, plated in chondrogenic micromass cultures, and transduced with adenoviruses expressing Cre recombinase (Ad-Cre) or green fluorescent protein (Ad-GFP). HDAC3 abundance was reduced more than 50% in the Ad-Cre–transduced (HDAC3-depleted) cultures, and the lysine acetylation of histones H3 and H4 (H3K9/K14ac, H3K56ac, H3K27ac, and H4K5/K8/K12/K16ac) was increased in Hdac3-depleted chondrocytes (Fig. 2A). Ad-Cre–transduced IMC micromasses produced fewer proteoglycans, indicating delays in chondrocyte maturation (Fig. 2B). Transcriptional profiling by RNA sequencing (RNA-seq), functional clustering analysis, and subsequent quantitative polymerase chain reaction (qPCR) validation experiments confirmed that the expression of many markers of chondrocyte maturation (including Col2a1, Acan, Ihh, and Col10a1) was suppressed in Hdac3-depleted cultures (Fig. 2C and figs. S4 and S6). On the other hand, HDAC3-deficient IMCs highly expressed numerous genes encoding cytokines and related signaling pathways that are classically linked to inflammation (Fig. 2D and figs. S5 and S6). Notable among the genes induced in HDAC3-depleted chondrocytes were cytokines and chemokines (Il-6, Cxcl1, and Saa3), as well as factors contributing to cartilage catabolism [matrix metal-loproteinase (MMP)–encoding genes Mmp3 and Mmp13] (Fig. 2D). Genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) showed that the abundance of H3K27ac was greater within and near genes that were induced in Hdac3-depleted IMC micromasses (Fig. 2, E to G) but was unchanged in genes that were suppressed (Fig. 2, H and I).
Fig. 2. Hdac3 deletion alters global gene expression and histone acetylation in chondrocytes.
(A) Western blotting for the indicated proteins performed 48 hours after adenoviral transduction in primary IMCs isolated from 1-week-old Hdac3fl/fl;Cre− pups, plated in micromass cultures, and transduced with Ad-GFP or Ad-Cre on day 3 of culture. (B) Micromasses described in (A) were fixed 48 hours after adenoviral transduction and stained with Alcian blue. (C and D) qPCR to validate RNA-seq results from Hdac3-depleted IMCs described in (A). Differential expression of representative genes with decreased (C) or increased (D) expression in HDAC3-deficient micromasses (Ad-Cre) is shown relative to control (Ad-GFP) IMCs, which was set to 1 (n = 3). (E to I) ChIP-seq was performed for H3K27ac in HDAC3-deficient micromasses described in (A). Average binding profiles of H3K27ac 5 kb upstream or downstream of the transcriptional start site (TSS) in control (blue) or HDAC3-deficient (green) micromasses: all genes (E), genes with increased expression after Hdac3 depletion (F), and genes with decreased expression after Hdac3 depletion (H). Representative distribution of H3K27ac in select genes with corresponding increased (G) or decreased (I) expression. *P = 0.05, Student’s t test.
Increased abundance of interleukin-6 (IL-6) and MMPs was confirmed in Hdac3-depleted IMC supernatants by enzyme-linked immunosorbent assay (ELISA) and a fluorescence resonance energy transfer–based fluorometric MMP assay, respectively (Fig. 3, A and B). Il-6 transcripts were also induced in immortalized chondrocytes (ATDC5 cells) plated in either monolayer (Fig. 3C) or micromass (Fig. 3D) and in the presence of either the pan-HDAC inhibitor SAHA or the HDAC3-specific inhibitor RGFP966. RGFP966 also induced the expression of Mmp13 transcripts in ATDC5 cells (Fig. 3, E and F). Together, these results demonstrate that HDAC3 deficiency suppresses chondrocyte maturation and increases the acetylation and expression of cytokine and matrix remodeling genes.
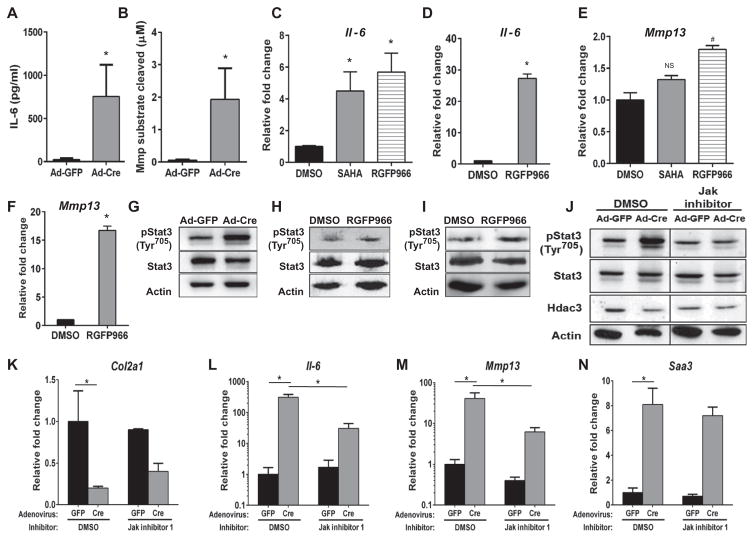
Fig. 3. HDAC3 deficiency in chondrocytes increases proinflammatory response signals through increased IL-6–STAT3 signaling and is rescued with JAK inhibition.
(A, B, G, and J to O) Primary IMCs were harvested from 1-week-old Hdac3fl/fl pups, plated in micromasses, and transduced with Ad-GFP or Ad-Cre on day 3. (A and B) Conditioned media (CM) from micromasses were analyzed with an ELISA to measure mouse IL-6 (A) or a fluorometric MMP enzymatic assay (B) (n = 3). (C to F) ATDC5 cells in monolayer (C and E) or micromass (D and F) were treated with the broad HDAC inhibitor SAHA (1 μM) or the HDAC3-specific inhibitor RGFP966 (10 μM) for 24 hours. qPCR for expression of Il-6 (C and D) and Mmp13 (E and F) (n = 3). (G to I) Western blotting for the indicated proteins in HDAC3-deficient chondrocytes in micromass (G), ATDC5 cells in micromass and treated with RGFP966 (H), or wild-type (WT) IMCs treated with RGFP966 (I) (10 μM) for 24 hours. (J to N) HDAC3-deficient chondrocytes in micromasses were treated with JAK Inhibitor I (1 μM) for 24 hours, at which point Western blotting (J) for the indicated proteins and qPCR (K to N) for the indicated mRNA transcripts were performed (n = 3). *P =0.05, #P = 0.06, Student’s t test.
Hdac3 deletion induces cytokine signaling in chondrocytes
Gene set enrichment analysis (GSEA) (50, 51) revealed that STAT and NF-κB transcription factor–binding motifs were significantly enriched in genes with increased expression in Hdac3-depleted IMCs (fig. S7). To determine whether increased production of cytokines (IL-6) initiated autocrine signaling events in HDAC3-deficient chondrocytes, JAK-STAT signaling was examined. Phosphorylation of Tyr705 in STAT3 was increased in Hdac3-depleted IMC micromasses, as well as in ATDC5 micromasses and primary wild-type murine chondrocytes cultured in micromasses treated with RGFP966 (Fig. 3, G to I). JAK inhibitors reduced the phosphorylation of STAT3 in HDAC3-deficient IMC micromasses (Fig. 3J and figs. S8 and S9). JAK inhibition slightly increased Col2a1 mRNA expression (Fig. 3K) and substantially reduced Il-6 and Mmp13 gene expression (note log scales), but not Saa3 transcripts in HDAC3-depleted chondrocytes (Fig. 3, L to N, and figs. S8 and S9). These results demonstrate that increased autocrine expression of cytokines suppresses maturation of HDAC3-depleted chondrocytes. However, the incomplete rescue of chondrogenesis and gene expression patterns suggests that other mechanisms are also operative.
Given the enrichment for NF-κB factor binding sites in induced genes, we next examined NF-κB signaling. The p65 (RelA) subunit of the NF-κB complex is an HDAC3 substrate (22, 23). Consistent with these results from other cell types (22, 23, 52, 53), deletion of HDAC3 in chondrocytes increased p65 acetylation at K310 (Fig. 4A). In control chondrocytes, p65 was primarily cytoplasmic in both cellular fractionation and immunofluorescence experiments (Fig. 4, B to D). However, in HDAC3-depleted chondrocytes, p65 was more prevalent in the nucleus.
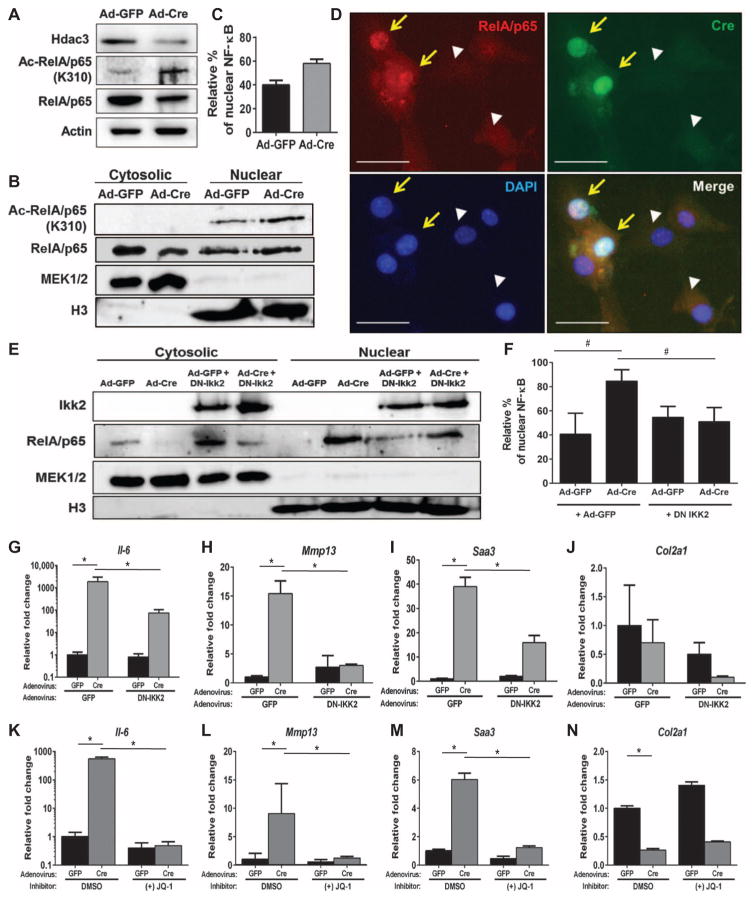
Fig. 4. HDAC3 depletion in chondrocytes increases NF-κB acetylation.
(A to D) Primary IMCs were harvested from 1-week-old Hdac3fl/fl pups, plated in micromass, and treated with Ad-GFP or Ad-Cre on day 3 of culture. (A) Western blotting was performed after 48 hours. (B) Western blotting for the indicated proteins in cytosolic and nuclear fractions collected from micromasses. (C) Quantification of the percentage of nuclear NF-κB relative to the total amount of NF-κB present from n = 3 experiments. (D) Primary immature mouse chondrocytes from 1-week-old Hdac3fl/fl mice were plated in monolayer and transduced with Ad-GFP or Ad-Cre. Immunofluorescence for total NF-κB/p65 and Cre recombinase was performed 48 hours after viral transduction. The yellow arrows indicate cells expressing Cre, and white arrowheads indicate control cells (not expressing Cre). Scale bars, 20 μm. (E to J) Micromass cultures were simultaneously transduced with Ad-GFP or Ad-Cre and either Ad-GFP or Ad-dnIKK2. (E) Western blotting for the indicated proteins was performed on the cellular fractions 48 hours after transduction. (F) Quantification of nuclear NF-κB relative to the total amount of NF-κB present (n = 3). (G to J) qPCR for the indicated mRNAs in HDAC3-deficient IMCs transduced with Ad-dnIKK2: Il-6 (G), Mmp13 (H), Saa3 (I), and Col2a1 (J) (n = 3). (K to N) HDAC3-deficient micromasses were treated with JQ-1 (10 μM) for 24 hours, and qPCR was performed for the indicated transcripts: Il-6 (K), Mmp13 (L), Saa3 (M), and Col2a1 (N) (n = 3). *P = 0.05, Student’s t test.
To modulate the effects of increased nuclear p65 on HDAC3-depleted chondrocytes, micromasses were simultaneously transduced with Ad-GFP or Ad-Cre and an adenovirus expressing a dominant-negative inhibitor of NF-κB subunit β (IKKβ) (dnIKK2). As expected, dnIKK2 increased cytoplasmic abundance and reduced nuclear abundance of p65 (Fig. 4, E and F). HDAC3-depleted chondrocytes expressing dnIKK2 produced fewer Il-6, Mmp13, and Saa3 transcripts (Fig. 4, G to I), but Col2a1 mRNA expression was not changed (Fig. 4J). Combining the JAK Inhibitor I with dnIKK2 or an IKK2 inhibitor reduced Il-6 expression below those observed with either inhibitor alone, but did not affect Mmp13 expression (fig. S10).
Given the large increases in H3K27 acetylation observed in induced genes (Fig. 3) and the increased acetylation of p65, we next examined whether inhibiting readers of acetylated lysines could affect gene expression in HDAC3-deficient chondrocytes. The BET family of proteins recognizes acetylated lysines on histones, p65, and other proteins and promotes transcriptional elongation (54). The BET small-molecule inhibitor JQ-1 completely reduced Il-6, Mmp13, and Saa3 transcripts in HDAC3-deficient chondrocytes but did not affect Col2a1 levels (Fig. 4, K to N). Together, these results demonstrate that HDAC3 is essential for Col2a1 gene expression and also facilitates chondrocyte maturation by suppressing histone acetylation of genes encoding cytokines and matrix remodeling enzymes, and repressing NF-κB and STAT signaling.
HDAC3-deficient chondrocytes increase osteoclast proliferation and differentiation in culture
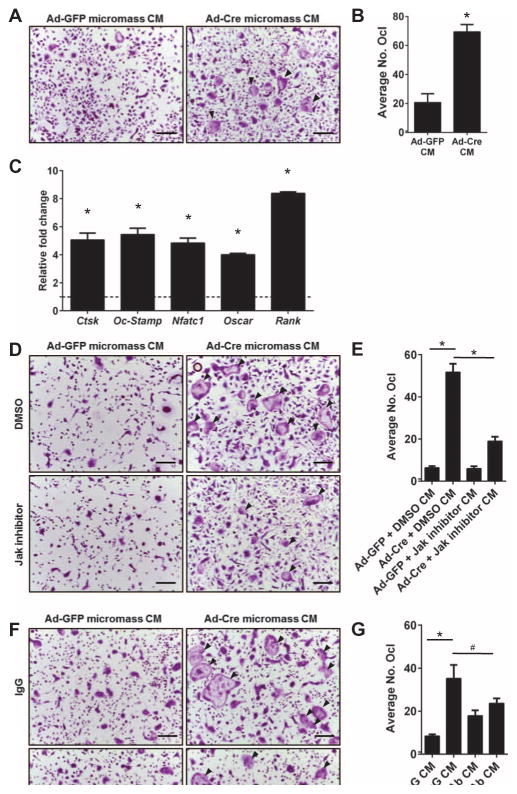
To determine whether the secretome of HDAC3-deficient chondrocytes influenced bone resorption, in vitro osteoclastogenesis assays were performed. Osteoclast and chondroclast recruitment, proliferation, and differentiation are essential for the resorption of the type II collagen–rich cartilaginous matrix and the recruitment of osteoblasts that replace it with a type I collagen–rich matrix that can be mineralized. Many factors influence osteoclast formation and function (55, 56). In particular, IL-6 stimulates the proliferation and differentiation of bone marrow monocytes into osteoclasts (57, 58). Thus, bone marrow monocytes from wild-type C57BL/6 mice were incubated overnight with 15% (v/v) CM from control (Ad-GFP) or HDAC3-deficient (Ad-Cre) IMC micromass cultures before adding osteoclast differentiation medium. Bone marrow cells treated with CM from HDAC3-depleted IMC micromasses had more mature, tartrate-resistant acid phosphatase–positive (TRAP+) osteoclasts by day 4 of differentiation, compared to cultures treated with CM from control chondrocytes (Fig. 5, A and B). Expression of osteoclast genes (Ctsk, Oc-Stamp, Nfatc1, Oscar, and Rank) was also increased in osteoclast cultures incubated with the CM of HDAC3-deficient micromasses (Fig. 5C). JAK inhibitors blocked osteoclastogenesis induced by CM from HDAC3-depleted micromasses (Fig. 5, D and E). A neutralizing antibody that binds to IL-6 also reduced osteoclastogenesis from CM of HDAC3-deficient micromasses (Fig. 5, F and G). Thus, an essential role of HDAC3 in chondrocytes is to suppress the expression of secreted factors that promote premature osteoclast activation and matrix degradation during endochondral bone development.
Fig. 5. Chondrocytic HDAC3 deficiency increases osteoclastogenesis in vitro.
(A to G) Osteoclast precursors were collected from 8-week-old WT mice and incubated overnight with 15% CM from HDAC3-deficient or control chondrocytes. Nonadherent cells were plated in normal osteoclast differentiation medium. (A) Day 4 TRAP-stained mature osteoclasts. Scale bars, 100 μm. (B) Quantification of TRAP+ mature osteoclasts (n = 3). No. Ocl, total number of osteoclasts. (C) RNA was isolated from mature osteoclast cultures, and qPCR was performed for the indicated genes. Relative fold changes were normalized to osteoclast cultures treated with CM from Ad-GFP–treated chondrocytes, indicated by the dotted line set to 1 (n = 3). (D and E) CM were collected from HDAC3-deficient chondrocytes treated with 1 μM JAK inhibitor for 24 hours. WT osteoclast precursors were preincubated with the CM from JAK inhibitor–treated HDAC3-deficient chondrocytes. (D) Day 4 TRAP-stained mature osteoclasts. Scale bars, 100 μm. (E) Quantification of TRAP+ osteoclasts (n = 3). (F and G) CM from HDAC3-deficient chondrocytes was pretreated with a neutralizing antibody recognizing IL-6 or an immunoglobulin G (IgG) control antibody (Ab). WT osteoclast precursors were collected and primed with the neutralized CM. (F) Day 4 TRAP-stained mature osteoclasts. (G) Quantification of TRAP+ osteoclasts (n = 3). *P = 0.05, #P = 0.08, Student’s t test. Black arrowheads indicate multinucleated osteoclasts.
IL-6, MMP13, and osteoclast activity are increased in Hdac3-CKOCol2ERT mice
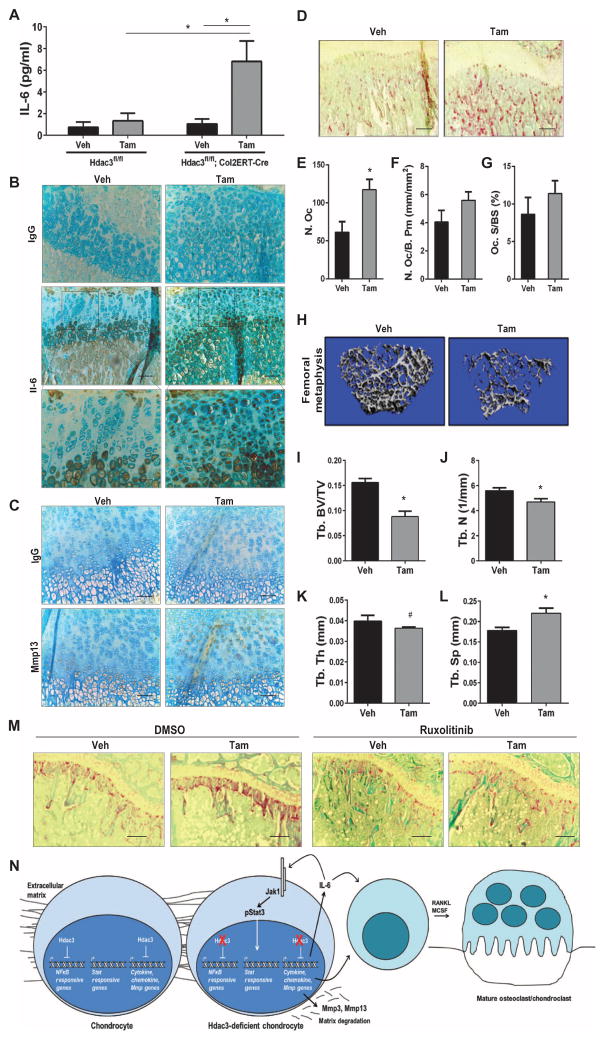
To determine whether the increased expression of Il-6 observed in HDAC3-depleted chondrocyte micromasses also occurred in vivo, IL-6 protein was measured in sera from Hdac3-CKOCol2ERT mice at 2 weeks after birth. IL-6 abundance was increased 6.5-fold in Hdac3-CKOCol2ERT mice compared to vehicle-injected mice in which IL-6 concentrations were near or below detection limits of the ELISA (Fig. 6A). Increases in circulating IL-6 were due to Col2-Cre–driven HDAC3 depletion alone and not tamoxifen, because Hdac3fl/fl animals injected with tamoxifen did not have increased abundance of circulating IL-6 (Fig. 6A). Immunohistochemistry revealed that IL-6 protein abundance was drastically increased in the growth plate chondrocytes of Hdac3-CKOCol2ERT mice. In particular, increased IL-6 abundance was detected prematurely in resting, proliferating, and prehypertrophic chondrocytes compared to control mice where IL-6 is restricted to the hypertrophic chondrocytes (Fig. 6B). We and others have observed that, in addition to being highly active in cartilage, the Col2ERT-Cre driver is also active in a subset of bone marrow mesenchymal progenitor cells, lining cells, and osteocytes (fig. S11A) (59–61). IL-6 is normally produced by cells in these regions but did not appear to be as drastically increased there in Hdac3-CKOCol2ERT mice as compared to its aberrant presence in the growth plate (fig. S11B). Although the growth plate is avascular, there is a region in the midplate that is more permissive for the diffusion of paracrine factors to the perichondrium and the vasculature (62). Thus, HDAC3 depletion in growth plate chondrocytes, particularly those in the midplate, likely contributes to elevated serum IL-6 levels.
Fig. 6. HDAC3-deficient chondrocytes increase inflammatory responses and osteoclast proliferation in vivo.
(A) ELISAs for IL-6 on sera collected from 2-week-old Hdac3f/.fl;Cre− or Col2ERT-Cre+ mice. (B) Immunohistochemistry for IL-6 on sections from 4-week-old tibiae. Scale bars, 100 μm. (C) Immunohistochemistry for MMP13 on tibia sections from P9 Hdac3-CKOCol2ERT or control mice. (D) Tibiae from 4-week-old Hdac3-CKOCol2ERT and control animals were stained for TRAP. Shown are representative images of the primary spongiosa. Scale bars, 200 μm. (E to G) Histomorphometric quantification of osteoclast parameters from 4-week-old Hdac3-CKOCol2ERT mice: (E) total number of osteoclasts (N. Oc), (F) number of osteoclast per bone perimeter (N. Oc/B. Pm), and (G) osteoclast surface per bone surface (Oc. S/BS) (n = 3 for control animals and n = 4 for CKO animals). (H to L) Micro-CT analysis of bone architecture from 8-week-old Hdac3-CKOCol2 mice. (H) Micro-CT reconstructions of the cancellous bone. (I to L) Quantification of micro-CT data on trabecular bone volume fraction (Tb. BV/TV) (I), trabecular number (Tb. N) (J), trabecular thickness (Tb. Th) (K), and trabecular separation (Tb. Sp) (L) (n = 5 mice for each group). (M) Tibiae from 6-week-old Hdac3-CKOCol2ERT mice were treated with ruxolitinib for 2 weeks and were TRAP-stained. Shown are representative images of the primary spongiosa. Scale bars, 200 μm (n = 3 vehicle/vehicle and tamoxifen/vehicle mice, 5 vehicle/ruxolitinib mice, and 4 tamoxifen/ ruxolitinib mice). In (A) to (M), *P = 0.05 and #P = 0.14, Student’s t test. (N) Working model. HDAC3 suppresses production of cytokines and catabolic matrix enzymes in chondrocytes during bone development.
MMP13 protein abundance was also evaluated by immunohistochemistry in the tibiae of Hdac3-CKOCol2ERT and control mice at P9 (Fig. 6C). MMP13 protein was restricted to hypertrophic chondrocytes in control animals, but was detected prematurely in proliferating and prehypertrophic chondrocytes, and at higher amounts in hypertrophic chondrocytes in Hdac3-CKOCol2ERT mice. These results further support the importance of HDAC3 in controlling temporal and spatial gene activation, particularly of extracellular matrix remodeling factors during chondrocyte maturation.
To assess chondroclast and osteoclast presence in the growth plates of HDAC3-deficient mice, TRAP activity was measured in tibial sections from 4-week-old Hdac3-CKOCol2ERT mice and corresponding controls. The Hdac3-CKOCol2ERT animals displayed increased TRAP activity with-in the primary spongiosa at 4 weeks of age (Fig. 6D). Total numbers of TRAP+ cells within the primary spongiosa were significantly greater in Hdac3-CKOCol2ERT mice compared to control mice (Fig. 6E). Hdac3-CKOCol2ERT animals also had increased numbers of TRAP+ cells per bone perimeter and elevated TRAP+ surface areas (Fig. 6, F and G).
To understand the consequences of chondrocytic HDAC3 deficiency in chondrocytes on bone structure, femurs from 2-month-old Hdac3-CKOCol2ERT mice animals were subjected to micro-CT. Cancellous bone density was lower in Hdac3-CKOCol2ERT mice (Fig. 6, H and I) due to reductions in trabecular bone number (Fig. 6J) and thickness (Fig. 6K), which increased trabecular spacing (Fig. 6L).
Finally, Hdac3-CKOCol2ERT mice were treated with ruxolitinib, a clinically approved and orally available JAK inhibitor. Daily JAK inhibitor treatments for 2 weeks between 6 and 8 weeks of age reduced TRAP+ osteoclast numbers in the primary spongiosa in Hdac3-CKOCol2ERT mice to amounts observed in control mice (Fig. 6M). This short treatment regimen modestly increased cancellous bone density and trabecular bone number and thickness in control and Hdac3-CKOCol2ERT mice (fig. S12). These results suggest that Hdac3 deletion in chondrocytes not only compromises terminal chondrocyte maturation, matrix remodeling, and subsequent ossification but also influences other cell types involved in endochondral bone formation, such as osteoclasts and chondroclasts (Fig. 6N).
DISCUSSION
This study elucidates the essential functions for autologous HDAC3 in proper growth plate chondrocyte maturation and long bone development. Mice in which Hdac3 is deleted in chondrocytes (Hdac3-CKOCol2ERT) also exhibit a number of cartilage-extrinsic phenotypes including delayed angiogenesis, accelerated bone resorption, and severely reduced bone mineral density. These results demonstrate that HDAC3 controls cell-cell communication between chondrocytes and cell types required for the integrated development of cartilage, bone, and blood vessels during skeletal development. In particular, we observed that HDAC3 loss in chondrocytes deregulates the cell-autonomous expression of many cytokines and MMPs that appear to be required for skeletal matrix modeling during endochondral bone formation.
Cytokines like IL-6 are classically thought of as proinflammatory or anti-inflammatory factors based on their potent biological effects as systemic factors. Yet, our results indicate that these ligands also have key roles as mediators of cell communication during normal bone development. Although cartilage lacks major blood vessels, chondrocytes communicate with each other and other cell types in neighboring bone and periosteal tissues including osteoclast and chondroclast precursors and endothelial cells through several mechanisms. Factors produced by growth plate chondrocytes can diffuse through a permissive region in the mid-plane of the growth plate near the prehypertrophic zone to the periosteum at a rate inversely proportional to their size (62). MMPs that digest matrix components and fluid flow could further facilitate the movement and release of molecules that may be stored in the matrix. Our results indicate that HDAC3 specifically controls the expression of MMPs as well as important cytokines in growth plate chondrocytes, and this may have systemic effects. Together, we propose that HDAC3-mediated suppression of cytokines and matrix remodeling genes in chondrocytes may facilitate coordination of cartilage development and bone formation during normal endochondral ossification, as well as perhaps in cartilaginous callous formation during bone fracture repair.
Transcriptome data and clinical findings support this interpretation that originates from skeletal phenotyping of our Hdac3 conditional null mice. Our RNA-seq data show that normal primary mouse chondrocytes expressed a number of cytokines and chemokines, but their abundances were significantly increased when HDAC3 was inactivated. The physiological production of these soluble factors in growth plate and articular chondrocytes suggests that these ligands have a normal role during mouse skeletal formation and maintenance when produced at appropriate levels. Chronic inflammation has direct and indirect effects on chondrocytes and interrupts endochondral ossification during development, resulting in many clinical manifestations, including permanently shortened limbs (3–6). Because HDAC3-mediated control of cytokines appears to be an essential physiological mechanism for skeletogenesis, as well as normal cartilage formation and maintenance, these clinical findings can perhaps be reevaluated as a potential perturbation of normal cytokine-mediated signaling pathways.
Our data show that deletion of Hdac3 derepressed proinflammatory and matrix degrading signaling pathways by hyperacetylating histones and NF-κB in chondrocytes. Robust NF-κB activation and expression of cytokines are also observed when Hdac3 was deleted in T regulatory lymphocytes (63). Among the many genes that were highly induced and hyperacetylated, Il-6 and Mmp13 were validated in vivo. The increased production of IL-6 and MMP13 had autocrine effects on HDAC3-depleted chondrocyte cultures. IL-6 activated the JAK-STAT signaling pathway in the HDAC3-depleted micromasses, and a JAK inhibitor partially reduced Il-6 and Mmp13 expression but did not increase Col2a1 expression or decrease Saa3 expression. These reductions in Il-6 and Mmp13 are consistent with the ability of JAK inhibitors to suppress inflammatory cytokine production in models of aging and frailty (64). In comparison, blockade of NF-κB signaling suppressed Mmp13 and Saa3 expression but also did not affect Col2a1 expression. Additionally, preventing BET proteins from recognizing the acetylated marks significantly suppressed Il-6, Mmp13, and Saa3 expression. In summary, our study provides a molecular mechanistic explanation for the physiological deregulation of cytokine signaling in chondrocytes, because deacetylation of histones and NF-κB promotes gene induction as predicted and Hdac3 deletion affects signaling pathways in chondrocytes that control the expression of JAK-STAT– and NF-κB–dependent cytokines and MMPs.
This work is of biomedical importance because HDAC inhibitors are increasingly recognized and clinically tested as potential therapies for many conditions and diseases including cancer and neurological disorders due to their epigenetic reprogramming capabilities (24–26). These conditions include several forms of arthritis (26, 65, 66). In animal models of osteoarthritis, which typically occurs after the skeleton is mature, HDAC inhibitors slowed disease progression (67–70). Moreover, in models of rheumatoid arthritis and periodontitis, HDAC inhibitors blocked bone resorption and osteoclast activation (65, 71–73). These favorable outcomes may be due to the ability of HDAC inhibitors to neutralize leukocyte populations that are major sources of inflammatory cytokines and chemokines (74–78). However, evidence that HDAC inhibitors have detrimental effects on the developing skeleton also exists (27–31). This study demonstrates that endochondral ossification was unfavorably affected in a number of ways by HDAC3 depletion in growth plate chondrocytes. Additional studies will need to be performed on osteoclasts and articular chondrocytes in adult mice to fully comprehend the role of HDAC3 in joint health and disease.
In summary, HDAC3 plays a crucial role in regulating the chondrocyte transcriptome by modifying the chromatin landscape and controlling non-histone protein activity, both of which allow for necessary temporal and spatial control of gene expression during the highly orchestrated process of endochondral bone formation. Loss of HDAC3 in culture models and in vivo delays ossification, alters terminal chondrocyte hypertrophy, and disrupts the coupling of chondrocyte maturation to ossification, by decreasing angiogenesis and increasing osteoclast activity. Furthermore, our study provides both physiological and molecular mechanistic evidence that these skeletal phenotypes are linked to HDAC3 control of histone acetylation, as well as the JAK-STAT and NF-κB signaling in response to the production of cytokines, chemokines, and MMPs in chondrocytes. The developmental effects of perturbing HDAC3-controlled pathways have lasting consequences by compromising bone quality, architecture, and overall skeletal health.
MATERIALS AND METHODS
Generation of Hdac3-CKO animals
C57BL/6 mice harboring Hdac3 alleles with loxP sites in introns flanking exon 7 (Hdac3fl/fl) were crossed to C57BL/6 mice expressing noninducible Col2a1-Cre or a tamoxifen-inducible Col2a1-Cre (Col2a1ERT-Cre; The Jackson Laboratory) to generate Hdac3-CKOCol2 and Hdac3-CKOCol2ERT mice, respectively. Hdac3-CKOCol2ERT mice pups were administered a single injection of tamoxifen (1 mg; Sigma-Aldrich) or its solvent, corn oil (control), on P5 as previously described to promote recombination (49). To assess the localization of cells producing the Col2a1ERT-Cre transgene, Col2a1ERT-Cre mice were crossed to Rosa26 reporter mice [R26R, B6.129S4-Gt (ROSA) 26Sortm1Sor/J; The Jackson Laboratory, strain no. 003474] as previously described (59). Animals were housed in an accredited facility under a 12-hour light/dark cycle and provided water and food (PicoLab Rodent Diet20, LabDiet) ad libitum. Animal research was conducted according to guidelines provided by the National Institutes of Health and the Institute of Laboratory Animal Resources, National Research Council. The Mayo Clinic Institutional Animal Care and Use Committee approved all animal studies.
Immunohistochemistry
Limbs were collected from animals at the ages indicated in the figure legends, fixed in formalin for 48 hours, and decalcified in 15% EDTA for 7 days. Decalcified bones were embedded in paraffin and sectioned to a thickness of 5 μm. Immunohistochemistry was performed with antibodies [diluted in 1% bovine serum albumin (BSA) in tris-buffered saline (TBS)] directed to Hdac3 (Millipore, 06-890; 1:50 dilution), PECAM-1 (BD Pharmingen, 550274; 1:50), phosphorylated γH2A.X (Abcam, ab289; 1:100 dilution), IL-6 (Abcam, ab6672; 1:50 dilution), or MMP13 (Abcam, ab39012; 1:50 dilution), or with a nonspecific IgG (control). Chromogens were detected with a polyvalent secondary HRP kit (Abcam, ab93697) and 3,3′-diaminobenzidine (DAB) (Sigma-Aldrich, D3939). Sections were counterstained with 0.5% Alcian blue.
Micro–computed tomography
Bone architecture and mineralization were evaluated by ex vivo micro-CT. The central portion of the femoral diaphysis and secondary spongiosa in the distal femoral metaphysis of each bone were scanned in 70% ethanol on a μCT 35 scanner (Scanco Medical AG). Trabecular bone scans were performed at 7-μm voxel size using an energy setting of 70 kilovolt peaks (kVp) and an integration time of 300 ms. For distal femoral metaphysis scans, a region of interest (ROI) spanning from 17 to 21% (Col2a1ERT-Cre mice) or from 20 to 25% of total bone length (Col2a1-Cre mice), as measured relative to the medial epiphysis, was analyzed in each mouse (threshold, 220). Trabecular bone volume fraction (%), trabecular number (mm−1), trabecular thickness (mm), and trabecular separation (mm) were computed using the manufacturer’s software.
Equilibrium partitioning of an ionic contrast agent–computed tomography
Growth plate cartilage thickness was examined in 4-week-old mice by EPIC-CT as previously described (79, 80). Briefly, femurs were carefully cleaned of soft tissues and submerged in 20% Hexabrix solution [in phosphate-buffered saline (PBS)] for 60 min at room temperature. The epiphysis of each femur was scanned in air on a μCT 35 scanner (Scanco Medical AG) at 7-μm voxels (scanner settings: energy, 45 kVp; intensity, 88 μA; integration time, 800 ms). Cartilage was selected for visualization by manual ROI segmentation (σ = 2.0; support, 4.0; threshold, 90 to 315).
TRAP staining
Tibiae from 4- and 8-week-old mice were fixed in formalin for 48 hours and decalcified in 15% EDTA for 7 and 14 days, respectively. Decalcified bones were embedded in paraffin and sectioned to a thickness of 5 μm. Deparaffinized and rehydrated tissue sections were incubated in 10 mM tris-HCl (pH 7.8) for 30 min at 37°C, rinsed with water, and then incubated in TRAP stain (Pararosaniline, Naphthol AS-TR phosphate) for 30 min at 37°C. Samples were counterstained with fast green.
X-gal staining
Femurs from 4-week-old Col2ERT/Rosa26 mice were fixed in 0.2% glutaraldehyde, cryoprotected in 30% sucrose [dissolved in PBS (pH 7.4)] at 4°C for 48 hours, frozen in embedding medium (Tissue-Tek O.C.T.), and sectioned to 8-μm thickness on a cryotome using CryoJane Tape Transfer System (Leica Biosystems). Sections were incubated in X-gal reaction buffer [5-bromo-4-chloro-3-indolyl-β-δ-galactopyranoside (1 mg/ml) in PBS (pH 7.4), 5 mM potassium ferrocyanide, 2 mM magnesium chloride, and 0.1% Triton X-100] overnight at 37°C in the dark, rinsed in PBS, counterstained with eosin, dehydrated through ethanol grades and xylenes, and mounted with Permount medium on glass slides.
Image quantification
Images were digitally scanned or collected with phase-contrast microscopy. Densitometry for Western blots was calculated by measuring the mean gray values with ImageJ software. The percent area of the SOC was determined by measuring the area of the SOC relative to the total epiphyseal area in each image using ImageJ software. The growth plate depths were determined by taking 15 measurements in medial, interior, and lateral regions of the growth plate for each mouse using ImageJ software.
Isolation and culture of IMCs
IMCs were isolated and cultured as previously described (81). Briefly, the femoral heads and tibial plateaus were harvested from 1-week-old Hdac3fl/fl pups. Cartilage tissue pieces were digested in collagenase (3 mg/ml) for 1 hour and then overnight in collagenase (0.5 mg/ml). The resulting IMC suspensions were brought to a concentration of 2 × 107 cells/ml and plated in micromasses with 10-μl drops, each containing 2 × 105 cells in Dulbecco’s modified Eagle’s medium (DMEM). Three to four micromasses were plated together in 35-mm plates. After 1 hour, micromasses were covered with 3 ml of DMEM and 5% fetal bovine serum (FBS). Micromasses were transduced with Ad-Cre (Vector Biolabs) or Ad-GFP (Vector Biolabs) at 1000 multiplicity of infection (MOI) on day 3 of culture. For dnIKK2 experiments, IMC micromass cultures were transduced simultaneously with Ad-Cre or Ad-GFP (1000 MOI) and either Ad-GFP or Ad-dnIKK2 (Vector Biolabs) (60 MOI). For experiments without inhibitors, cells were collected 48 hours after transduction for RNA and protein extraction and matrix staining with Alcian blue. JAK Inhibitor I (1 μM; Millipore, 420097), JQ-1 (1 μM; BPS Bioscience, 27402), IKK2 Inhibitor V (1 μM; Calbiochem, 401481), or vehicle [0.01% (v/v) dimethyl sulfoxide (DMSO)] was added to IMC micromass cultures 48 hours after adenoviral transduction as indicated. RNA and protein extracts were collected 24 hours later.
Alcian blue staining
IMC micromasses were fixed with 10% neutral buffered formalin for 48 hours and stained with 0.5% Alcian blue, 3% acetic acid for 2 hours. Tibiae were fixed in 10% neutral buffered formalin, decalcified in 15% EDTA for 7 days, paraffin-embedded, sectioned to a thickness of 5 μm, stained with Alcian blue for 1 min, and counterstained with eosin for 1 min.
Cytosolic and nuclear fractionation
IMC micromasses were washed and collected in PBS. Cells were homogenized by pipetting, collected in PBS by microcentrifugation, and incubated with 10 mM tris-HCl (pH 7.84), 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, and 1× protease inhibitor mixture (Roche) for 5 min on ice. The resulting lysate was cleared by centrifugation at 2000g for 5 min at 4°C. Supernatants (cytosolic extracts) were collected for SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. The remaining nuclear pellets were washed with PBS; suspended in 20 mM Hepes (pH 7.9), 1 M NaCl, 0.2 mM EDTA, 0.1 mM EGTA, 1.5 mM MgCl2, 1.2 mM phenyl-methylsulfonyl fluoride, and 20% glycerol; and incubated on ice for 5 min. These nuclear lysates were cleared by centrifugation at 2000g for 5 min at 4°C. The resulting supernatant was collected as the nuclear fraction for SDS-PAGE and Western blotting.
Immunofluorescence
IMCs were plated in monolayer on coverslips in a 24-well plate. On day 3 of culture, cells were transduced with Ad-GFP or Ad-Cre. After the 48-hour transduction, cells were rinsed with 1× PBS and fixed in 4% paraformaldehyde in 1× PBS for 15 min at room temperature. Coverslips were rinsed three times in 1× PBS for 5 min each and then blocked in blocking buffer (1× PBS, 5% normal serum, and 0.3% Triton X-100) for 1 hour at room temperature. Coverslips were then incubated with primary antibodies (diluted 1:300 in 1× PBS, 1% BSA, 0.3% Triton X-100) directed toward NF-κB/p65 (Cell Signaling, 8242P) and Cre recombinase (Millipore, MAB3120) overnight at 4°C in a humidified chamber. Cells were rinsed in 1× PBS and incubated with corresponding fluorochrome-conjugated secondary antibodies (Invitrogen; Alexa Fluor 488, A31619; Alexa Fluor 555, A31629) (diluted 1:1000 in 1× PBS, 1% BSA, 0.3% Triton X-100) for 1 hour at room temperature in the dark. Coverslips were rinsed in 1× PBS before mounting onto glass slides with Vectashield Mounting Medium with DAPI (Vector Laboratories, H-1200).
ATDC5 cell culture and HDAC inhibitor treatments
ATDC5 cells were plated in 10 μl drops, with each micromass containing 2 × 105 cells in DMEM and 5% FBS. Three to four micromasses were plated together in wells of a six-well plate. After 1 hour, micromasses were covered with 3 ml of DMEM, 5% FBS, and 1× ITS (insulin-selenium-transferrin). On day 3 of culture, ATDC5 micromasses were treated with 10 μM RGFP966 or vehicle (0.03% DMSO). RNA and protein extracts were collected 24 hours later.
Western blotting
IMC or ATDC5 cultures were lysed in SDS sample buffer [0.1% glycerol, 0.01% SDS, 0.1 M tris (pH 6.8)] for 10 min on ice. Total protein concentrations were determined with the Bio-Rad DC Protein Assay. Proteins (20 μg) were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were blotted using antibodies for Hdac3 (Abcam, ab7030), acetylated H3K9K14 (H3K9K14ac; Millipore, 06-599), H3K56ac (Abcam, ab76307), H3 (Millipore, 05-928), H4ac (Millipore, 06-866), H4 (Millipore, 04-858), phosphorylated (p-Tyr705) and total Stat3 (Cell Signaling Technology, 9145S and 12640S, respectively), β-actin (Sigma-Aldrich, A5316), acetylated (K310ac) and total NF-κB/p65 (Abcam, ab52175; Cell Signaling Technology, 8242P), MEK1/2 (Cell Signaling Technology, 9126S), IKKβ (Cell Signaling Technology, 2370P), and corresponding secondary antibodies (Santa Cruz Biotechnology). Chemiluminescent detection was performed with SuperSignal West Femto Maximum Sensitivity Substrate reagent (Pierce).
RNA isolation and qPCR
Total RNA was isolated from IMC micromass cultures and xiphoid processes of 8-week-old mice with TRIzol reagent (Invitrogen) and phenol/ chloroform. RNA (3 μg) was reverse-transcribed to complementary DNA (cDNA) with the SuperScript III First-Strand Synthesis cDNA kit (Invitrogen) for real-time semiquantitative PCR (qPCR) with gene-specific primers (table S1). Transcript levels were normalized to the reference gene Gapdh. Transcript abundance and relative fold changes in gene expression were quantified using the 2−ΔΔCt method relative to control.
High-throughput RNA-seq and bioinformatics analysis
High-throughput RNA-seq and bioinformatics analyses were performed on RNA from Ad-GFP– or Ad-Cre–transduced IMC micromasses as previously reported, following the MAP-RSeq pipeline (82, 83). Briefly, after read alignment, paired-end reads are aligned by TopHat 2.0.6 (84) against the mm10 genome using the bowtie1 aligner option (85). Gene expression is expressed in reads per kilobase per million mapped reads (RPKM). Complete RNA-seq data sets are available at the Gene Expression Omnibus (GSE75549). RPKM values for gene lists were filtered for anything ≥0.1 and a fold change ≥2, excluding microRNAs and small nuclear RNAs. The resulting gene lists (850 genes that had decreased gene expression with Hdac3 deficiency and 1062 genes that had increased gene expression with Hdac3 depletion) were analyzed with DAVID (Database for Annotation, Visualization, and Integrated Discovery) v6.7 for functional annotation clustering of genes. To identify common transcription factor–binding motifs, GSEA was performed with the C3 module (motif gene sets) from the Molecular Signatures Database (MSigDB_v5.0; http://software.broadinstitute.org/gsea/msigdb/collections.jsp#C3) (50).
Chromatin immunoprecipitation sequencing
Micromasses transduced with Ad-GFP or Ad-Cre were cross-linked with 1% formaldehyde (Pierce, catalog no. 28906) for 10 min and then incubated with 125 mM glycine for 5 min at room temperature to quench the fixation. Cells were washed twice with TBS, suspended in cell lysis buffer [10 mM tris-HCl (pH 7.5), 10 mM NaCl, 0.5% NP-40], and incubated on ice for 10 min. The lysates were washed with micrococcal nuclease (MNase) digestion buffer [20 mM tris-HCl (pH 7.5), 15 mM NaCl, 60 mM KCl, 1 mM CaCl2] and incubated for 20 min at 37°C with continuous mixing in a thermal mixer (Fisher Scientific) in the presence of MNase (New England Biolabs, catalog no. M0247). After adding the same volume of sonication buffer [100 mM tris-HCl (pH 8.1), 20 mM EDTA, 200 mM NaCl, 2% Triton X-100, 0.2% sodium deoxycholate], lysates were sonicated for 15 min (30 s on/30 s off) in Bioruptor Twin (UCD-400) (Diagenode Inc.) and centrifuged at 12,000g for 10 min. The cleared supernatant from 4 × 106 cells was incubated with 2 μg of anti-H3K27ac (Abcam, ab4729) on a rocker overnight. Prewashed protein G agarose beads (30 μl) were added to the reactions for 4 hours. The beads were extensively washed with ChIP buffer [50 mM tris-HCl (pH 8.1), 10 mM EDTA, 100 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate], high salt buffer [50 mM tris-HCl (pH 8.1), 10 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate], LiCl2 buffer [10 mM tris-HCl (pH 8.0), 0.25 M LiCl2, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA], and tris-EDTA buffer. Bound chromatin was eluted and reverse cross-linked at 65°C overnight. DNA was purified using MinElute PCR purification kit (Qiagen) after the treatment of ribonuclease A and proteinase K. ChIP enrichment was validated by performing qPCR in the genomic loci targeting the transcription start sites of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and T1 genes. Primers used for ChIP qPCR were as follows: mGAPDH-TSS-F (5′-CTCATCCCCGCAAAGGCGGA-3′), mGAPDH-TSS-R (5′-TCGGAC-CTGGCGATGGCTCG-3′), mT1-TSS-F (5′-GAGACGCCGATCCG-CCGAAG-3′), and mT1-TSS-R (5′-ACTCTCCACTCCCACGCGCT-3′). ChIP-seq libraries were prepared from 10 ng of ChIP and input DNA using the Ovation Ultralow DR Multiplex Kit (NuGEN). The ChIP-seq libraries were sequenced to 51 base pairs (bp) from both ends on the Illumina HiSeq 2000 in the Medical Genomics Facility within the Mayo Clinic Center for Individualized Medicine. H3K27ac data sets are available at the Gene Expression Omnibus (GSE75547). Data were analyzed using the HiChIP pipeline (86). Briefly, paired-end reads were mapped by Burrows-Wheeler Aligner (87), and pairs with one or both ends uniquely mapped were retained. Binding sites (13,131) were identified for H3K27ac in the Ad-Cre–treated micromasses, and 12,874 binding sites were identified for H3K27ac in the Ad-GFP–treated micromasses using the MACS2 software at a false discovery rate of ≤1% (88). For data visualization, BEDTools (89) was used in combination with scripts to generate normalized tag density profile at a window size of 200 bp and step size of 20 bp. For RNA-seq and ChIP-seq integration, the raw gene count file from MAP-RSeq was used to identify differentially expressed genes by DESeq2 (90). During this analysis, the RNA-seq gene list was filtered to remove genes with read counts lower than 30 in either of these samples. The differential gene list was divided into up-regulated and down-regulated genes by using a fold change of 2. ngs.plot was used to obtain binding profiles for H3K27ac in Ad-Cre and Ad-GFP micromasses across all the genes (91). The average binding profiles for 94 up-regulated and 9 down-regulated genes were also obtained for these samples.
IL-6 ELISA
Sera were collected from peripheral blood of 2-week-old Hdac3-CKOCol2ERT mice or Hdac3fl/fl mice that had been injected with tamoxifen or vehicle using serum separator tubes. Samples were incubated at room temperature for 30 min to allow for clotting and then centrifuged for 10 min at 2000g. CM were collected from Ad-GFP or Ad-Cre IMC micromass cultures 48 hours after viral transduction and centrifuged briefly to remove debris before analysis. IL-6 was quantified from samples using a mouse IL-6 ELISA kit (EMD Millipore, EZMIL6). Each sample was run in duplicate (n = 16 Hdac3-CKOCol2ERT mice per treatment group, 13 vehicle-injected Hdac3fl/fl mice, and 10 tamoxifen-injected Hdac3fl/fl mice).
MMP activity assay
CM from Ad-GFP or Ad-Cre IMC micromass cultures were collected 48 hours after viral transduction and centrifuged briefly to remove debris before analysis. MMP activity was measured in CM with the SensoLyte 520 Generic MMP Assay Kit, following the manufacturer’s instructions. Briefly, CM samples were incubated with 1 mM p-aminophenylmercuric acetate for 40 min at 37°C to activate the pro-MMP enzymes within the samples. Generic MMP substrate was added to each sample and the fluorescence reference standards. Samples were mixed gently for 30 s before incubation at room temperature for 60 min. Stop solution was added to all samples, and the fluorescence reference standard before the fluorescence intensity was measured using a Tecan Infinite F200 Pro plate reader with excitation and emission wavelengths of 490 nm/520 nm. The fluorescence reference standard, which serves as an indicator of the amount of MMP enzymatic reaction final product, was used to determine the amount of MMP substrate cleaved in the CM, relative to their respective fluorescence intensity readings. Each sample was run in duplicate.
Osteoclast assays
Bone marrow monocytes were isolated as previously described (92). Briefly, long bones were dissected free of soft tissues, and the epiphyses were removed from both ends of the femurs and tibiae. Marrow was flushed with PBS and treated with 1× Red Blood Cell Lysis Buffer (eBioscience Inc.). Resulting cell cultures were plated overnight in α-MEM (minimum essential medium) supplemented with 10% FBS (HyClone Laboratories) and 15% (v/v) CM from IMC micromass cultures treated with either Ad-GFP or Ad-Cre, CM from HDAC3-deficient micromass cultures treated with 1 μM JAK Inhibitor I (Millipore, 420097), or CM from HDAC3-deficient micromass cultures incubated with a neutralizing αIL-6 antibody (0.75 μg/ml; R&D Systems, MAB406) or an IgG control for 30 min with agitation at room temperature. After 24 hours, nonadherent cells were collected and plated in 24-well plates or on coverslips in a 24-well plate at a density of 4 × 105 cells per well with α-MEM supplemented with 10% FBS, receptor activator of NF-κB ligand (100 ng/ml) (R&D Systems), and macrophage colony-stimulating factor (25 ng/ml) (differentiation medium) for 3 days. To evaluate mature osteoclasts, fresh differentiation medium was added and the cells were cultured for an additional 24 hours. RNA was harvested 3 to 4 days later. Coverslips were collected on days 3 and 4, and TRAP staining was performed with the Acid Phosphatase, Leukocyte (TRAP) Kit (Sigma-Aldrich, 387A-1KT) per the manufacturer’s instructions.
In vivo JAK inhibitor treatments
JAK signaling was inhibited in 6-week-old Hdac3-CKOCol2ERT mice as previously described (64). Briefly, ruxolitinib (INCB018424, ChemieTek no. 3) was dissolved in DMSO to 30 mg/ml and then mixed into high-fat chow (60% fat, 20% protein, and 20% carbohydrate; Research Diets, D12492). Each mouse was given a 125-mg pellet of food containing ruxolitinib (30 mg/kg body weight) or vehicle daily for 2 weeks and was observed until they had eaten the entire pellet. After each treatment, mice were returned to original housing and had ad libitum access to normal chow (12% fat, 24% protein, and 64% carbohydrate) and water.
Statistical analysis
Data shown are means ± SEM from the number of samples or experiments indicated in the figure legends. All assays were repeated at least three times with independent samples. P values were determined with Student’s t tests.
Supplementary Material
Fig. S1. Expression pattern of Hdac3 during embryonic and postnatal long bone development.
Fig. S2. Hdac3 knockout in chondrocytes during embryogenesis is lethal.
Fig. S3. Tamoxifen does not affect growth plate maturation in wild-type mice.
Fig. S4. Top 30 annotation clusters of genes with decreased gene expression in Hdac3-depleted IMCs.
Fig. S5. Top 30 annotation clusters of genes with increased gene expression in Hdac3-depleted IMCs.
Fig. S6. Gene lists associated with top annotation clusters from DAVID pathway analysis.
Fig. S7. GSEA on differentially expressed genes in Hdac3-depleted IMCs.
Fig. S8. JAK Inhibitor I reduces STAT3 phosphorylation and Il-6 and Mmp13 gene expression in a concentration-dependent manner in Hdac3-depleted chondrocytes.
Fig. S9. JAK-STAT pathway inhibitors reduce STAT3 phosphorylation and Il-6 and Mmp13 gene expression in Hdac3-depleted chondrocytes.
Fig. S10. Combining JAK-STAT pathway and NF-κB inhibitors does not further reduce Il-6 and Mmp13 gene expression in Hdac3-depleted chondrocytes.
Fig. S11. The Col2a1ERT-Cre driver is expressed in chondrocytes and osteoblasts but does not increase IL-6 abundance in osteoblasts.
Fig. S12. JAK inhibition partially restores the bone phenotype of Hdac3-CKOCol2ERT mice.
Table S1. Primer sequences.
Acknowledgments
We would like to thank J.-H. Lee, X. Li, O. Pichurin, D. Razidlo, and B. Stensgard for technical assistance. We are grateful to K. Gaonka for bioinformatics assistance and D. Larson for statistical help. We thank T. Ordog for helpful discussions.
Funding: This work was supported by research and training grants from the NIH (R01 AR68103, R01 AR065402, R01 AR049069, R01 AR067129, R01 066101, R01 AG048388, R01 AG013925, T32 AR56950, F31 AR067646, K01 AR65397, and F32 AR066508), the Ted Nash Foundation, the Connor Group, the charitable foundation of William and Karen Eby, and Mayo Clinic (Kogod Center for Aging, Center for Biomedical Discovery, and Center for Regenerative Medicine).
Footnotes
Author contributions: L.R.C., E.W.B., and J.W.W. conceived and designed experiments. L.R.C., E.W.B., M.E.M.-L., and D.D.P. performed experiments. L.R.C., E.W.B., M.E.M.-L., M.M.W., D.D.P., A.D., M.X., T.T., J.L.K., A.J.v.W., M.J.O., and J.J.W. analyzed data and contributed to critical discussions. L.R.C., A.J.v.W., and J.J.W. wrote and edited the article.
Competing interests: The authors declare that they have no competing interests.
References
- 1.Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 3.Sederquist B, Fernandez-Vojvodich P, Zaman F, Sävendahl L. Recent research on the growth plate: Impact of inflammatory cytokines on longitudinal bone growth. J Mol Endocrinol. 2014;53:T35–T44. doi: 10.1530/JME-14-0006. [DOI] [PubMed] [Google Scholar]
- 4.MacRae VE, Farquharson C, Ahmed SF. The restricted potential for recovery of growth plate chondrogenesis and longitudinal bone growth following exposure to pro-inflammatory cytokines. J Endocrinol. 2006;189:319–328. doi: 10.1677/joe.1.06609. [DOI] [PubMed] [Google Scholar]
- 5.MacRae VE, Farquharson C, Ahmed SF. The pathophysiology of the growth plate in juvenile idiopathic arthritis. Rheumatology. 2006;45:11–19. doi: 10.1093/rheumatology/kei091. [DOI] [PubMed] [Google Scholar]
- 6.De Luca F. Impaired growth plate chondrogenesis in children with chronic illnesses. Pediatr Res. 2006;59:625–629. doi: 10.1203/01.pdr.0000214966.60416.1b. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Vojvodich P, Palmblad K, Karimian E, Andersson U, Sävendahl L. Pro-inflammatory cytokines produced by growth plate chondrocytes may act locally to modulate longitudinal bone growth. Horm Res Paediatr. 2012;77:180–187. doi: 10.1159/000337569. [DOI] [PubMed] [Google Scholar]
- 8.Mazzetti I, Magagnoli G, Paoletti S, Uguccioni M, Olivotto E, Vitellozzi R, Cattini L, Facchini A, Borzi RM. A role for chemokines in the induction of chondrocyte phenotype modulation. Arthritis Rheum. 2004;50:112–122. doi: 10.1002/art.11474. [DOI] [PubMed] [Google Scholar]
- 9.Claes L, Recknagel S, Ignatius A. Fracture healing under healthy and inflammatory conditions. Nat Rev Rheumatol. 2012;8:133–143. doi: 10.1038/nrrheum.2012.1. [DOI] [PubMed] [Google Scholar]
- 10.Gerstenfeld LC, Cullinane DM, Barnes GL, Graves DT, Einhorn TA. Fracture healing as a post-natal developmental process: Molecular, spatial, and temporal aspects of its regulation. J Cell Biochem. 2003;88:873–884. doi: 10.1002/jcb.10435. [DOI] [PubMed] [Google Scholar]
- 11.Kon T, Cho TJ, Aizawa T, Yamazaki M, Nooh N, Graves D, Gerstenfeld LC, Einhorn TA. Expression of osteoprotegerin, receptor activator of NF-κB ligand (osteo-protegerin ligand) and related proinflammatory cytokines during fracture healing. J Bone Miner Res. 2001;16:1004–1014. doi: 10.1359/jbmr.2001.16.6.1004. [DOI] [PubMed] [Google Scholar]
- 12.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 13.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 14.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Longworth MS, Laimins LA. Histone deacetylase 3 localizes to the plasma membrane and is a substrate of Src. Oncogene. 2006;25:4495–4500. doi: 10.1038/sj.onc.1209473. [DOI] [PubMed] [Google Scholar]
- 16.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 18.Jeon EJ, Lee KY, Choi NS, Lee MH, Kim HN, Jin YH, Ryoo HM, Choi JY, Yoshida M, Nishino N, Oh BC, Lee KS, Lee YH, Bae SC. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J Biol Chem. 2006;281:16502–16511. doi: 10.1074/jbc.M512494200. [DOI] [PubMed] [Google Scholar]
- 19.Juan LJ, Shia WJ, Chen MH, Yang WM, Seto E, Lin YS, Wu CW. Histone de-acetylases specifically down-regulate p53-dependent gene activation. J Biol Chem. 2000;275:20436–20443. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- 20.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 21.Yuan Z-l, Guan Y-j, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 22.Chen L-f, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 23.Perkins ND. Integrating cell-signalling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 24.Bradley EW, Carpio LR, van Wijnen AJ, McGee-Lawrence ME, Westendorf JJ. Histone deacetylases in bone development and skeletal disorders. Physiol Rev. 2015;95:1359–1381. doi: 10.1152/physrev.00004.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakshmaiah KC, Jacob LA, Aparna S, Lokanatha D, Saldanha SC. Epigenetic therapy of cancer with histone deacetylase inhibitors. J Cancer Res Ther. 2014;10:469–478. doi: 10.4103/0973-1482.137937. [DOI] [PubMed] [Google Scholar]
- 26.Dinarello CA, Fossati G, Mascagni P. Histone deacetylase inhibitors for treating a spectrum of diseases not related to cancer. Mol Med. 2011;17:333–352. doi: 10.2119/molmed.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Renzo F, Broccia ML, Giavini E, Menegola E. Relationship between embryonic histonic hyperacetylation and axial skeletal defects in mouse exposed to the three HDAC inhibitors apicidin, MS-275, and sodium butyrate. Toxicol Sci. 2007;98:582–588. doi: 10.1093/toxsci/kfm115. [DOI] [PubMed] [Google Scholar]
- 28.Di Renzo F, Cappelletti G, Broccia ML, Giavini E, Menegola E. Boric acid inhibits embryonic histone deacetylases: A suggested mechanism to explain boric acid-related teratogenicity. Toxicol Appl Pharmacol. 2007;220:178–185. doi: 10.1016/j.taap.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Menegola E, Di Renzo F, Broccia ML, Prudenziati M, Minucci S, Massa V, Giavini E. Inhibition of histone deacetylase activity on specific embryonic tissues as a new mechanism for teratogenicity. Birth Defects Res B Dev Reprod Toxicol. 2005;74:392–398. doi: 10.1002/bdrb.20053. [DOI] [PubMed] [Google Scholar]
- 30.Eikel D, Lampen A, Nau H. Teratogenic effects mediated by inhibition of histone de-acetylases: Evidence from quantitative structure activity relationships of 20 valproic acid derivatives. Chem Res Toxicol. 2006;19:272–278. doi: 10.1021/tx0502241. [DOI] [PubMed] [Google Scholar]
- 31.Paradis FH, Hales BF. The effects of class-specific histone deacetylase inhibitors on the development of limbs during organogenesis. Toxicol Sci. 2015;148:220–228. doi: 10.1093/toxsci/kfv174. [DOI] [PubMed] [Google Scholar]
- 32.McGee-Lawrence ME, McCleary-Wheeler AL, Secreto FJ, Razidlo DF, Zhang M, Stensgard BA, Li X, Stein GS, Lian JB, Westendorf JJ. Suberoylanilide hydroxamic acid (SAHA; vorinostat) causes bone loss by inhibiting immature osteoblasts. Bone. 2011;48:1117–1126. doi: 10.1016/j.bone.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pratap J, Akech J, Wixted JJ, Szabo G, Hussain S, McGee-Lawrence ME, Li X, Bedard K, Dhillon RJ, van Wijnen AJ, Stein JL, Stein GS, Westendorf JJ, Lian JB. The histone deacetylase inhibitor, vorinostat, reduces tumor growth at the metastatic bone site and associated osteolysis, but promotes normal bone loss. Mol Cancer Ther. 2010;9:3210–3220. doi: 10.1158/1535-7163.MCT-10-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato Y, Kondo I, Ishida S, Motooka H, Takayama K, Tomita Y, Maeda H, Satoh K. Decreased bone mass and increased bone turnover with valproate therapy in adults with epilepsy. Neurology. 2001;57:445–449. doi: 10.1212/wnl.57.3.445. [DOI] [PubMed] [Google Scholar]
- 35.Vestergaard P, Rejnmark L, Mosekilde L. Fracture risk associated with use of anti-epileptic drugs. Epilepsia. 2004;45:1330–1337. doi: 10.1111/j.0013-9580.2004.18804.x. [DOI] [PubMed] [Google Scholar]
- 36.Guo CY, Ronen GM, Atkinson SA. Long-term valproate and lamotrigine treatment may be a marker for reduced growth and bone mass in children with epilepsy. Epilepsia. 2001;42:1141–1147. doi: 10.1046/j.1528-1157.2001.416800.x. [DOI] [PubMed] [Google Scholar]
- 37.Gordon JA, Stein JL, Westendorf JJ, van Wijnen AJ. Chromatin modifiers and histone modifications in bone formation, regeneration, and therapeutic intervention for bone-related disease. Bone. 2015;81:739–745. doi: 10.1016/j.bone.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone deacetylase 3 interacts with Runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem. 2004;279:41998–42007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- 39.Hesse E, Saito H, Kiviranta R, Correa D, Yamana K, Neff L, Toben D, Duda G, Atfi A, Geoffroy V, Horne WC, Baron R. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J Cell Biol. 2010;191:1271–1283. doi: 10.1083/jcb.201009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 41.Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, Hiebert SW. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Razidlo DF, Whitney TJ, Casper ME, McGee-Lawrence ME, Stensgard BA, Li X, Secreto FJ, Knutson SK, Hiebert SW, Westendorf JJ. Histone deacetylase 3 depletion in osteo/chondroprogenitor cells decreases bone density and increases marrow fat. PLOS One. 2010;5:e11492. doi: 10.1371/journal.pone.0011492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGee-Lawrence ME, Bradley EW, Dudakovic A, Carlson SW, Ryan ZC, Kumar R, Dadsetan M, Yaszemski MJ, Chen Q, An KN, Westendorf JJ. Histone deacetylase 3 is required for maintenance of bone mass during aging. Bone. 2013;52:296–307. doi: 10.1016/j.bone.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bradley EW, Carpio LR, Westendorf JJ. Histone deacetylase 3 suppression increases PH domain and leucine-rich repeat phosphatase (Phlpp)1 expression in chondrocytes to suppress Akt signaling and matrix secretion. J Biol Chem. 2013;288:9572–9582. doi: 10.1074/jbc.M112.423723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh N, Gupta M, Trivedi CM, Singh MK, Li L, Epstein JA. Murine craniofacial development requires Hdac3-mediated repression of Msx gene expression. Dev Biol. 2013;377:333–344. doi: 10.1016/j.ydbio.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh N, Trivedi CM, Lu M, Mullican SE, Lazar MA, Epstein JA. Histone deacetylase 3 regulates smooth muscle differentiation in neural crest cells and development of the cardiac outflow tract. Circ Res. 2011;109:1240–1249. doi: 10.1161/CIRCRESAHA.111.255067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakai K, Hiripi L, Glumoff V, Brandau O, Eerola R, Vuorio E, Bösze Z, Fässler R, Aszódi A. Stage-and tissue-specific expression of a Col2a1-Cre fusion gene in transgenic mice. Matrix Biol. 2001;19:761–767. doi: 10.1016/s0945-053x(00)00122-0. [DOI] [PubMed] [Google Scholar]
- 48.Chen M, Lichtler AC, Sheu TJ, Xie C, Zhang X, O’Keefe RJ, Chen D. Generation of a transgenic mouse model with chondrocyte-specific and tamoxifen-inducible expression of Cre recombinase. Genesis. 2007;45:44–50. doi: 10.1002/dvg.20261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakamura E, Nguyen MT, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreERT to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn. 2006;235:2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsusaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Transcription factors NF-IL6 and NF-κB synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci USA. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DiNatale BC, Schroeder JC, Francey LJ, Kusnadi A, Perdew GH. Mechanistic insights into the events that lead to synergistic induction of interleukin 6 transcription upon activation of the aryl hydrocarbon receptor and inflammatory signaling. J Biol Chem. 2010;285:24388–24397. doi: 10.1074/jbc.M110.118570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SK, Lorenzo J. Cytokines regulating osteoclast formation and function. Curr Opin Rheumatol. 2006;18:411–418. doi: 10.1097/01.bor.0000231911.42666.78. [DOI] [PubMed] [Google Scholar]
- 56.Boyce BF, Schwarz EM, Xing L. Osteoclast precursors: Cytokine-stimulated immunomodulators of inflammatory bone disease. Curr Opin Rheumatol. 2006;18:427–432. doi: 10.1097/01.bor.0000231913.32364.32. [DOI] [PubMed] [Google Scholar]
- 57.Roodman GD. Cell biology of the osteoclast. Exp Hematol. 1999;27:1229–1241. doi: 10.1016/s0301-472x(99)00061-2. [DOI] [PubMed] [Google Scholar]
- 58.Otsuka T, Thacker JD, Hogge DE. The effects of interleukin 6 and interleukin 3 on early hematopoietic events in long-term cultures of human marrow. Exp Hematol. 1991;19:1042–1048. [PubMed] [Google Scholar]
- 59.McGee-Lawrence ME, Carpio LR, Schulze RJ, Pierce JL, McNiven MA, Farr JN, Khosla S, Oursler MJ, Westendorf JJ. Hdac3 deficiency increases marrow adiposity and induces lipid storage and glucocorticoid metabolism in osteochondroprogenitor cells. J Bone Miner Res. 2015;31:116–128. doi: 10.1002/jbmr.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, Carmeliet G, Kronenberg HM. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ono N, Ono W, Nagasawa T, Kronenberg HM. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol. 2014;16:1157–1167. doi: 10.1038/ncb3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams RM, Zipfel WR, Tinsley ML, Farnum CE. Solute transport in growth plate cartilage: In vitro and in vivo. Biophys J. 2007;93:1039–1050. doi: 10.1529/biophysj.106.097675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L, Liu Y, Han R, Beier UH, Bhatti TR, Akimova T, Greene MI, Hiebert SW, Hancock WW. FOXP3+ regulatory T cell development and function require histone/ protein deacetylase 3. J Clin Invest. 2015;125:1111–1123. doi: 10.1172/JCI77088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu M, Tchkonia T, Ding H, Ogrodnik M, Lubbers ER, Pirtskhalava T, White TA, Johnson KO, Stout MB, Mezera V, Giorgadze N, Jensen MD, LeBrasseur NK, Kirkland JL. JAK inhibition alleviates the cellular senescence-associated secretory phenotype and frailty in old age. Proc Natl Acad Sci USA. 2015;112:E6301–E6310. doi: 10.1073/pnas.1515386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cantley MD, Bartold PM, Fairlie DP, Rainsford KD, Haynes DR. Histone deacetylase inhibitors as suppressors of bone destruction in inflammatory diseases. J Pharm Pharmacol. 2012;64:763–774. doi: 10.1111/j.2042-7158.2011.01421.x. [DOI] [PubMed] [Google Scholar]
- 66.Blanchard F, Chipoy C. Histone deacetylase inhibitors: New drugs for the treatment of inflammatory diseases? Drug Discov Today. 2005;10:197–204. doi: 10.1016/S1359-6446(04)03309-4. [DOI] [PubMed] [Google Scholar]
- 67.Chen W-P, Bao J-P, Hu P-F, Feng J, Wu L-D. Alleviation of osteoarthritis by Trichostatin A, a histone deacetylase inhibitor, in experimental osteoarthritis. Mol Biol Rep. 2010;37:3967–3972. doi: 10.1007/s11033-010-0055-9. [DOI] [PubMed] [Google Scholar]
- 68.Culley KL, Hui W, Barter MJ, Davidson RK, Swingler TE, Destrument APM, Scott JL, Donell ST, Fenwick S, Rowan AD, Young DA, Clark IM. Class I histone deacetylase inhibition modulates metalloproteinase expression and blocks cytokine-induced cartilage degradation. Arthritis Rheum. 2013;65:1822–1830. doi: 10.1002/art.37965. [DOI] [PubMed] [Google Scholar]
- 69.Leah E. Osteoarthritis: Chondroprotection by histone deacetylase inhibition. Nat Rev Rheumatol. 2013;9:321. doi: 10.1038/nrrheum.2013.63. [DOI] [PubMed] [Google Scholar]
- 70.Wang JH, Shih KS, Wu YW, Wang AW, Yang CR. Histone deacetylase inhibitors increase microRNA-146a expression and enhance negative regulation of interleukin-1β signaling in osteoarthritis fibroblast-like synoviocytes. Osteoarthritis Cartilage. 2013;21:1987–1996. doi: 10.1016/j.joca.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 71.Takada Y, Gillenwater A, Ichikawa H, Aggarwal BB. Suberoylanilide hydroxamic acid potentiates apoptosis, inhibits invasion, and abolishes osteoclastogenesis by suppressing nuclear factor-κB activation. J Biol Chem. 2006;281:5612–5622. doi: 10.1074/jbc.M507213200. [DOI] [PubMed] [Google Scholar]
- 72.Kim HN, Ha H, Lee JH, Jung K, Yang D, Woo KM, Lee ZH. Trichostatin A inhibits osteoclastogenesis and bone resorption by suppressing the induction of c-Fos by RANKL. Eur J Pharmacol. 2009;623:22–29. doi: 10.1016/j.ejphar.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 73.Nakamura T, Kukita T, Shobuike T, Nagata K, Wu Z, Ogawa K, Hotokebuchi T, Kohashi O, Kukita A. Inhibition of histone deacetylase suppresses osteoclastogenesis and bone destruction by inducing IFN-β production. J Immunol. 2005;175:5809–5816. doi: 10.4049/jimmunol.175.9.5809. [DOI] [PubMed] [Google Scholar]
- 74.Dinarello CA. Anti-inflammatory agents: Present and future. Cell. 2010;140:935–950. doi: 10.1016/j.cell.2010.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adcock IM. HDAC inhibitors as anti-inflammatory agents. Br J Pharmacol. 2007;150:829–831. doi: 10.1038/sj.bjp.0707166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Halili MA, Andrews MR, Labzin LI, Schroder K, Matthias G, Cao C, Lovelace E, Reid RC, Le GT, Hume DA, Irvine KM, Matthias P, Fairlie DP, Sweet MJ. Differential effects of selective HDAC inhibitors on macrophage inflammatory responses to the Toll-like receptor 4 agonist LPS. J Leukoc Biol. 2010;87:1103–1114. doi: 10.1189/jlb.0509363. [DOI] [PubMed] [Google Scholar]
- 77.Sweet MJ, Shakespear MR, Kamal NA, Fairlie DP. HDAC inhibitors: Modulating leukocyte differentiation, survival, proliferation and inflammation. Immunol Cell Biol. 2012;90:14–22. doi: 10.1038/icb.2011.88. [DOI] [PubMed] [Google Scholar]
- 78.Grabiec AM, Krausz S, de Jager W, Burakowski T, Groot D, Sanders ME, Prakken BJ, Maslinski W, Eldering E, Tak PP, Reedquist KA. Histone deacetylase inhibitors suppress inflammatory activation of rheumatoid arthritis patient synovial macrophages and tissue. J Immunol. 2010;184:2718–2728. doi: 10.4049/jimmunol.0901467. [DOI] [PubMed] [Google Scholar]
- 79.Kotwal N, Li J, Sandy J, Plaas A, Sumner DR. Initial application of EPIC-μCT to assess mouse articular cartilage morphology and composition: Effects of aging and treadmill running. Osteoarthritis Cartilage. 2012;20:887–895. doi: 10.1016/j.joca.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palmer AW, Guldberg RE, Levenston ME. Analysis of cartilage matrix fixed charge density and three-dimensional morphology via contrast-enhanced microcomputed tomography. Proc Natl Acad Sci USA. 2006;103:19255–19260. doi: 10.1073/pnas.0606406103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gosset M, Berenbaum F, Thirion S, Jacques C. Primary culture and phenotyping of murine chondrocytes. Nat Protoc. 2008;3:1253–1260. doi: 10.1038/nprot.2008.95. [DOI] [PubMed] [Google Scholar]
- 82.Dudakovic A, Camilleri E, Riester SM, Lewallen EA, Kvasha S, Chen X, Radel DJ, Anderson JM, Nair AA, Evans JM, Krych AJ, Smith J, Deyle DR, Stein JL, Stein GS, Im HJ, Cool SM, Westendorf JJ, Kakar S, Dietz AB, van Wijnen AJ. High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J Cell Biochem. 2014;115:1816–1828. doi: 10.1002/jcb.24852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kalari KR, Nair AA, Bhavsar JD, O’Brien DR, Davila JI, Bockol MA, Nie J, Tang X, Baheti S, Doughty JB, Middha S, Sicotte H, Thompson AE, Asmann YW, Kocher JP. MAP-RSeq: Mayo analysis pipeline for RNA sequencing. BMC Bioinformatics. 2014;15:224. doi: 10.1186/1471-2105-15-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang L, Wang S, Li W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 85.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yan H, Evans J, Kalmbach M, Moore R, Middha S, Luban S, Wang L, Bhagwate A, Li Y, Sun Z, Chen X, Kocher JPA. HiChIP: A high-throughput pipeline for integrative analysis of ChIP-Seq data. BMC Bioinformatics. 2014;15:280. doi: 10.1186/1471-2105-15-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weivoda MM, Ruan M, Pederson L, Hachfeld C, Davey RA, Zajac JD, Westendorf JJ, Khosla S, Oursler MJ. Osteoclast TGF-β receptor signaling induces Wnt1 secretion and couples bone resorption to bone formation. J Bone Miner Res. 2016;31:76–85. doi: 10.1002/jbmr.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression pattern of Hdac3 during embryonic and postnatal long bone development.
Fig. S2. Hdac3 knockout in chondrocytes during embryogenesis is lethal.
Fig. S3. Tamoxifen does not affect growth plate maturation in wild-type mice.
Fig. S4. Top 30 annotation clusters of genes with decreased gene expression in Hdac3-depleted IMCs.
Fig. S5. Top 30 annotation clusters of genes with increased gene expression in Hdac3-depleted IMCs.
Fig. S6. Gene lists associated with top annotation clusters from DAVID pathway analysis.
Fig. S7. GSEA on differentially expressed genes in Hdac3-depleted IMCs.
Fig. S8. JAK Inhibitor I reduces STAT3 phosphorylation and Il-6 and Mmp13 gene expression in a concentration-dependent manner in Hdac3-depleted chondrocytes.
Fig. S9. JAK-STAT pathway inhibitors reduce STAT3 phosphorylation and Il-6 and Mmp13 gene expression in Hdac3-depleted chondrocytes.
Fig. S10. Combining JAK-STAT pathway and NF-κB inhibitors does not further reduce Il-6 and Mmp13 gene expression in Hdac3-depleted chondrocytes.
Fig. S11. The Col2a1ERT-Cre driver is expressed in chondrocytes and osteoblasts but does not increase IL-6 abundance in osteoblasts.
Fig. S12. JAK inhibition partially restores the bone phenotype of Hdac3-CKOCol2ERT mice.
Table S1. Primer sequences.