Abstract
There is a need to better understand and handle the “dark matter” of proteomics – the vast diversity of post-translational and chemical modifications that are unaccounted in a typical analysis and thus remain unidentified. We present a novel fragment-ion indexing method, and its implementation in peptide identification tool MSFragger, that enables an over 100-fold improvement in speed over most existing tools. Using some of the largest proteomic datasets to date, we demonstrate how MSFragger empowers the open database search concept for comprehensive identification of peptides and all their modified forms, uncovering dramatic differences in the modification rates across experimental samples and conditions. We further illustrate its utility using protein-RNA crosslinked peptide data, and using affinity purification experiments where we observe on average a 300% increase in the number of identified spectra for enriched proteins. We also discuss the benefits of open searching for improved false discovery rate estimation in proteomics.
INTRODUCTION
Peptide identification algorithms have served as a cornerstone of shotgun proteomics for several decades 1. The most commonly used computational strategy is based on searching acquired tandem mass (MS/MS) spectra against a protein sequence database using database search algorithms 2. However, even given significant improvements in the quality of MS/MS data acquired on modern mass spectrometers, a very significant fraction of spectra remains unexplained.
We and others have been fascinated by the underlying complexity of the “dark matter” in shotgun proteomics 3 – including the vast diversity of post-translational modifications (PTMs) as well as novel sequences (e.g. mutations and splice isoforms) – that are unaccounted for in traditional database search and thus remain unidentified 4–8. A number of computational strategies emerged for the detection of such peptides including multi-step database search 9, 10, curated modifications search9, 11, spectral-pair based methods screening for modified versions of peptides initially identified in unmodified form 12–15, sequence tagging 16–20, and spectral alignment 21, 22 (reviewed in 23). However, the proteomics community continues to search for practical computational tools for this task. These efforts are exemplified by a recent report 4 exploring the feasibility of “open” (i.e. using wide precursor mass tolerance of hundreds of Daltons allowing for identification of modified peptides) searches using conventional database search tools.
In our quest to develop a broadly applicable and fast computational strategy for open database search we designed a novel fragment ion indexing method that provides orders of magnitude improvement in speed over existing tools. We implemented this method in a new database search tool MSFragger. MSFragger makes open searches feasible even for very large datasets containing millions of MS/MS spectra, helping to reconstruct modification profiles and to uncover dramatic differences in the modification rates across different experiments. It is capable of performing open searches with variable modifications, making it applicable to data from labeling-based quantitative proteomics experiments. We further demonstrate MSFragger’s utility in the analysis of protein-RNA crosslinked peptides and affinity purification mass spectrometry (AP-MS) data. Finally, open searching uncovers, and provides a potential solution to the problem of inaccurate false discovery rate (FDR) estimates in traditional narrow window searches due to unaccounted peptide modifications. MSFragger is platform independent, not limited to data from a particular MS instrument, and can be easily incorporated into most existing data analysis pipelines.
RESULTS
Novel fragment ion index enables ultrafast database search
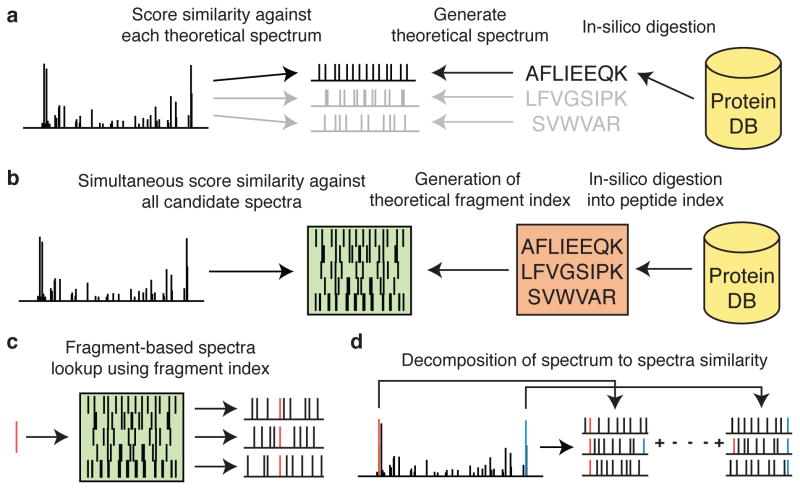
MSFragger begins by performing an in-silico digestion of the protein database (Fig 1a). It then removes redundant peptides and orders them by their theoretical mass (including any modified peptides generated as a result of variable modifications), creating a peptide index. While peptide indexing has been described previously as a way to accelerate database search24–26, this step alone has little impact on spectrum similarity calculations, which is the most time consuming step. MSFragger addresses this bottleneck by creating a novel theoretical fragment index. This key computational advance enables highly efficient and simultaneous scoring of an experimental spectrum against all candidate peptides (Online Methods; Fig. 1b; Supplementary Fig. 1).
Figure 1. Database search strategies and the MSFragger algorithm.
(a) Conventional database search involves in-silico digestion of a protein database into candidate peptides from which theoretical spectra are sequentially generated and compared against experimental spectra one at a time. (b) MSFragger digests a protein database and generates a non-redundant set of peptides that are arranged in a peptide index. This index is then used as a reference to generate a fragment index that allows for rapid retrieval of theoretical spectra that contains a fragment of a query mass. This fragment index is then used for the efficient and simultaneous scoring of an experimental spectrum against all candidate spectra. (c) Mass binning and precursor mass ordering within the fragment index allows for rapid retrieval of candidate spectra that matches a given experimental fragment ion. Scores of candidate peptides corresponding to retrieved spectra are incremented. (d) Processing of all experimental fragment ions results in the identification of all matching fragments between experimental spectrum and all candidate theoretical spectra, decomposing spectrum to spectra matches to fragment to spectra matches. Matched fragments can then be used to compute a similarity score.
We first evaluated the performance of the MSFragger algorithm on a deep HEK2934 dataset, and compared it to that of commonly used search engines Comet 27 and X! Tandem 9. The scores and error rates of modified peptides are likely to be different than those of unmodified peptides, prompting class-specific FDR estimation28, 29. To account for these differences we adopted an extended mass model when computing peptide probabilities, ensuring mass-shift dependent FDR estimation and filtering (Online Methods). Note that in open searches the term ‘modifications’ is used interchangeably with ‘mass shifts’ and includes in-source fragmentation events, missed cleavages, and isotope errors. Overall, all search engines performed similarly when run using similar search parameters (Table 1). In the traditional (narrow window) search, MSFragger and Comet respectively identified 9795 and 9757 protein groups (1% protein FDR), and 456548 and 461806 PSMs (1% protein and PSM FDR). MSFragger also identified similar numbers as X! Tandem, when accounting for the innate variable modifications that X! Tandem specifies by default. In open search, which represent the primary motivation for the development of MSFragger, we observed a dramatic increase in the number of identified PSMs across all search engines, in line with the earlier report using SEQUEST 4. For example, MSFragger identified 609897 PSMs using open search, an increase of 33.6% compared to narrow window search, with a minimal loss of 1.4% in the number of protein identifications. Note that when performing protein inference using open search results, we took a conservative approach of using unmodified peptides and peptides with specified variable modifications only (Online Methods). When all modified peptides were included, the number of protein identifications from open searches exceeded that of narrow window searches (e.g. by 4.4% for MSFragger). However, additional work is necessary to carefully evaluate the accuracy of the protein inference step when using all peptides identified in open search.
Table 1. Identification rates and analysis time for HEK293 dataset.
Identification numbers are for the entire 24 LC-MS/MS run dataset, filtered at 1% FDR at both protein and PSM levels. Search times given for a single LC-MS/MS run consisting of 41820 MS/MS spectra analyzed on a quad core workstation.
| Search Engine | Time (minutes) | Proteins | PSMs | Peptides |
|---|---|---|---|---|
| Narrow window search | ||||
| SEQUEST* | 9.3 | 9,513 | 396,736 | 110,262 |
| Comet | 1.7 | 9,757 | 461,806 | 115,612 |
| X! Tandem** | 1.7 | 10,182 | 466,701 | 119,304 |
| MSFragger | 0.4 | 9,795 | 456,548 | 115,755 |
| Open search (500 Da) | ||||
| SEQUEST* | 673.0 | 9,178 | 510,139 | 111,205 |
| Comet | 815.4 | 9,545 | 584,218 | 123,679 |
| X! Tandem | 976.0 | 9,830 | 638,052 | 133,318 |
| MSFragger | 5.4 | 9,656 | 609,897 | 126,037 |
For time estimation, SEQUEST searches were performed using Proteome Discoverer 2.1 (SEQUEST HT) on a more powerful 8-core workstation. Narrow window searches were done using 100 ppm precursor mass window except for SEQUEST (5 ppm). SEQUEST identification rates were taken from 4;
X! Tandem searches include several variable modifications that cannot be turned off.
Open searches using conventional database search tools are slow, given the vastly expanded search space. Comet and X! Tandem took 13.6 and 16.3 hours respectively to analyze a single LC-MS/MS run using a quad core workstation. In stark contrast, MSFragger took only 5.4 minutes, making it over 150 times faster than these commonly used tools. We have also compared MSFragger with tools that employ peptide indexing such as Tide 30 and SEQUEST HT (Supplementary Table 1). Tide, which only allows 100Da precursor windows and does not take advantage of multiple processor cores, took 176.7 minutes (compared to 9.8 minutes with MSFragger when subjected to the same constraints). SEQUEST HT (Proteome Discoverer 2.1) took over 11 hours on a more powerful octa-core workstation. The speed and scalability (Supplementary Fig. 2) of MSFragger allowed open searching of the entire HEK293 dataset (24 LC-MS/MS runs) in less than 30 minutes on a single powerful workstation compared to days or even weeks required to search these data using existing tools on the same machine.
We also sought to compare MSFragger to algorithms specifically designed for comprehensive PTM analysis. We chose MODa 19, which has been established as an effective tool for blind PTM search. Using comparable settings, both tools produced very similar PTM profiles (Supplementary Fig. 3), except MSFragger was notably faster (Supplementary Table 1) and identified a larger number of PSMs than MODa at a FDR of 1%: 622857 (MSFragger, tryptic search) vs. 522812 (MODa, semi-tryptic search) and 439216 (MODa, tryptic search). The difference between MODa and MSFragger results can be explained, in part, by the fact that MODa is required to localize the mass shift to a particular amino acid, whereas open searching only identifies the peptide sequence and the mass shift (which may be the result of multiple modifications).
Refinement of the open database search strategy
The development of MSFragger algorithm not only makes open searching practical, but also presents an opportunity to further investigate and refine this computational strategy. It is often assumed that the number of identified unique peptide sequences would be greatly reduced in open search compared to narrow window search due to the vastly expanded search space. However, our results using multiple search engines (Table 1) demonstrate that this is generally not the case. At the same time, it is true that not all unmodified, tryptic peptides found in narrow window search are found in open search. To see if those peptides can also be recovered, we implemented a boosting feature within MSFragger that preferentially ranks unmodified peptides over modified peptides when performing open search (Online Methods). However, such a strategy, while implemented as an option in MSFragger, has not been found to significantly improve the results (Supplementary Fig. 4).
Open searching does not account for modified fragment ions, resulting in reduced sensitivity for common modifications when compared to traditional variable modification searching. The speed of MSFragger allows us to combine the two approaches to successfully recover peptides with specified variable modifications at rates similar to unmodified peptides (Supplementary Fig. 5). Furthermore, we attempted to address the weakness of the open search strategy for C-terminal modifications (as y-ions are the most abundant and commonly observed in CID/HCD fragmentation) by inserting complementary ions6, 31. While we observed an increase in the recovery of peptides with modifications near the C-terminus, the addition of complementary ions failed to increase the overall number of identifications (Online Methods; Supplementary Fig. 6). A more effective strategy is to add complementary ions to the theoretical spectrum rather than the experimental spectrum, which we plan to pursue in future work via extension of the fragment ion indexing scheme.
The problem of co-isolating multiple co-eluting peptides and the resulting chimera MS/MS spectra is well established1, 32 and manifests itself in unique ways in open searches. When a co-fragmented peptide is identified with a higher score, an artefactual (not attributed to any modification) mass shift is produced that can either be small (within several Daltons) or large (hundreds of Daltons) depending on whether the co-fragmented peptide ions are of the same or different charge state, respectively. Such cases can be identified using linked MS1 and MS/MS spectral viewers (Supplementary Fig 7a,c), and further evaluated using tools such as BatMass33 (Supplementary Fig 7b,d). While the number of such cases is small, in future work chimeric spectra can be dealt with more accurately in open searches via MS1 feature detection of co-isolated peptides25 within MSFragger or using external tools34.
High prevalence of peptides identified in modified form only
To investigate the peptides (distinct sequences) that were found in narrow window search but not open search, we looked at the intersection of search results in the HEK293 dataset (both searches were done without variable modifications). We subdivided the peptides based on their estimated confidence (Fig. 2a) and examined the group specific FDR. As expected, peptides that were accepted at 1% FDR in both searches (101138 in total) were of high confidence with an estimated FDR of 0.15%. Peptides found in both searches, but only accepted at 1% FDR in one of the two searches were of lower confidence as evidenced by the increased group FDR. Of greatest intrigue, however, were the peptides that were confidently identified in one search but not identified at all in the other.
Figure 2. Peptide identifications across traditional narrow window and open searches demonstrate false discovery rates underestimation.
(a) Peptides passing a 1% FDR filter in both narrow window and open searches are compared. Common peptides are of high confidence. Peptides found only in open search are also of high confidence suggesting that many peptides are only found in its modified form. High FDR was observed for peptides unique to narrow window search. (b) Profile of peptides that were only found in modified forms is similar to that of all modified peptides. PSMs from peptides that were only found in narrow window search were mapped to their higher scoring matches in open search and generated a profile devoid of modifications that can be easily represented as some series of amino acid insertions and deletions. These PSMs may be false positive events that arise due to unaccounted for modifications in narrow window search. (c) PSMs supporting peptides only found in narrow window search are commonly matched to peptides with greater counts in open search, giving greater confidences to their open assignment. (d) Peptides suspected to be false positives as a result of unaccounted for modifications in narrow window search are plotted across peptide confidences. Their numbers exceeds the number of decoys and are prevalent in ranges of high peptide confidences, suggesting that are not well estimated by the target-decoy strategy and cannot be eliminated using any scoring threshold. (e) Confirmation of target-decoy violation by examining PSMs with common modifications in narrow window search.
There were 12622 peptides confidently identified in open search but not in narrow window search. The relatively low group FDR of these peptides (4.15%) suggests that most of these are bona fide examples of peptides that were only detected in modified forms. The substantial number of such peptides is problematic for ‘dependent peptide’ approaches for PTM identification 14 (including spectral library-based methods12, 15) that rely on co-identification of the unmodified peptide. A comparison of the modification profile of these peptides to one that is generated from all modified peptides shows high similarity (Fig. 2b), suggesting that most of these identifications correspond to constitutive or highly abundant modifications.
Open searching uncovers FDR problem in traditional narrow window searches
In contrast, the 3773 peptides identified in narrow window search but not in open search had a much higher group FDR of 14.68%. We mapped the spectra supporting these identifications to their results in open search. Of particular interest were spectra that were assigned to unmodified peptides in narrow window search but reassigned, due to an improved match, as modified peptides (with different sequence) in open search. These cases represent potential instances of false positives in narrow window search that are caused by chemical or biological modifications6, 35. In each such instance - a pair of peptides whose masses differ by the mass of the modification detected in the open search - we compared the total number of supporting PSMs associated with the peptide sequence matched in narrow window search to that in open search (Fig. 2c). Under the assumption that peptides that are supported by a greater number of PSMs are more likely to be true identifications, there was significantly higher support for the peptides identified in open search. Only 17% of the spectra were assigned to peptides that had greater support in narrow window search while 68% have greater support for their open search assignment.
We called peptide identifications that were only found in narrow window search to be “suspect” (a potential false positive) if there was greater support for the open search assignment for each supporting PSM. Of the 3773 peptides that were only found in narrow window search, 1139 were suspect. This is significantly more than the number of decoys (554) in the same group, and even more than the total number of decoys in the entire narrow window search at 1% FDR (1091 decoys in total). This suggests that false positives in narrow window search are not correctly estimated by decoy peptides. It is particularly worrisome that some of these suspect peptides have very high scores (Fig. 2d).
Due to high significance of this observation, we sought to verify our findings that the target-decoy strategy fails to effectively capture false positives that are due to unaccounted modifications. We selected high scoring peptide identifications in open search that were observed in both its unmodified form and with a mass shift corresponding to a common modification (oxidation or carbamylation). As we did not specify any variable modifications, the target-decoy assumption is that spectra from these modified peptides would match equally (and incorrectly) to both targets and decoys in narrow window search. However, that was not the case as the rate of matching to target sequences was roughly 6 times that to decoys for carbamylated peptide spectra, and over 9 times for oxidized peptide spectra (Fig. 2e). The violation of the target-decoy assumption is likely due to homology between true peptide sequences and other peptides in the target space, which we previously noted in the context of proteogenomics6, 36. Further supporting this, the modification profile of peptides identified in open search and whose spectra produced suspect identifications in narrow window search markedly lacked phosphorylation and aminoethylbenzenesulfonylation (Fig. 2b). These two mass shifts (79.97 and 183.04 Da) are difficult to represent as some sequence of amino acid addition and deletion. Overall, our analysis using HEK293 dataset (similar trends were observed for other datasets) demonstrates that accounting for all modified peptide forms using the open search strategy of MSFragger is important for confident peptide identifications and accurate FDR estimation, even when the identification of modified peptides is not a primary interest on its own.
Ultrafast open search enables large-scale modification profiling
MSFragger’s ultrafast performance enables comprehensive profiling of chemical and biological modifications across large-scale proteomics datasets. To demonstrate this, we probed three large proteome-wide studies using open searches and compared their modification profiles. In addition to the HEK293 dataset used to benchmark MSFragger, a HeLa 37 and a triple-negative breast cancer (TNBC) 38 datasets were used (Online Methods; Supplementary Table 2). We performed MS1-based correction of precursor masses followed by identification-based mass recalibration to improve the delineation of modifications having close masses across disparate experiments and labs (Online Methods; Supplementary Fig. 8). The list of 500 most abundance mass shifts (excluding modifications specified as variable modifications in the search), is shown in Supplementary Table 3. We confirmed that in all datasets FDR estimates for modified peptides were well controlled and not inflated relative to unmodified peptides. For example, in HEK293 dataset, peptide-level FDR was 0.18%, 0.11%, and 0.11% for peptides with top 500 most abundant mass shifts, top 100 mass most abundant shifts, and for unmodified peptides, respectively (Supplementary Table 3).
We first interrogated several common chemical modifications (Fig. 3a). While the localization profiles were largely concordant (Supplementary Fig. 9), their normalized abundances (modification rates) across the datasets were quite dissimilar. For example, the rate of phosphorylation in the HeLa dataset was over 14 times than that in the TNBC dataset. Furthermore, some of these modifications were found on amino acids that are generally not considered in traditional workflows, such as tryptophan oxidation.
Figure 3. Analysis of large-scale shotgun proteomics experiments reveals differences in modification profiles.
Mass difference features are identified with high mass accuracy aligned across multiple experiments. Features are characterized by their localization rates and amino acid propensities. (a) Common modifications are present across different experiments with vastly different modification rates. Modifications are sometimes localized to amino acids that are unaccounted for in traditional workflows. (b) Large numbers of abundant features were found unique to particular experiments. Localization information assisted in characterizing these unknown modifications. (c) Highly abundant mass features were observed in which the mass difference could not be effectively localized.
We observed many highly abundant modifications that lacked annotations in Unimod and were unique (or of significantly greater abundance) to a particular dataset (Supplementary Table 3). To help decipher these unannotated modifications, we performed site localization analyses (Online Methods; Supplementary Table 4). For example, the HeLa dataset contained over 23,000 PSMs with a modification mass of 52.913 Da that was often localized to aspartic acid or glutamic acid, characteristic of metal ion adducts. This is likely to be iron displacing three protons (Unimod annotates ‘Replacement of 2 protons by iron’ modification only). While deducing identities of unannotated modifications was outside the scope of this work, it was easy to notice that many occurred on cysteines (Fig. 3b).
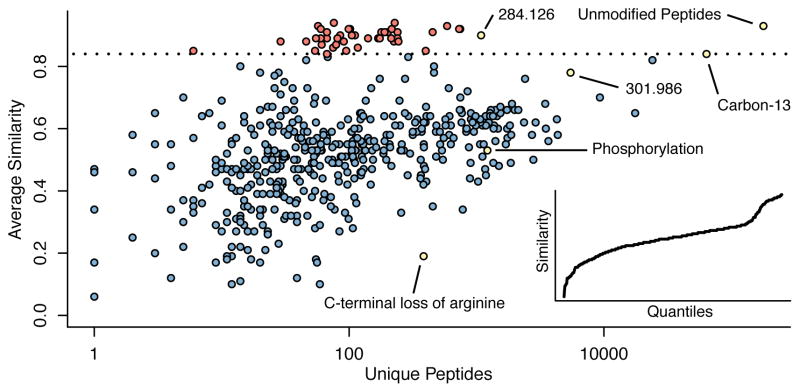
For some modifications, we were unable to localize the mass shift on the peptide (Fig. 3c). This suggests that there are few fragments that support the modification mass or that the detected modification mass is the result of multiple modifications found on the same peptide. To investigate such cases, for each modification mass we computed a spectral similarity score between peptides containing that modification and their corresponding unmodified forms (Online Methods; Fig. 4). Most modifications possessed a similarity score between 0.4 and 0.6, including known modifications such as phosphorylation. However, we observed a large number of modifications (e.g. 3417 PSMs with mass shift 301.986 Da in HEK293 dataset, 3068 PSMs with mass shift 284.126 Da in HeLa dataset) with similarity scores close to 1, indicating that spectra for peptides with these modifications were largely unchanged from that of the unmodified peptide (Supplementary Fig. 10). The lack of differences in the spectra, and significant differences in the modification rates across the datasets (Supplementary Table 3), suggest sample preparation protocol specific labile modifications that are lost during fragmentation.
Figure 4. Open searching detects modified peptides containing labile modifications.
Spectral similarity scores for each mass bin were computed to capture the spectral similarity between a modified peptide and its unmodified counterpart. Most modifications, such as phosphorylation, have average similarities between 0.4 and 0.6. Modifications that are localized to peptide C-terminus disrupt the intense y-ion series and have lower similarity scores. Few mass bins contain low similarity scores as these modified peptides would otherwise be impossible to detect using open searching. Interestingly, there exists a population of mass bins that have similarity scores exceeding that of carbon-13 (which leaves a largely unaltered spectrum). These modifications may represent labile modifications that are lost during peptide fragmentation.
Utility of MSFragger in various proteomics applications
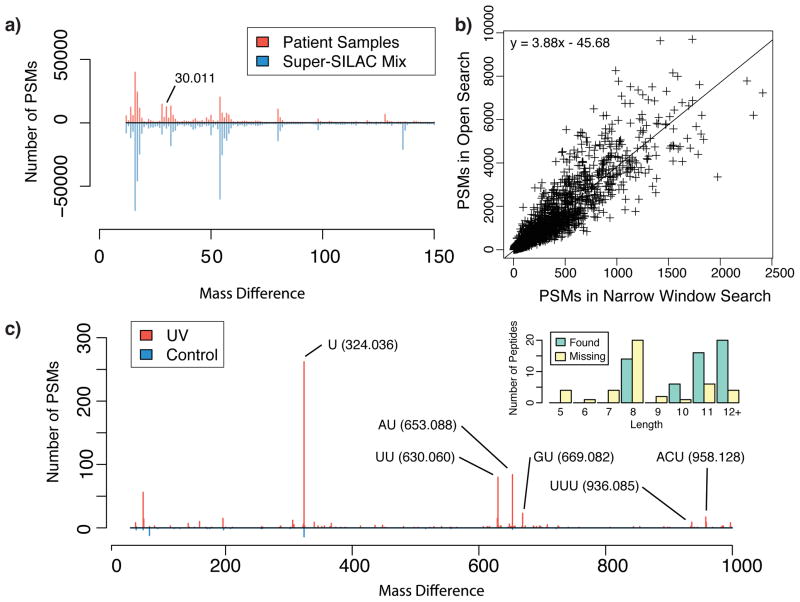
MSFragger enables a wide range of analyses beyond interrogation of unlabeled proteomes. First, we are able to perform open searches using spectra from labeling-based experiments (e.g. SILAC or TMT) by specifying the labeled amino acids as a variable modification, thus allowing quantitative comparison of the modification states of proteins en masse. To test this, we examined a breast cancer dataset consisting of 442 LC-MS/MS runs representing 88 formalin-fixed paraffin-embedded (FFPE) patient samples that were analyzed together with a heavy labeled super-SILAC mix39. Examination of the modification profiles revealed a wide range of abundant modifications in these samples, as well as uncovered differences in modification abundances between the breast cancer samples and the super-SILAC mix, including a 30.011 Da mass shift that likely represents a methylol adduct which is characteristic of FFPE proteomes 40 (Fig. 5a).
Figure 5. Application of MSFragger to diverse proteomics experiments.
(a) The speed of MSFragger allows for reasonable analysis times even when the SILAC labels are specified as variable modifications in conjunction with open searching. In this comparison between a panel of breast tissues and a heavy labeled super-SILAC mix, we observe differences in their modification profiles with certain modifications unique to the super-SILAC mix. (b) Low sample complexities in affinity purification mass spectrometry experiments allow lower abundance modified peptides to be more effectively sampled. On average, across a dataset consisting of 2594 bait proteins, the number of bait PSMs identified in open search was 3.88 times that of narrow window search. (c) Open searching of a RNA-protein crosslinking dataset using MSFragger successfully identifies RNA crosslinked peptides. 134 of the 189 originally reported crosslinked peptides were recovered. Shorter crosslinked peptides are unlikely to have sufficient non-shifted fragment ions for detection in open searching and account for the majority of peptides not recovered.
Next, we applied MSFragger to a large-scale protein interaction study using an affinity purification mass spectrometry (AP-MS) experimental workflow that consisted of 2,594 baits analyzed in technical duplicates41. We reasoned that lowered sample complexity in AP-MS experiments provides an opportunity to examine in-depth the modification state of enriched proteins, most notably the proteins used as baits. We performed both narrow window and open searches on over 64.6 million MS/MS spectra across 5,188 LC-MS/MS runs (Online Methods). Open search increased the total number of PSMs by 32%, similar to the increases observed for data from whole cell lysates. For the bait proteins, however, the number of identified PSMs increased, on average, by almost 300% (Fig. 5b; Supplementary Table 5). For some bait proteins the increase in the number of identifications was astonishing. For example, the mitochondrial persulfide dioxygenase protein ETHE1 - a key member of the sulfur oxidation pathway that is itself involved in reactive oxidation of cysteine residues 42 - was identified by 48 and 2474 peptide ions in narrow window and open search, respectively (Supplementary Table 6). A significant fraction of this increase was attributed to cysteine modifications. When we subjected the top 100 bait proteins having the largest increase in the number of identified peptide ions to functional enrichment analysis using DAVID, the top enriched GO: Biological Process category (p-value 0.00007) was ‘small molecule metabolic process’ containing 23 proteins from the selected list, including ETHE1 (Supplementary Table 7). Proteins within this category are involved in catalyzing modification processes and small molecule adducts, which may be linked to significantly higher number of modifications observed on these proteins themselves. These results suggest that application of MSFragger to affinity purification experiments can provide insights into a wide array of modifications, including rare and low abundance ones, on highly enriched proteins. Furthermore, open searching may offer better accounting of protein abundances using spectral counts in AP-MS experiments and improve the quality of recovered interaction networks derived using interaction scoring tools 43, 44.
Finally, we applied MSFragger to a RNA-protein crosslinking study 45. Computational analysis for such studies can be challenging due to the need to determine a priori a list of potential crosslinked products. As open search allows for the identification of peptides with unknown modifications, no such list is required. Using a 1,000 Da precursor mass window, we performed open search on a run comprising of human UV-crosslinked RNA-protein complexes and a control non-irradiated run. We observed highly visible mass shifts associated with peptides crosslinked to mono, di, and tri-nucleotides in the irradiated sample that were largely absent from the control sample (Fig. 5c). We compared our results to that of the RNPxl computational strategy described in the original study and found that open search confidently identified 163 crosslinked species, compared to 189 reported by RNPxl, with 134 identifications in common. As expected, the open search strategy failed to identify some of the crosslinked species containing very short peptides due to an insufficient number of unmodified fragment ions (Fig. 5c inset). On the other hand, MSFragger identified 29 additional crosslinked species, most of which (all except 4) were from proteins containing other crosslinked peptides already identified by RNPxl. Furthermore, MSFragger also identified a number of modified peptides from various RNA-binding proteins (including some not identified by RNPxl) with mass shifts that approximate the RNA crosslinks (Supplementary Table 8). These peptides are likely crosslinked peptides that also contain some other chemical modification or adduct and are thus undetectable by the RNPxl strategy. Examples include the peptides YGRPPDSHHSR and SYGRPPPDVEGMTSLK from the protein SRSF2 (which was not identified by RNPxl despite identifying 5 other proteins from the SRSF family). This shows that MSFragger provides a simple but highly effective analysis workflow for identification of protein-RNA crosslinked peptides, and demonstrates the added insights gained through open searching in any experimental setup.
DISCUSSION
The vast array of chemical and biological modifications brings another dimension to proteomics that is not fully explored in most studies, in part due to additional bioinformatics challenges associated with comprehensive PTM searches. The advantage of open database searching, made practical using MSFragger, lies in its simplicity. In less time than what it currently takes to run narrow window searches, we can, in conjunction with existing workflows, simultaneously and comprehensively identify both modified and unmodified peptide forms. Given the fast growth of public repositories of MS data 46, MSFragger can be used to search for rare (including novel) biological modifications across many biological samples and experimental conditions, adding to the list of earlier successful examples of such discoveries 47. Open searching could be advantageous for characterization of neo-antigens and other endogenous peptides 48, 49, many of which are present in modified forms. Monitoring the rates of common chemical modifications, and changes in modification rates across datasets and sample preparation protocols, is important for reproducibility in quantitative proteomics experiments, especially when relying on quantification of selected peptides as proxies for estimating abundance of their corresponding protein 50. Comprehensive modification searches must be performed as part of any proteogenomics study, where confident identification of novel peptides (especially peptides containing polymorphisms) requires ruling out the possibility that their supporting spectra are due to common chemical or post-translational modifications of known peptides 36. Furthermore, considering a wide range of peptide modifications may be necessary for obtaining accurate FDR estimations using the target-decoy strategy even in narrow window searches that are only concerned with the identification of unmodified, tryptic peptides due to high scoring false positives from spectra of modified peptides. Thus, we believe that the open search strategy, made practical by MSFragger, has the potential to become a valuable option even for routine analysis of shotgun proteomics data.
ONLINE METHODS
Mass spectrometry datasets and file conversion
We used six publicly available datasets for evaluating MSFragger (see Supplementary Table 2 for the list of files and the number of MS/MS spectra in each file). All data were acquired using Thermo Scientific Q Exactive mass spectrometer (except data from the RNA-protein cross-linking study which used a Thermo Scientific LTQ-Orbitrap mass spectrometer, with MS/MS spectra acquired in the Orbitrap analyzer). Thermo .raw files were converted to either mzXML or mzML formats using the msconvert.exe tool from ProteoWizard (3.0.7398 64-bit version). Conversion was performed using vendor provided centroiding and default parameters.
MSFragger algorithm
(1) MSFragger spectra input and pre-processing
MSFragger accesses mzXML and mzML files using MSFTBX, the Data Access Library provided as part of the BatMass project 33 and Mascot Generic File (MGF) files using an internal parser. These data input paths allow MS/MS spectra stored in any of the three file formats (mzXML/mzML/MGF) to be analyzed by MSFragger. Spectra pre-processing begins with linear scaling of peak intensities so that the most intense peak within each spectrum is set to 100,000. Resultant scaled intensities are rounded and stored as integers for fast arithmetic operations. The top N peaks from each spectrum are retained and are then filtered based on the minimum intensity ratio and the m/z range specified in the search parameters file. In this study, the top 100 peaks with a minimum intensity ratio of 0.01 (relative to the base peak) were used with no m/z range filter.
(2) In-silico protein digestion and peptide indexing in MSFragger
MSFragger allows for fully enzymatic, semi-enzymatic, and non-enzymatic digestion to be specified as search parameters. It also allows for limits on missed cleavages, peptide lengths and masses to be specified. For a given protein database and a fixed set of digestion parameters, a peptide index is generated to form a necessary reference for the fragment index. Peptide indexing takes just a few minutes on a typical computer. Furthermore, MSFragger caches the peptide indices it generates on disk and attempts to find and use a compatible peptide index on subsequent invocations. As the first step of in-silico digestion, all proteins are concatenated into one long amino acid sequence with proteins separated by delimiter characters. MSFragger then partitions this long amino acid sequence into chunks for parallel in-silico digestion into peptide sequences based on the specified digestion parameters. Efficient memory allocation methods and compact representations of peptides (as offsets in the concatenated amino acid sequence and length) allow for fast in-silico digestion. The digested peptide sequences are then sorted using a parallel least significant digit radix sort and redundant peptides are flagged by comparing adjacent peptide sequences in the sorted list of peptides.
Modified versions of the digested peptide sequences are then generated based on the user-specified variable modifications. Combinatorial bitmasks that specify the positions of modified residues are pre-computed so that the set of variably modified residues can be specified as a single integer. These sequence numbers are then combinatorially combined across all variable modifications to generate a single integer that represents the variable modification state of a peptide sequence. A 12-byte entry containing the offset, length, modification sequence number, and the modified mass is generated for each such modified peptide. These modified peptides are then sorted in parallel by their modified mass forming the MSFragger peptide index.
(3) Fragment index generation
The fragment index used in MSFragger consists of all theoretical b and y-ions up to a specified charge state from each peptide in the peptide index. For efficient fragment index searching, the fragment bin width used for the fragment index must be proportional to the desired fragment tolerance specified in the search and to the expected number of candidate peptides encountered per experimental spectrum. Hence, MSFragger dynamically computes an appropriate bin width, in Daltons, that allows for efficient fragment index searching based on the user specified precursor mass tolerance and the fragment mass tolerance. Each peptide entry in the peptide index, consisting of both unmodified and variably modified peptides, can be referenced by a single 32-bit integer identification number (ID), imposing a current limit of approximately 2 billion peptide entries. Within each peptide entry, the theoretical fragments are generated and binned based on their masses using the determined bin width. The theoretical fragments are stored within the fragment index as an 8-byte entry that references the parent peptide ID, the mass offset within the bin, the charge state, and the fragment ion identity (e.g. b-5 or y-2). Fragments within each bin are stored in order of their parent IDs (and hence the parent precursor mass) as the fragment index is generated in the order of the peptide index. The memory consumption of the fragment index is modest. For a tryptic digestion (with 1 missed cleavage) of the human UniprotKB database (with reversed decoys) used in the study, the fragment index is only 1.6GB. Adding methionine oxidation and N-terminal acetylation of proteins as variable modifications boosted the index size to 2.9GB. Examples of fragment index sizes (which includes the above common variable modifications) for larger search spaces include HLA peptides (non-enzymatic digest of 9-11 amino acids; 22.6GB), semi-tryptic peptides (55.8GB) and variably phosphorylated peptides (86.5GB). MSFragger identifies the amount of memory available to it via the Java Virtual Machine and automatically partitions the fragment index generation and search into multiple iterations based on projected memory required for the fragment index, storing intermediate results on disk before merging and outputting the results in the final pass. This enables MSFragger to perform searches on computers that do not have sufficient memory to store the full fragment index, although at reduced speeds. In addition to the fragment index, MSFragger requires additional memory for storing the peptide index, spectra data, results, and intermediate data structures during search that is roughly 1GB in most use cases.
(4) Fragment index searching
In database search, the similarity scores are computed between each experimental spectrum and the theoretical spectra of all candidate peptides within a precursor mass range. These scores are heavily dependent on the number of shared fragment ions between the experimental spectrum and theoretical spectra. The major computational advance presented by MSFragger lies in its ability to rapidly identify these shared fragment ions and thus compute spectrum-spectra scores with near optimal efficiency. MSFragger first identifies the number of candidate peptides using the precursor mass window and the computed peptide index. It then allocates a scoring table for each candidate peptide where the number and summed intensities of matched b and y-ions can be stored. It then performs spectrum to spectra scoring using the fragment index in the following manner. Consider a fragment ion with mass M within an experimental spectrum with precursor mass P. Using the fragment index, the algorithm can identify the theoretical spectra that contain a fragment with a matching mass by examining the fragment bins that overlap the interval [M − dF, M + dF], where dF is the fragment mass tolerance specified in Daltons or otherwise computed from M and the specified tolerance in parts per million (Figure 1c).
For each overlapping fragment bin, a binary search (recall that the fragments within each bin are ordered by their parent precursor masses) is used to identify the fragment within the bin that corresponds to precursor mass P − dP, where dP is the precursor mass tolerance. The bin is then traversed and the theoretical fragments within the bins are compared to determine whether they truly lie within the fragment mass tolerance window, and if the theoretical fragment charge state is compatible. If a match is identified, the scores of the parent peptide (recall that each theoretical fragment contains a reference to its parent) are then incremented in the scoring table. This traversal continues until the end of the bin or upon arrival at a fragment with parent precursor mass greater than P + dP. The process is then repeated for each overlapping fragment bin. At completion, this process using a single experimental fragment ion represents the contribution of that fragment ion to all spectrum-spectra scores. This process is repeated for each experimental fragment ion (Figure 1d), in essence, decomposing many spectrum-spectrum matches into multiple fragment-spectra matches. After processing all experimental fragment ions, the scoring table of candidate peptides contains the number of matching ions (and intensities) and is used to generate a similarity score for each candidate peptide.
The efficiency of this process lies in its ability to only examine fragments with a high likelihood of contributing to the similarity score. In conventional strategies, performing a comparison between an experimental spectrum and a theoretical spectrum can take tens or hundreds of operations, even in cases where they share no common fragments. In the MSFragger strategy, theoretical spectra that share no common fragments are effectively bypassed (apart from reading a score of 0 from the scoring table) as mostly relevant fragments are compared. In the case of open window searching, approximately 1.5 comparisons are performed on average per candidate peptide and over 80% of fragment comparisons within the fragment index contribute to a similarity score (Supplementary Figure 1). This algorithmic advantage that allows MSFragger to perform so few comparisons in similarity calculations is the reason why it performs over 100 times faster than conventional search tools.
Fragment index searching in MSFragger is highly optimized. Tradeoffs between the number of bins to traverse (cost of binary searching and other overhead) and hit efficiency (percentage of fragments that fall within the fragment mass tolerance) is weighted and considered in fragment bin width selection (Supplementary Figure 1). The traversal algorithm is optimized for modern CPU cache sizes to reduce main memory accesses using a simultaneous traversal scheme for all experimental fragment ions. This allows for overall improved performance and reduces memory bottlenecks in multi-core systems.
(5) Scoring and results reporting
MSFragger computes a hyperscore similar to that of X!:
where Nb is the number of matched b-ions, Ny is the number of matched y-ions, Ib,i are the intensities of matched b-ions, and Iy,i are the intensities of matched b-ions. While the theoretical fragment index can be adapted to include other fragment ion types, only b and y ions are included and used for scoring at this time. Expectation calculation is also performed in a similar manner as X! Tandem through linear regression of the survival function to estimate the expectation of a given hyperscore 51. The top N results, as specified by the search parameters, are reported in a XML file in the pepXML format, which can then be processed using the tools from the Trans-Proteomics Pipeline (TPP) 52. For use in other computational workflows, converters exist that can convert pepXML results into other standard data output formats. Alternatively, a simple tab separated values output of the results can be obtained instead of the pepXML.
(6) Boosting unmodified peptides
MSFragger implements an unmodified peptide boosting feature. When invoked, PSMs that have an absolute value of the mass shift dM (defined as the difference between the theoretical and observed precursor peptide mass) less than the true precursor tolerance threshold specified by the search parameters ‘precursor_true_tolerance’/‘precursor_true_units’ are placed into a different scoring heap that only contains such unmodified peptides. After the calculation of expectations for all PSMs in both the regular and unmodified scoring heap, a ranking expectation is generated for all PSMs. For entries in the regular scoring heap, containing both modified and unmodified PSMs, the ranking expectation is the same as the computed expectation. The ranking expectation for entries in the unmodified peptides heap are modified based on the specified search parameters (multiplied by the specified expectation boost or an arbitrary small value for those that pass the ‘zero_bin_accept_expect’ expectation) and recorded as the ranking expectation. All PSMs are then merged and ordered by their ranking expectations prior to results reporting. It is important to note that the original expectations are reported rather than the ranking expectation.
(7) Complementary ions for the recovery of C-terminal modifications
The addition of complementary ions follows the basic spectra pre-processing described previously31, 53. The top N observed fragment ions, as specified by the ‘add_topN_complementary’ ions parameter are selected and are assumed to be either a singly charged y-ion for all spectra and a doubly charged y-ions for spectra with an identified charge state of 3+ or higher. The m/z of the complementary singly charged b-ion is then calculated from the calculated neutral mass of the assumed y-ion and the observed precursor mass. A complementary ion with this m/z and intensity equal to the y-ion from which it was derived is then inserted into the spectrum. Note that complementary ions are generated for both the singly charged and the doubly charged assumption of the observed fragment ion so that N complementary ions are inserted for spectra with charge state 2+ and 2N complementary ions are inserted for spectra with charge state 3+ or higher. These modified experimental spectra are then subjected to open database searching. As the original experimental fragment ion (from which the complementary ions are generated) is retained in the spectrum, it is possible that a single experimental observation can be incorrectly interpreted as multiple fragmentation events. Future work involving the addition of complementary ions to the theoretical spectrum instead will eliminate this problem and improve localization of modifications.
MS1-based precursor mass correction and identification based calibration
Instrument recorded precursor mass values for MS/MS spectra can be inaccurate while repeated observations of a precursor in survey (MS1) scans can be highly precise. A supplementary tool was developed as part of the MSFragger pipeline that, for each MS/MS event, takes the recorded m/z and retention time, examines the corresponding space in MS1 scans, and extracts the nearest peak feature by tracing the mass in retention time. The m/z is then calculated as a weighted average (by intensity) of all peaks in the trace. The precursor m/z for each MS/MS event is then updated with this value. For certain MS/MS events in which it was not possible to reconstruct the associated peak feature, no changes to the recorded m/z are made. Following precursor mass correction, identification-based mass recalibration of the MS/MS run is performed. In order to compare modification profiles that are resolved at sub-ppm levels across disparate experiments and labs, this calibration step is critical as slight deviations can cause broadening of features in the profile and loss of power in recovering modifications. To perform this calibration, unmodified peptide identifications with observed mass difference dM less than 20ppm are selected. As instrument bias may drift over time and varies across m/z, a two-dimensional calibration grid is constructed using a retention time width of 5 minutes and an m/z width of 200 m/z. For each unmodified peptide, the corresponding cell in the grid is found. A weighted ppm bias, based on the proximity to each point, is added to each of the four points corresponding to that cell. The weighted averages on the calibration grid are then used to adjust the precursor m/z for all observed MS/MS events in the run. The corrected and calibrated m/z values are then written to a calibration file that is incorporated in downstream analysis.
Statistical modeling of MS/MS search results and protein inference
X! Tandem, Comet, and MSFragger output files were uniformly processed by PeptideProphet 54 via the Trans-Proteomic Pipeline (TPP v4.8.0), followed by ProteinProphet 55 analysis to assemble peptides into proteins/protein groups. The results from the narrow window searches were processed using the following settings: PeptideProphet was run using ‘P’ (semi-parametric modeling), ‘d’ (report decoy hits), ‘E’ (calculation of posterior probabilities using search engine computed expectation values as primary peptide identification scores), and ‘A’ (high mass accuracy model), ‘PPM’ (use parts per million instead of Daltons in accurate mass binning), and the ProteinProphet was run using default settings. For open searches, several custom modifications were made to these downstream processing tools. First, PeptideProphet was run without ‘A’ and ‘PPM’ options, and using a mass accuracy model extended to cover the entire (−1000Da to 1000Da) range (see Extended mass model below). Second, in ProteinProphet, we did not want to incorporate modified peptides in the determination of protein groups or the establishment of protein identities. Thus, ProteinProphet was adjusted to ignore any modified peptides, while being careful to retain peptide identifications that are likely triggered from C13 isotope peaks of unmodified peptides.
Extended mass model in PeptideProphet
For open searches, the mass model of PeptideProphet was extended to effectively adjust for different likelihoods of obtaining a correct identification among unmodified peptides and peptides with different types of modifications (mass shifts). In brief, PeptideProphet models the distribution of scores observed in each data set as a mixture of two component distributions representing correct and incorrect identification, respectively. The key underlying assumption is a multivariate mixture distribution of the database search score (here, the expectation values produced by the search tools) and other parameters (most notably, the mass shift dM), which leads to the calculation of the probability of correct identification for individual peptide assignments by the Bayes rule. The mass shift parameter dM (which in the context of narrow window searches is referred to as mass accuracy) is computed for each PSM as the difference between the calculated and measured precursor peptide masses 56. Unlike narrow window searches, in open searches the range of possible dM values is extended, e.g. to cover (−1000 Da to 1000 Da) range. The dM values are discretized into bins of 1 Da in size (centered at integer values). The distributions of database search scores and dM mass shifts are modeled simultaneously, resulting in the joint probability model and computation of posterior peptide probabilities. In doing so, the mass shift dM model is estimated from the data, defining likelihoods of observing a correct vs. incorrect identification among all PSMs belonging to a particular dM bin. As the main outcome, two PSMs with identical expectation values but having different binned dM values (e.g. 0 and 135) would receive very different probability scores, reflecting the fact that the estimated fraction of correct identifications in the dM ~ 0 bin (i.e. unmodified peptides) is much higher than that among peptides with a dM value around 135 Da (rare modification). Note that while the mass model helps to account for the differences in the likelihoods of observing unmodified peptides and different modified forms, coarse single Dalton binning fails to account for the parts per million (ppm) levels of accuracy present in these data from high mass accuracy instruments, and thus the model can further benefit from future revisions.
False Discovery Rate estimation
For the benchmarking analysis of the HEK293 dataset, protein groups assembled by ProteinProphet were first filtered to remove proteins with protein probability below 0.9. The list was then filtered to 1% FDR using the maximum peptide probability as the ranking metric (maximum peptide probability was found to be more discriminative score than the protein probability in large datasets 57). PSMs, and peptides belonging to this set of filtered proteins that also passed a 1% FDR (within their respective class) were counted and their FDRs were reported. In all other analyses, protein level FDR was not estimated and filtering was performed at the peptide and PSM levels. PSMs were assembled into unique peptide sequences and the maximum PSM probability was used as the peptide probability. Peptides were then filtered at a 1% peptide FDR and PSMs that passed both a 1% peptide level FDR and a 1% PSM level FDR were retained for downstream analysis.
Benchmarking analysis using HEK293 dataset
For extensive benchmarking and comparison between MSFragger and other tools using HEK293 dataset, all spectra were searched using MSFragger, X! Tandem (Piledriver 2015.04.01.1), and Comet (2015.02 rev.1). Analysis was done using all files (24 LC-MS/MS runs, ~1.1 million spectra) for identification rate benchmarking, or one representative file for timing benchmarks (run b1906, 41820 spectra). The searched sequence database was created from the human protein sequences of Ensembl version 78 appended with reversed protein sequences as decoys and common contaminants (cRAP proteins sequences from gpmDB and contaminants from MaxQuant). All searches were done considering only y- and b- ions in scoring, allowing tryptic peptides only, up to 1 missed cleavage, and with cysteine carbamidomethylation specified as a fixed modification. Data were searched using either 100 ppm (narrow windows searches) or 500 Da (open searches) precursor mass tolerances. X! Tandem search engine used the following algorithm-specific parameters: select top 50 peaks for fragment matching, 20 ppm fragment ion mass tolerance, and requiring at least 4 matched fragment ions for a PSM to be reported. Note that X! Tandem automatically considers three additional modifications (conversion to pyroglutamate from glutamine or glutamic acid, and N-terminal acetylation). Comet searches were performed using recommended settings for high mass accuracy fragment data (precursor mass binning of 0.02 Da, 0 mass offset). MSFragger searches were performed using described parameters. To enable more accurate comparison with X! Tandem results, MSFragger searches (both narrow window and open) were also performed allowing same common modifications as those mentioned above for X! Tandem specified as variable modifications. For comparison with SEQUEST, the identification numbers (as listed in Table 1), i.e. the numbers of PSMs, unique peptide sequences, and proteins, were taken from the original publication 4.
For benchmarking the computational time (as listed in Table 1), MSFragger, Comet, and X! Tandem were also run using the single representative file referenced above on a quad core Linux workstation (Intel Xeon E3-1230v2). In addition, the data were searched using Tide (Crux version 2.1.16838), which only allows a maximum of 100Da mass tolerance and is single threaded. The run time for Tide, and for MSFragger run under the same constrains as Tide, are shown in Supplementary Table 1. For SEQUEST, the computational time listed in Table 1 was obtained by searching the data using the SEQUEST-HT version as implemented as part of the Proteome Discoverer v. 2.1 software, operated on a octa-core workstation (2x Intel Xeon E5-2609v2). The search parameters for SEQUEST-HT were as above, except the mass tolerance in the narrow window search was 5 ppm as in the original publication. All computational time benchmarking results can be found in Supplementary Table 1.
Comparison between MSFragger and MODa
MODa (v. 1.51) was run in single-blind mode with a maximum modification size of 500 Daltons and a fragment tolerance of 0.02 Daltons. Cysteine carbamidomethylation was specified as a static modification. High resolution MS/MS search was enabled. Tryptic digestion was specified with at most one missed cleavage. Both fully tryptic and semi-tryptic searches were performed using MODa. FDR filtering was performed using the “anal_moda.jar” tool bundled with the MODa tool to achieve a FDR of 1%. For comparison with MSFragger, we filtered the fully tryptic MSFragger open search results at 1% PSM FDR (without the 1% protein level filter that was used for the rest of the HEK293 benchmark comparison).
Large scale profiling of chemical modification
Large scale profiling of chemical modifications was performed using the sequence database created from the human sequences of UniprotKB (Download date: 2015-10-09) appended with reversed protein sequences as decoys and common contaminants (cRAP proteins sequences from gpmDB and contaminants from MaxQuant). A precursor mass tolerance of 500 Da was used with fragment tolerance of 20 ppm. Isotopic error correction was disabled and common variable modifications of methionine oxidation and N-terminal acetylation were enabled. Carbamidomethylation was specified as a static modification. PSMs and peptides that contain modifications that were specified in our search parameters were not considered to have a mass shift for the tabulation of mass shifts. Fully tryptic digestion was specified allowing up to 1 missed cleavage. Complementary ions and boosting features were disabled and other MSFragger options were left as default.
MSFragger search results from each LC-MS/MS run were subjected to peptide validation as described above. Peptide probability was determined by the highest supporting PSM probability. Results for each experiment were aggregated and filtered at 1% peptide FDR. PSMs were separately filtered at 1% PSM FDR and only PSMs that passed both the 1% PSM FDR and 1% peptide FDR were retained for downstream analysis.
Modeling of observed modification profiles and detection of modification peaks
Normalized density profiles for each experiment were generated for comparison across different experiments. Corrected mass differences, with random noise on the order of +/− 5 uDa added to break ties, were binned using 0.0002 Da bins to form an initial counts histogram. These counts were then distributed to adjacent bins using the weights 0.23 (bin to left), 0.49 (same bin), 0.23 (bin to right) to smooth the histogram and improve the monotonicity of peak shapes. These histograms were then normalized by dividing each bin by the total number of spectra (in millions) acquired in the respective experiments. Averaging the counts in each bin generated an average profile of the three experiments. Mixture modeling of the average profile failed to precisely capture known modifications. Examination of the profile revealed peaks of varying broadness and further examination revealed the peak shape to be a complex function of the charge state and m/z of the underlying PSMs. Instead, a prominence based peak detection method was used that found features on the histogram by requiring that the peak prominence was at least 0.3 times that of the peak height. As known modifications were observed to have a peak width of approximately 0.004 Da (given current instrument accuracies and the correction/calibration method applied as described above), these features were ordered by the rise in density compared to the 0.003 Da flanking regions. It should be noted that some of the detected features (mass bins) could be artifacts of the peak picking algorithm, or may correspond to various combinations of multiple modifications.
Mass shift annotation using Unimod
The Unimod repository was downloaded (on 2016-04-22) in XML format and was parsed to extract modification names and mass shifts. Mass shifts associated with the addition or deletions of the twenty amino acids were appended to this list. Multiples of the mass difference between carbon-13 and carbon-12 were added as ‘First isotopic peak’ and ‘Second isotopic peak’ to account for isotopic peak picking errors. Entries that represent a single mass shift in this list were concatenated into a single entry so that a single text identifier represented each mass shift. Annotation of the list of mass shifts proceeded in decreasing order of abundance. For each mass shift, the mass is queried against the described database of annotations with a mass tolerance of 0.002 Da. If a match is found, the mass shift is annotated with the entry from the database. If the mass shift cannot be matched to a single entry in the database, we attempt to compose multiple (up to 3) previously observed (in the order of annotation) mass shifts to account for compound modifications. If the mass shift remains unexplained, we add it to our list of annotations as a new un-annotated mass shift.
Localization of detected mass differences
For each PSM, including unmodified peptides, the observed mass difference is evaluated to see if it can be attributable to a modification of a specific site (position in the peptide). For each MS/MS run, the list of identified spectra (which includes the spectrum ID, peptide sequence, list of variably modified amino acids, and observed mass difference) is obtained from the MSFragger analysis pipeline, and the corresponding MS/MS spectra are extracted from the original mass spectrometry data file. The number of matched fragment ions is then re-computed using the same hyperscore function as originally done in MSFragger. The observed mass difference is iteratively placed on each amino acid, and for each position the spectrum similarity is computed to derive the number of matching fragment ions, and then the hyperscore. A PSM is called localizable if there is at least one position that generates a higher number of matched fragments than the rest. As there may be insufficient fragments to support an unambiguous localization in the peptide sequence, all positions that share the highest hyperscore are marked as a possible localization site. A PSM is called to be localized to the N-terminal if the localized positions form an uninterrupted stretch of amino acids from the N-terminal.
The localization results are then aggregated for each identified mass bin, and their localization characteristics examined. For each bin, the overall localization rate (the percentage of PSMs within that bin that are localizable), the N-terminal localization rate (the percentage of PSMs within that bin that are localizable and the localization is N-terminal), and the amino acid enrichment are computed. The amino acid enrichment is determined by first computing the amino acid composition of all peptides within the mass bin. Then, the number of localization sites attributable to each amino acid is summed across all localizable PSMs (for a PSM with multiple localization sites, each site gains a weight equal to 1/number of localized sites). The total localization count for each amino acid is then normalized to form the localization rate. Amino acid enrichment is then determined by the ratio of localization rate to composition rate. It should be noted that while this metric is informative in many cases, it may be misleading in bins containing few PSMs or bins that are dominated by several abundant peptides that skews the counts and normalization factors.
Spectral similarity scores for modifications
For each modified PSM, we identify corresponding PSMs of the same charge state that identifies the same peptide but with a mass difference of less than 0.001 Da (indicating an unmodified peptide). We compute the average cosine similarity between the spectrum of the modified PSM and spectra corresponding to the unmodified peptide (if there are more than 50 such spectra, 50 are chosen at random). We then normalize for variations within unmodified spectra by dividing the average cosine similarity within the set of unmodified spectra to obtain a similarity score for the modified PSM. For each modification mass, its similarity score is determined by averaging the similarity scores calculated for each modified PSM within its mass tolerance.
Analysis of SILAC datasets
The breast cancer SILAC dataset was analyzed using the same search settings as the large-scale modification profiling described above with the exception that two variable modifications were added for the heavy labeled residues: 8.0142 Da at lysine and 10.00827 Da at arginine. Precursor mass correction/calibration and peptide validation were performed on each file and the aggregated files from the experiment were subjected to a 1% peptide and PSM FDR filter (each retained PSM passed 1% PSM FDR and matched a peptide that passed 1% peptide FDR). Each PSM in the resultant list was then examined for the presence of a heavy labeled residue (as determined by identification with a heavy labeled variable modification). Unlabeled PSMs were considered to have originated from the patient samples while labeled PSMs were considered to have originated from the super-SILAC mix.
Analysis of AP-MS dataset
Open search parameters for the AP-MS dataset were also similar to the settings used for large-scale modification profiling with one exception. As iodoacetamide treatment of samples was not used, no static modification was specified for cysteine. Each of the 5,188 runs was subjected to peptide validation and mass correction individually. FDR filtering was performed for each run individually, filtering the data at 1% FDR (at both peptide and PSM levels). Narrow window searches were performed using the same parameters with the exception of a 20 ppm precursor tolerance window and isotope selection errors of 0/1/2 was enabled.
For each LC-MS/MS run, all PSMs that were matched to a UniProt accession associated with the bait protein were considered to have originated from the bait protein (including any shared peptides). The number of unique sequences was determined by examining the set of unique peptides represented by the PSMs. Total counts for a particular bait protein across the replicates were determined by summing bait PSMs across the two replicates and determining the number of unique peptide sequences. Average fold change between narrow window and open searches was determined by linear regression in R.
Analysis of RNA-protein crosslink dataset
Open searching for the crosslinking dataset was performed similar to the large-scale modification profiling searches. The precursor mass window was enlarged to +/− 1000 Da to accommodate heavier crosslinked fragments. Carbamidomethylation was not specified as a fixed modification on cysteine. Comparison of results obtained by RNPxl and MSFragger was performed using the peptide sequence and mass difference. Identifications from RNPxl were translated into a peptide sequence and a total RNA-peptide mass. An identification from MSFragger was considered to be a match if it shared the same peptide sequence and had a total mass that differed from the RNPxl-based identification by no more than 0.05 Da.
Code Availability
MSFragger was developed in the cross-platform Java language and can be accessed at www.nesvilab.org/software. A protocol for using MSFragger to perform database search can be found at http://dx.doi.org/xx.xxxx/protex.yyyy.xxx58.
Data Availability
All raw files are available as described in Supplementary Table 2. Processed data files that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank R. Beavis for helpful discussions, N. Bandeira and S. Na for help with MODa software, and E. Huttlin for assisting with the transfer of raw mass spectrometry data from the AP-MS study. This work was funded in part by grants from the NIH (R01GM94231 and U24CA210967 to A.I.N.).
ABBREVIATIONS
- LC
Liquid chromatography
- MS
Mass spectrometry
- MS/MS
Tandem mass spectrometry
- PSM
Peptide to spectrum match
- PTM
Post-translational modification
- FDR
False Discovery Rate
- TNBC
Triple negative breast cancers
- SILAC
Stable isotope labeling with amino acids in cell culture
- FFPE
Formalin-fixed paraffin-embedded
- AP-MS
Affinity Purification followed by Mass Spectrometry
Footnotes
AUTHOR CONTRIBUTIONS
A.T.K. and A.I.N. conceived the project. A.T.K. developed the algorithm, wrote the software, and analyzed the results. A.I.N. assisted with the algorithm development and software design, analyzed the results, and supervised the entire project; F.V.L., D.M.A., and D.M. contributed to software development and data analysis; A.T.K. and A.I.N. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. Journal of Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eng JK, Searle BC, Clauser KR, Tabb DL. A Face in the Crowd: Recognizing Peptides Through Database Search. Molecular & Cellular Proteomics: MCP. 2011;10:R111.009522. doi: 10.1074/mcp.R111.009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skinner OS, Kelleher NL. Illuminating the dark matter of shotgun proteomics. Nat Biotech. 2015;33:717–718. doi: 10.1038/nbt.3287. [DOI] [PubMed] [Google Scholar]
- 4.Chick JM, et al. A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat Biotech. 2015;33:743–749. doi: 10.1038/nbt.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griss J, et al. Recognizing millions of consistently unidentified spectra across hundreds of shotgun proteomics datasets. Nat Meth. 2016;13:651–656. doi: 10.1038/nmeth.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nesvizhskii AI, et al. Dynamic Spectrum Quality Assessment and Iterative Computational Analysis of Shotgun Proteomic Data: Toward More Efficient Identification of Post-translational Modifications, Sequence Polymorphisms, and Novel Peptides. Molecular & Cellular Proteomics. 2006;5:652–670. doi: 10.1074/mcp.M500319-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen ML, Savitski MM, Zubarev RA. Extent of Modifications in Human Proteome Samples and Their Effect on Dynamic Range of Analysis in Shotgun Proteomics. Molecular & Cellular Proteomics. 2006;5:2384–2391. doi: 10.1074/mcp.M600248-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Ning K, Fermin D, Nesvizhskii AI. Computational analysis of unassigned high-quality MS/MS spectra in proteomic data sets. PROTEOMICS. 2010;10:2712–2718. doi: 10.1002/pmic.200900473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 10.Creasy DM, Cottrell JS. Error tolerant searching of uninterpreted tandem mass spectrometry data. Proteomics. 2002;2:1426–1434. doi: 10.1002/1615-9861(200210)2:10<1426::AID-PROT1426>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 11.Shortreed MR, et al. Global Identification of Protein Post-translational Modifications in a Single-Pass Database Search. Journal of proteome research. 2015;14:4714–4720. doi: 10.1021/acs.jproteome.5b00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahrne E, Nikitin F, Lisacek F, Muller M. QuickMod: A Tool for Open Modification Spectrum Library Searches. Journal of proteome research. 2011;10:2913–2921. doi: 10.1021/pr200152g. [DOI] [PubMed] [Google Scholar]
- 13.Bandeira N, Tsur D, Frank A, Pevzner PA. Protein identification by spectral networks analysis. Proceedings of the National Academy of Sciences. 2007;104:6140–6145. doi: 10.1073/pnas.0701130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savitski MM, Nielsen ML, Zubarev RA. ModifiComb, a New Proteomic Tool for Mapping Substoichiometric Post-translational Modifications, Finding Novel Types of Modifications, and Fingerprinting Complex Protein Mixtures. Molecular & Cellular Proteomics. 2006;5:935–948. doi: 10.1074/mcp.T500034-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Ma CW, Lam H. Hunting for unexpected post-translational modifications by spectral library searching with tier-wise scoring. Journal of proteome research. 2014;13:2262–2271. doi: 10.1021/pr401006g. [DOI] [PubMed] [Google Scholar]
- 16.Tabb DL, Saraf A, Yates JR., 3rd GutenTag: high-throughput sequence tagging via an empirically derived fragmentation model. Analytical chemistry. 2003;75:6415–6421. doi: 10.1021/ac0347462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bern M, Cai Y, Goldberg D. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Analytical chemistry. 2007;79:1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- 18.Dasari S, et al. Sequence Tagging Reveals Unexpected Modifications in Toxicoproteomics. Chemical Research in Toxicology. 2011;24:204–216. doi: 10.1021/tx100275t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Na S, Bandeira N, Paek E. Fast multi-blind modification search through tandem mass spectrometry. Mol Cell Proteomics. 2012;11:M111.010199. doi: 10.1074/mcp.M111.010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Searle BC, et al. Identification of Protein Modifications Using MS/MS de Novo Sequencing and the OpenSea Alignment Algorithm. Journal of proteome research. 2005;4:546–554. doi: 10.1021/pr049781j. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Chen W, Cobb MH, Zhao Y. PTMap--a sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:761–766. doi: 10.1073/pnas.0811739106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanner S, Pevzner PA, Bafna V. Unrestrictive identification of post-translational modifications through peptide mass spectrometry. Nature Protocols. 2006;1:67–72. doi: 10.1038/nprot.2006.10. [DOI] [PubMed] [Google Scholar]
- 23.Fu Y. In: Statistical Analysis in Proteomics. Jung K, editor. Springer New York; New York, NY: 2016. pp. 265–275. [Google Scholar]
- 24.Chi H, et al. pFind-Alioth: A novel unrestricted database search algorithm to improve the interpretation of high-resolution MS/MS data. Journal of Proteomics. 2015;125:89–97. doi: 10.1016/j.jprot.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Cox J, et al. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. Journal of proteome research. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 26.McIlwain S, et al. Crux: Rapid Open Source Protein Tandem Mass Spectrometry Analysis. Journal of proteome research. 2014;13:4488–4491. doi: 10.1021/pr500741y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eng JK, Jahan TA, Hoopmann MR. Comet: An open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 28.Fu Y, Qian X. Transferred Subgroup False Discovery Rate for Rare Post-translational Modifications Detected by Mass Spectrometry. Molecular & Cellular Proteomics. 2014;13:1359–1368. doi: 10.1074/mcp.O113.030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaudel M, et al. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat Biotech. 2015;33:22–24. doi: 10.1038/nbt.3109. [DOI] [PubMed] [Google Scholar]
- 30.Diament BJ, Noble WS. Faster SEQUEST Searching for Peptide Identification from Tandem Mass Spectra. Journal of proteome research. 2011;10:3871–3879. doi: 10.1021/pr101196n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsou CC, et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat Meth. 2015;12:258–264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houel S, et al. Quantifying the impact of chimera MS/MS spectra on peptide identification in large-scale proteomics studies. Journal of proteome research. 2010;9:4152–4160. doi: 10.1021/pr1003856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avtonomov DM, Raskind A, Nesvizhskii AI. BatMass: a Java Software Platform for LC–MS Data Visualization in Proteomics and Metabolomics. Journal of proteome research. 2016;15:2500–2509. doi: 10.1021/acs.jproteome.6b00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B, Pirmoradian M, Chernobrovkin A, Zubarev RA. DeMix workflow for efficient identification of cofragmented peptides in high resolution data-dependent tandem mass spectrometry. Mol Cell Proteomics. 2014;13:3211–3223. doi: 10.1074/mcp.O114.038877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bogdanow B, Zauber H, Selbach M. Systematic Errors in Peptide and Protein Identification and Quantification by Modified Peptides. Mol Cell Proteomics. 2016;15:2791–2801. doi: 10.1074/mcp.M115.055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nesvizhskii AI. Proteogenomics: concepts, applications and computational strategies. Nat Meth. 2014;11:1114–1125. doi: 10.1038/nmeth.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma K, et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell reports. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 38.Lawrence RT, et al. The proteomic landscape of triple-negative breast cancer. Cell reports. 2015;11:630–644. doi: 10.1016/j.celrep.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pozniak Y, et al. System-wide Clinical Proteomics of Breast Cancer Reveals Global Remodeling of Tissue Homeostasis. Cell systems. 2016;2:172–184. doi: 10.1016/j.cels.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Metz B, et al. Identification of Formaldehyde-induced Modifications in Proteins: REACTIONS WITH MODEL PEPTIDES. Journal of Biological Chemistry. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- 41.Huttlin EL, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabil O, Banerjee R. Enzymology of H2S biogenesis, decay and signaling. Antioxidants & redox signaling. 2014;20:770–782. doi: 10.1089/ars.2013.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi H, et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Meth. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sardiu ME, Washburn MP. Construction of protein interaction networks based on the label-free quantitative proteomics. Methods in molecular biology (Clifton, NJ) 2011;781:71–85. doi: 10.1007/978-1-61779-276-2_5. [DOI] [PubMed] [Google Scholar]
- 45.Kramer K, et al. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat Meth. 2014;11:1064–1070. doi: 10.1038/nmeth.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez-Riverol Y, Alpi E, Wang R, Hermjakob H, Vizcaino JA. Making proteomics data accessible and reusable: current state of proteomics databases and repositories. Proteomics. 2015;15:930–949. doi: 10.1002/pmic.201400302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 49.Mommen GPM, et al. Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD) Proceedings of the National Academy of Sciences. 2014;111:4507–4512. doi: 10.1073/pnas.1321458111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van den Broek I, et al. Quantifying protein measurands by peptide measurements: where do errors arise? Journal of proteome research. 2015;14:928–942. doi: 10.1021/pr5011179. [DOI] [PubMed] [Google Scholar]
- 51.Fenyo D, Beavis RC. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Analytical chemistry. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 52.Deutsch EW, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kryuchkov F, Verano-Braga T, Hansen TA, Sprenger RR, Kjeldsen F. Deconvolution of mixture spectra and increased throughput of peptide identification by utilization of intensified complementary ions formed in tandem mass spectrometry. Journal of proteome research. 2013;12:3362–3371. doi: 10.1021/pr400210m. [DOI] [PubMed] [Google Scholar]
- 54.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 55.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 56.Choi H, Ghosh D, Nesvizhskii AI. Statistical validation of peptide identifications in large-scale proteomics using the target-decoy database search strategy and flexible mixture modeling. Journal of proteome research. 2008;7:286–292. doi: 10.1021/pr7006818. [DOI] [PubMed] [Google Scholar]
- 57.Shanmugam AK, Yocum AK, Nesvizhskii AI. Utility of RNA-seq and GPMDB protein observation frequency for improving the sensitivity of protein identification by tandem MS. Journal of proteome research. 2014;13:4113–4119. doi: 10.1021/pr500496p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, Nesvizhskii AI. Using MSFragger for ultrafast database searching. Protocol Exchange. 2017 doi: 10.1038/nmeth.4256. XX.XXXX/protex.YYYY.XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw files are available as described in Supplementary Table 2. Processed data files that support the findings of this study are available from the corresponding author upon request.