Abstract
Repression of repetitive elements is crucial to preserve genome integrity and has been traditionally ascribed to constitutive heterochromatin pathways. FacioScapuloHumeral Muscular Dystrophy (FSHD), one of the most common myopathies, is characterized by a complex interplay of genetic and epigenetic events. The main FSHD form is linked to a reduced copy number of the D4Z4 macrosatellite repeat on 4q35, causing loss of silencing and aberrant expression of the D4Z4-embedded DUX4 gene leading to disease. By an unknown mechanism, D4Z4 copy-number correlates with FSHD phenotype. Here we show that the DUX4 proximal promoter (DUX4p) is sufficient to nucleate the enrichment of both constitutive and facultative heterochromatin components and to mediate a copy-number dependent gene silencing. We found that both the CpG/GC dense DNA content and the repetitive nature of DUX4p arrays are important for their repressive ability. We showed that DUX4p mediates a copy number-dependent Polycomb Repressive Complex 1 (PRC1) recruitment, which is responsible for the copy-number dependent gene repression. Overall, we directly link genetic and epigenetic defects in FSHD by proposing a novel molecular explanation for the copy number-dependency in FSHD pathogenesis, and offer insight into the molecular functions of repeats in chromatin regulation.
Introduction
Despite repetitive elements representing the largest portion of the human genome (1–4), they are poorly characterized because of their intrinsic complexity. Nevertheless, increasing evidence suggests that DNA repeats play key roles in the regulation of gene expression at multiple levels (5–9). For example, mammalian genomic repeats have been shown to harbor the greatest proportion of histone modifications (1,10,11) and to provide binding sites for regulatory factors modulating the activity of tissue-specific promoters or enhancers (6,12–18).
Tandem repeats represent more than 20% of the human genome (1) and account for a significant source of genomic variation, since their copy-number is usually highly polymorphic among individuals (1,2,19,20). They can span from a few base pairs in microsatellites to several kilobases in macrosatellites (2,21,22), and they play crucial roles in key cellular processes. With centromeric and telomeric satellites, tandem repeats provide an essential contribution to genome integrity (23–27) and chromosome segregation (28). In addition, tandem repeats play a role in heterochromatin formation (27), X-chromosome inactivation (XCI) (14,29,30), but also in the evolution and generation of genetic diversity (31–33). Finally, with their intrinsic instability, they can contribute to chromosome rearrangements and diseases (34–36). Accordingly, to preserve genome integrity (11,37,38), tandem repeats can be targeted by multiple repressive pathways leading to the formation of constitutive or facultative heterochromatin (39). This unveiled an unexpected cross-talk between two apparently distinct mechanisms of repression, which can coordinate to regulate fundamental processes (40).
FacioScapuloHumeral muscular Dystrophy (FSHD) (MIM 158900) is one of the most prevalent neuromuscular disorders (41). In its major form, accounting for 95% of cases, the disease is linked to deletions reducing the copy-number of a macrosatellite repeat called D4Z4, located in 4q35 (42). While healthy subjects display 11-100 D4Z4 units, FSHD patients usually present with only 1-10 D4Z4 units (43–45). Interestingly, the residual D4Z4 copy number at the deleted 4q35 allele correlates with disease onset and progression. FSHD patients carrying 1-3 residual D4Z4 units tend to develop the disease earlier and with a more severe outcome than patients displaying 9-10 units (46–53).
Several epigenetic alterations have been described on the affected FSHD locus, including DNA hypomethylation (54) and reduction of heterochromatic histone marks H3K9me3 and H3K27me3 (tri-methylation of histone H3 Lysine 9 and Lysine 27) (55–58). Ultimately, this leads to aberrant expression at 4q35 (58–63) and in particular to the toxic overexpression of the D4Z4-embedded DUX4 retrogene, which is considered the major gene responsible for the disease (reviewed in (64,65)). Nevertheless, the molecular bases for the FSHD copy-number dependency are poorly known.
Given the importance of DUX4 in disease, we developed a reporter system to monitor the influence of repeat copy-number on gene expression driven by the DUX4 promoter. We found that high GC content and repeat copy number lead to efficient gene repression. We analyzed the main epigenetic pathways playing a role in FSHD and found that the Polycomb Repressive Complex 1 (PRC1) plays a major role in the repeat copy-number dependent regulation of gene expression. Our results provide a novel molecular explanation for the peculiar link between genetic and epigenetic alterations taking place in one of the most important neuromuscular disease.
Results
DUX4p mediates a copy-number dependent repression
The molecular characterisation of FSHD muscular dystrophy has revealed a highly complex interaction between genetic and epigenetic components (65). One of the prominent aspects of this disease is that patients carrying few copies of D4Z4 (1–3) tend to develop a more severe form of the disease and with earlier onset than patients with 8-10 residual copies (46–53,66). Consistent with this, the aberrant expression of DUX4, the main FSHD gene, inversely correlates with repeat copy number (67). In order to explore the molecular mechanism at the basis of the copy-number dependency in DUX4 gene regulation, we generated three different reporter gene constructs carrying an EGFP reporter gene under the control of the DUX4 proximal promoter (DUX4p) and displaying a copy number typically found either in healthy subjects (12), or in mild (5) and severe (1) FSHD forms (Supplementary Material, Fig. S1A–F). We focused on a 180 bp D4Z4 fragment containing the regulatory elements of the DUX4 proximal promoter (61) (Supplementary Material, Fig. S1G). To evaluate the effect of DUX4p copy-number on gene expression, the terminal DUX4p unit was prolonged at the 3’ end until the ATG of DUX4 ORF (68), driving the expression of the EGFP reporter (Supplementary Material, Fig. S1G).
Initially, we tested the constructs in transient transfection (Supplementary Material, Fig. S2, ). To this aim, we transfected equimolar quantities of the DUX4p constructs together with a plasmid constitutively expressing ECFP, as transfection control. The median EGFP fluorescence intensity (Supplementary Material, Fig. S2D) and the percentages of EGFP positive cells (Supplementary Material, Fig. S2E) were analysed in the total population of ECFP positive cells through flow cytometry and expressed as relative to 1-DUX4p. Intriguingly, we observed a copy-number dependent effect of DUX4p repeats on both EGFP intensity and percentage of positive cells (Supplementary Material, Fig. S2). In particular, cells transfected with the 12-DUX4p construct showed a significantly reduced relative EGFP intensity, when compared to both 1-DUX4p and 5-DUX4p constructs. Interestingly, the EGFP signal from the 5-DUX4p repeat construct stands on an intermediate level, significantly higher and significantly lower than those of 12-DUX4p and 1-DUX4p copies, respectively (Supplementary Material, Fig. S2D). Similarly, the number of DUX4p units has an effect on the relative number of EGFP expressing cells, since a significantly smaller percentage of EGFP positive cells were present in the 12-DUX4p and 5-DUX4p populations when compared to 1-DUX4p. Again, the effect increases as a function of the number of DUX4p units (Supplementary Material, Fig. S2E). Overall, an increase in the DUX4p copy-number correlates with a decreased reporter gene signal.
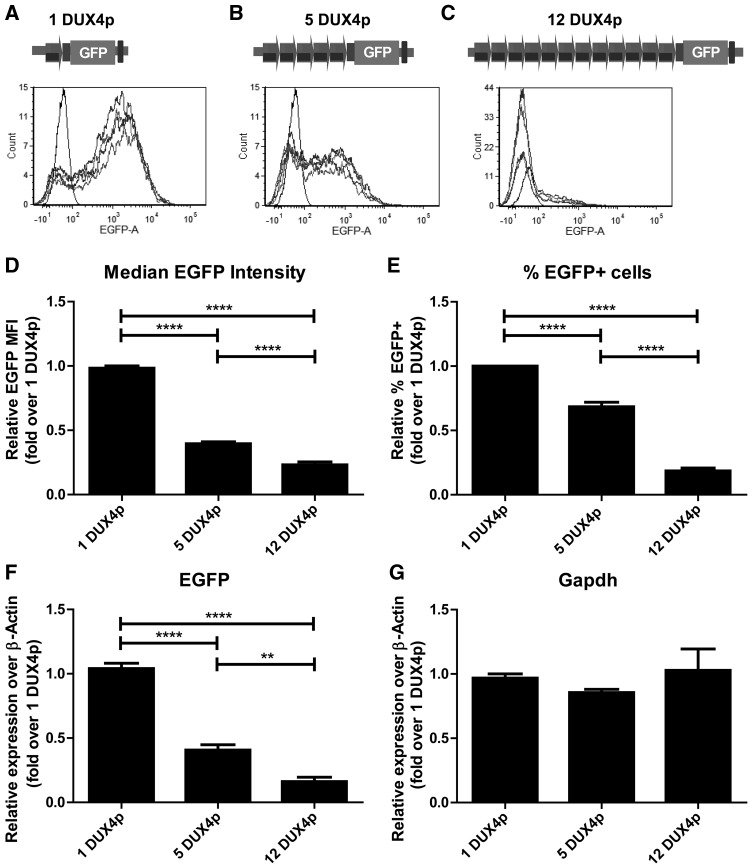
We decided to investigate the influence of repeat copy-number on the DUX4 promoter activity on a chromatinized template. Since in human samples the selective study of D4Z4 sequences is challenged by the presence of multiple copies of D4Z4 and D4Z4-like repeats, scattered in several human chromosomes (69–71), we exploited the fact that D4Z4 is a primate-specific repeat (72) and performed site-specific integration of DUX4p reporters within the same genomic locus of C2C12 murine muscle cells. Notably, DUX4 promoter activity in transgenic C2C12 or human muscle cells are equivalent (73). As a result, we generated isogenic cell lines varying only for the copy-number of DUX4p (Fig. 1A–C). As shown in Fig. 1, a DUX4p copy-number dependent repression of the reporter gene was observed. In particular, cells retaining 12 DUX4p units show a significant reduction in both relative median EGFP signal (Fig. 1D) and relative percentage of EGFP expressing cells in the total population (Fig. 1E), when compared to cells with 1 or 5 DUX4p units. Similarly, cells containing 5 DUX4p copies show significantly higher and significantly lower levels of both parameters compared to 12-DUX4p and 1-DUX4p cells, respectively (Fig. 1D and E). Finally, we investigated EGFP expression at the RNA level by RT-qPCR (Fig. 1F). In line with the flow cytometry analyses, we observed that higher levels of reporter gene transcription correlate with fewer DUX4p-copies, while Gapdh mRNA levels, used as control, remain unchanged (Fig. 1F and G). Altogether, these results show a copy-number dependent repressive activity on DUX4 promoter that is particularly evident for 12 DUX4p repeats, in line with the threshold copy number characteristic of FSHD pathogenesis (43–45).
Figure 1.
DUX4p repeats mediate copy-number dependent repression in isogenic muscle cells. A–C. Top, schematic view of constructs used for Flp-FRT-mediated recombination and carrying 1 (A), 5 (B), or 12 (C) DUX4p repeats and an EGFP reporter gene under the control of the DUX4 promoter. Upon site-specific integration in C2C12-frt muscle cells, the isogenic cells differing for DUX4p copy number (A–C) were tested for EGFP expression. For relative quantification of EGFP protein levels among samples, flow cytometry analyses of EGFP Median Fluorescence Intensity (MFI) (D) and of percentage of EGFP+ events (E) are shown. Examples of histogram analyses with EGFP (x axis) versus counts (y axis) are reported (A–C, bottom), compared to a non-fluorescent (wild type) cell line. For relative quantification of EGFP at the RNA level, RT-qPCR analyses of EGFP (F) and Gapdh negative control (G) are presented over β-Actin. Results are expressed over 1 DUX4p (D–G). The mean of the signals obtained from six (D,E) or four (F,G) independent experiments is shown. The error bars represent SEM of independent biological replicates. Asterisks indicate statistical significance (p value) as evaluated by One way ANOVA Bonferroni’s multiple comparison test. ****=P < 0.0001; **= P < 0.01.
A high CpG/GC density and the repetitive nature contribute to the repressive function of DUX4p
Analogously to other non-centromeric macrosatellites (74–76), D4Z4 is GC-rich (73%), as contrasted with the average genome frequency of 42% (77). Moreover, D4Z4 displays 10% CpG dinucleotide frequency, compared to the human average frequency of 1% (78). Similarly, each DUX4p unit displays 72% GC and 10 CpGs. To dissect the bases of their repressive activity, we wondered whether the high CpG/GC DNA content or the repeated nature of DUX4p arrays could play a role in DUX4p-mediated repression. To address whether a DNA sequence with CpG/GC features similar to 12-DUX4p but lacking repeats would show the same ability to influence gene expression, we generated a new isogenic muscle cell line in which 11 DUX4p units of the 12-DUX4p construct are replaced with a non-repetitive sequence from the B3gnt6 locus, with identical size, identical CpG content and similar GC richness (CGI B3gnt6; Fig. 2A and B). Importantly, this sequence is a known target of epigenetic repressors such as PRC1 and PRC2 complexes (79). In parallel, to investigate the role of repeats uncoupled from the CpG content, we generated a second group of isogenic muscle cells displaying variable copy number of repeats derived from the human β-Globin (HbG) locus and having 0 CpG and low GC content but identical size compared to each DUX4p unit (1-, 5- or 11-HbG; Fig. 2C). Like for the DUX4p cells, in all these new cell lines the EGFP reporter gene is driven by the DUX4 promoter (Supplementary Material, Fig. S1G).
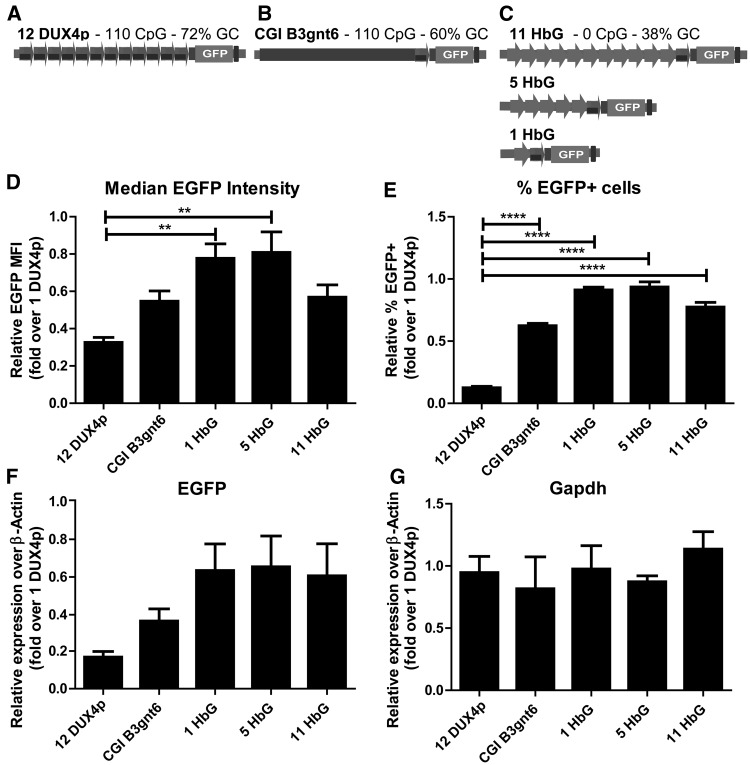
Figure 2.
Contribution of CpG/GC-rich and repetitive nature to copy-number dependent repression. A-C. Schematic view of constructs named: 12 DUX4p (A), CGI B3gnt6 (B), 11-, 5-, 1-HbG (C). The constructs carry: 11 copies of DUX4p repeats (overall, 110 CpG and 72% GC, A); a non-repetitive sequence containing a CpG island (CGI) from B3gnt6 locus (110 CpG and 60% GC, B); 11, 5 or 1 copies of HbG repeats (each with 0 CpG and 38% GC, C). Additionally, all constructs retain a DUX4p unit prolonged to include DUX4 ATG sequence, and an EGFP reporter gene (A-C). Upon site-specific integration of the constructs in C2C12-frt muscle cells, the isogenic cells (A–C) were tested for EGFP expression. For relative quantification of EGFP protein levels among samples, flow cytometry analyses of EGFP Median Fluorescence Intensity (MFI) (D) and of percentage of EGFP+ events (E) are shown. For relative quantification of EGFP at the RNA level, RT-qPCR analyses of EGFP (F) and Gapdh negative control (G) are presented over β-Actin. Results are expressed over 1 DUX4p (D-G). The mean of the signals obtained from five (D,E) or four (F,G) independent experiments is shown. The error bars represent SEM of independent biological replicates. Asterisks indicate statistical significance (p value) as evaluated by One way ANOVA Bonferroni’s multiple comparison test. ****= P < 0.0001; **= P < 0.01.
Significantly, flow cytometry analyses expressed as fold over 1-DUX4p showed that 12-DUX4p retains the strongest ability to silence EGFP expression (Fig. 2), since the 12-DUX4p cells display the least intense median EGFP signal (Fig. 2D). Moreover, 12-DUX4p showed a significantly lower percentage of EGFP positive cells compared to CGI B3gnt6 and 1-5-11 HbG (Fig. 2E). Similar results were observed at RNA level, since EGFP transcript levels were lower in 12-DUX4p than in all other cells lines (Fig. 2F), while mRNA levels of the Gapdh control remained constant in all the samples (Fig. 2G). Overall, these results indicate that a high CpG/GC DNA content combined with the presence of repeats results in the strongest silencing effect, suggesting that both features are important in the repression of DUX4.
DNA methylation and H3K9me3 constitutive heterochromatin marks are not copy-number dependent
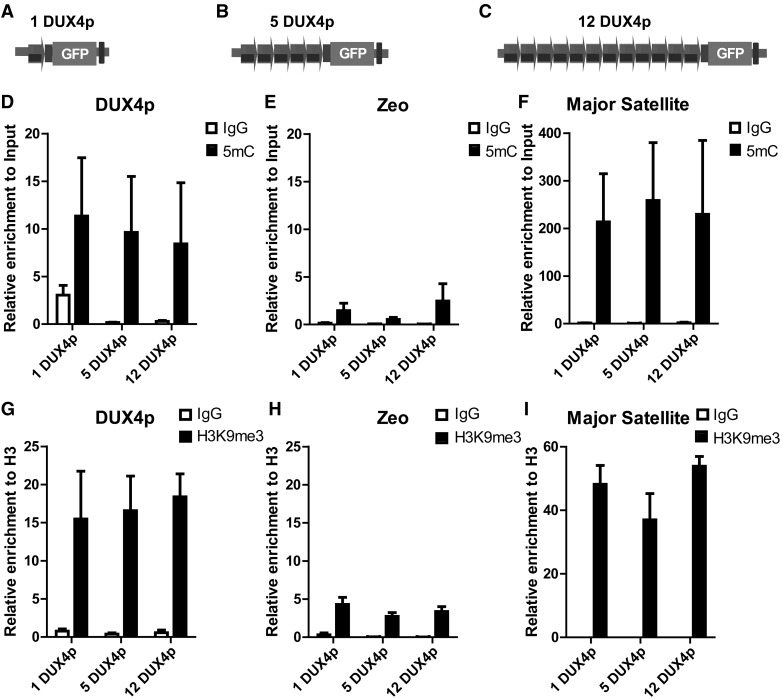
The constitutive heterochromatin marks DNA methylation (80– 82) and H3K9me3 (55) have been associated with repression of the FSHD locus. To determine whether these features could explain the copy-number dependent gene silencing effect of DUX4p arrays, we performed Methyl DNA Immunoprecipitation (MeDIP) analysis of 5-methyl-Cytosine (5mC) and Chromatin Immunoprecipitation (ChIP) of H3K9me3. As shown in Fig. 3, DNA methylation and H3K9me3 were higher on DUX4p than a negative control region with identical CpG density (Zeocine, or Zeo, present in C2C12-frt cells close to the FRT integration site) and equivalent to those observed on the endogenous FSHD locus in human cells (83,84). Nevertheless, DNA methylation or H3K9me3 enrichments were comparable between 1-, 5- and 12-DUX4p cells. Although DNA methylation levels are different between FSHD and healthy subjects (67) and a correlation with total D4Z4 repeat number has been reported in muscle cells from FSHD patients (85) (see discussion), we failed to observe a correlation between DNA methylation and repression of our reporter. Overall, with the limitations of our system, it appears that constitutive heterochromatin marks, although enriched at DUX4p arrays, do not play a major role in copy-number dependent DUX4 reporter gene silencing.
Figure 3.
DNA methylation and histone H3K9me3 enrichment does not correlate with DUX4p copy number. A-C. Schematic view of isogenic muscle cells carrying 1 (A), 5 (B), or 12 (C) DUX4p repeats and an EGFP reporter gene, used for MeDIP and ChIP analyses. D-I. Isogenic muscle cells differing for DUX4p copy-number were analyzed by MeDIP for DNA methylation (D-F) and by ChIP for H3K9me3 (G-I), on DUX4p (D, G), on negative (E, H) or positive (F, I) control regions, respectively, Zeocine gene (E, H) and Major Satellite repeats (F, I). ChIP-qPCR data are shown relative to total H3. The mean of the signals obtained from three independent experiments is shown (D-F, G-I). The error bars represent SEM of independent biological replicates.
DUX4p drives copy-number dependent PRC1 recruitment
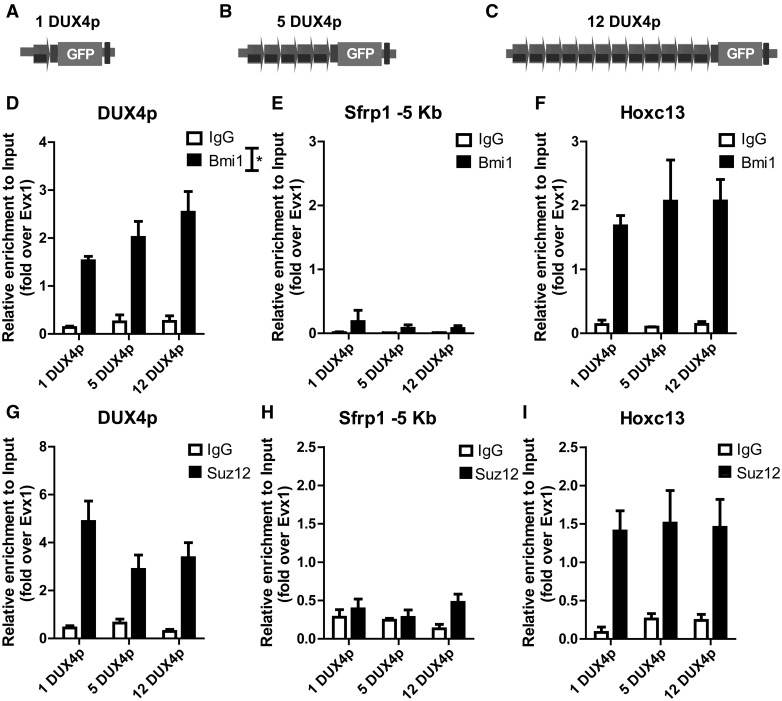
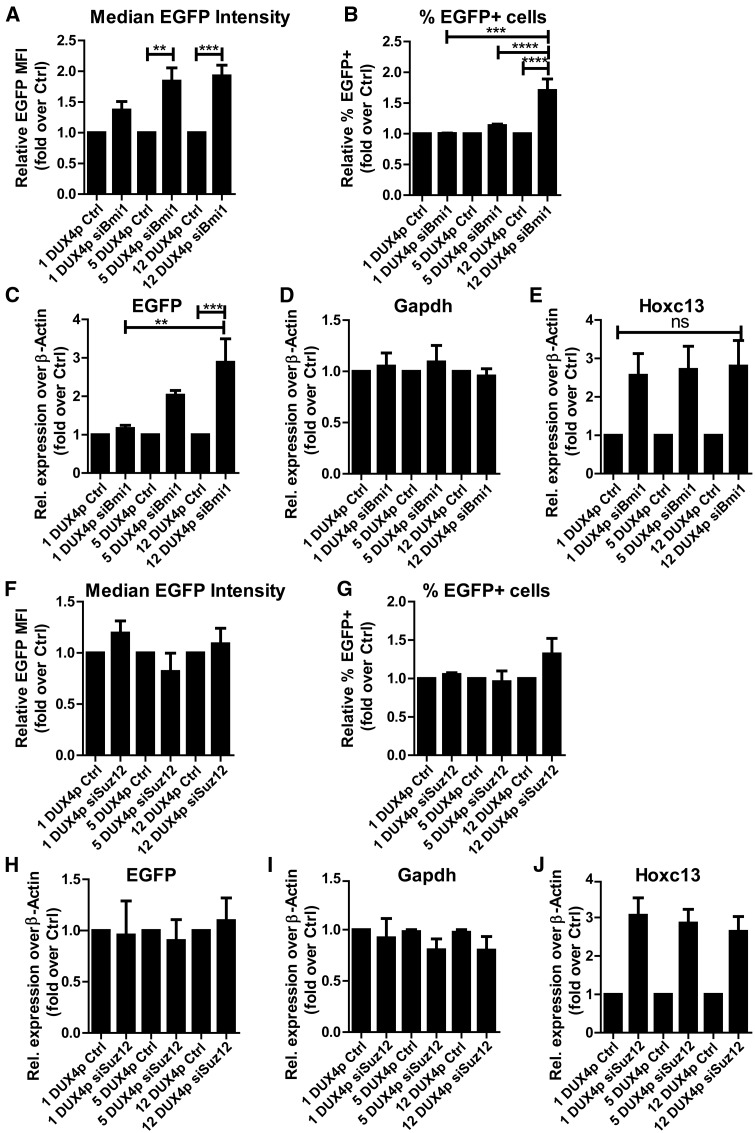
The FSHD locus displays also features of PcG targets (57,58). To address whether PcG proteins could be responsible of DUX4p-mediated repressive function, we compared Polycomb recruitment in isogenic muscle cells carrying 1-5-12 DUX4p copies (Fig. 4A–C) by focusing on Bmi1 and Suz12, respectively core components of PRC1 and PRC2 (86). ChIP-qPCR analyses revealed that a single DUX4p unit is sufficient for Bmi1 recruitment, as its enrichment on DUX4p is comparable to that of the bona fide PcG target gene Hoxc13 (Fig. 4D–F). More importantly, Bmi1 recruitment significantly increases with higher DUX4p copy-number (Fig. 4D). This copy number effect is specific, since no signal is present on a non-Polycomb target genomic sequence (Fig. 4E) (87) and no significant copy number-dependent difference is observed in Hoxc13 (Fig. 4F). On the contrary, while the PRC2 core component Suz12 is enriched on DUX4p similarly to the bona fide Polycomb target Hoxc13 (Figs 4G–I), its enrichment displays no correlation with DUX4p copy number (Fig. 4G). Overall, these results show that a single unit of DUX4p is sufficient to recruit both PRC1 and PRC2, but only PRC1 enrichment positively correlates with DUX4p copy-number.
Figure 4.
DUX4p arrays recruit Bmi1 (PRC1) and Suz12 (PRC2), but only Bmi1 shows copy-number dependent recruitment. A–C. Schematic view of isogenic muscle cells carrying 1 (A), 5 (B), or 12 (C) DUX4p repeats and an EGFP reporter gene, used for ChIP assays. D-I. Isogenic muscle cells differing for DUX4p copy-number were analyzed by ChIP for the recruitment of Bmi1 (D–F) and Suz12 (G–I), on DUX4p (D, G), and on genomic regions used for negative (E,H) or positive (F,I) controls, respectively: a non-target genomic region located 5 Kb upstream of the TSS of the Sfrp1 gene (87) (E,H) and the bona fide PcG target gene Hoxc13 (F, I). All ChIP-qPCR data are shown relative to the bona fide PcG target gene Evx1 (79). The mean of the signals obtained from three independent experiments is shown (D–F, G–I). The error bars represent SEM of independent biological replicates. Asterisk indicates statistical significance (p value) as evaluated by One way ANOVA analysis. *= P < 0.05.
PRC1 is required for DUX4p-induced gene silencing
In order to test whether Polycomb is directly involved in DUX4p-mediated silencing, we performed RNAi-depletion of PRC1 and PRC2 core components Bmi1 (siBmi1) and Suz12 (siSuz12) in muscle cells carrying 1-5-12 DUX4p copies (Supplementary Material, Fig. S3A–C). RT-qPCR and immunoblotting analyses showed that we obtained efficient and comparable knockdowns for both Bmi1 and Suz12 in all cells (Supplementary Material, Fig. S3D–K). Moreover, ChIP-qPCR experiments indicated that siRNA-treatments were effective in depleting Bmi1 and Suz12 from DUX4p (Supplementary Material, Fig. S4).
To determine whether PcG plays a role in the gene silencing effect mediated by DUX4p, we analyzed EGFP expression in siBmi1 and siSuz12 treated cells. With flow cytometry analyses, we measured EGFP reporter expression by quantifying median fluorescence intensity (Fig. 5A) and percentage of fluorescent cells (Fig. 5B) in knockdown cells compared to non-silencing controls. Remarkably, loss of the PRC1 core component Bmi1 was sufficient to cause a DUX4p copy-number dependent reporter gene de-repression, which was particularly evident in cells carrying 12-DUX4p units (Fig. 5A–B). Flow cytometry results were confirmed by RT-qPCR analyses of EGFP mRNA levels. Indeed, EGFP gene de-repression was significantly stronger in 12-DUX4p siBmi1 cells if compared with its non-silencing and with 1-DUX4p siBmi1 cells (Fig. 5C). Importantly, while Gapdh mRNA levels were constant in all populations (Fig. 5D), the bona fide PcG target Hoxc13 was equally de-repressed in 1-, 5-, or 12-DUX4p siBmi1 cells (Fig. 5E). Overall, these results indicate that PRC1 is playing a direct role in the copy-number dependent silencing mediated by DUX4p with a threshold effect analogous to that seen in FSHD (43–45).
Figure 5.
Bmi1 (PRC1) but not Suz12 (PRC2) is required for copy-number dependent gene repression. A–J. Muscle cells carrying 1, 5, or 12 DUX4p repeats and an EGFP reporter, knockdown for the PRC1 core subunit Bmi1 (siBmi, A–E), for the PRC2 core subunit Suz12 (siSuz12, F–J) or expressing a non-silencing control (Ctrl, A–J) were tested for EGFP expression. For relative quantification of EGFP protein levels among samples, flow cytometry results (A–B, F–G) of EGFP Median Fluorescence Intensity (MFI) (A,F) and of percentage of EGFP+ events (B,G) are shown. For relative quantification of EGFP at the RNA level, RT-qPCR analyses of EGFP (C, H), Gapdh negative control (D, I) and Hoxc13 positive control (E,J) are presented over β-Actin. Results are expressed over Ctrls (A–J). The mean of the signals obtained from five (A,B, F–J) or four (C–E) independent experiments is shown. The error bars represent SEM of independent biological replicates. Asterisks indicate statistical significance (p value) as evaluated by One way ANOVA Bonferroni’s multiple comparison test. ****= P < 0.0001; ***= P < 0.001; **= P < 0.01; ns = non significant.
Although Suz12 enrichment on DUX4p is not copy number-dependent (Fig. 4G), we investigated whether it plays a role in DUX4p-mediated gene silencing. To this aim, we analyzed EGFP expression upon treatment with siRNAs targeting Suz12. Differently to RNAi of the PRC1 subunit Bmi1, the EGFP reporter expression remained unchanged upon treatment with siRNAs against Suz12 compared with non-silencing controls (Fig. 5F–J). This was verified at the protein level through flow cytometry analyses of EGFP median intensity (Fig. 5F) and percentage of positive cells (Fig. 5G), but also at the transcriptional level by RT-qPCR quantification of EGFP mRNA (Fig. 5H). Importantly, while Gapdh transcript levels remain unchanged (Fig. 5I), Suz12 knockdown caused a good de-repression of the bona fide PcG target Hoxc13 (Fig. 5J), indicating that the Suz12 knockdown was effective in causing specific transcriptional de-repression of PRC2 target genes. Collectively, these results indicate that DUX4p-mediated copy-number dependent repression of gene expression requires PRC1 but it is independent from PRC2.
PRC1 is required to maintain repression of the endogenous DUX4 gene
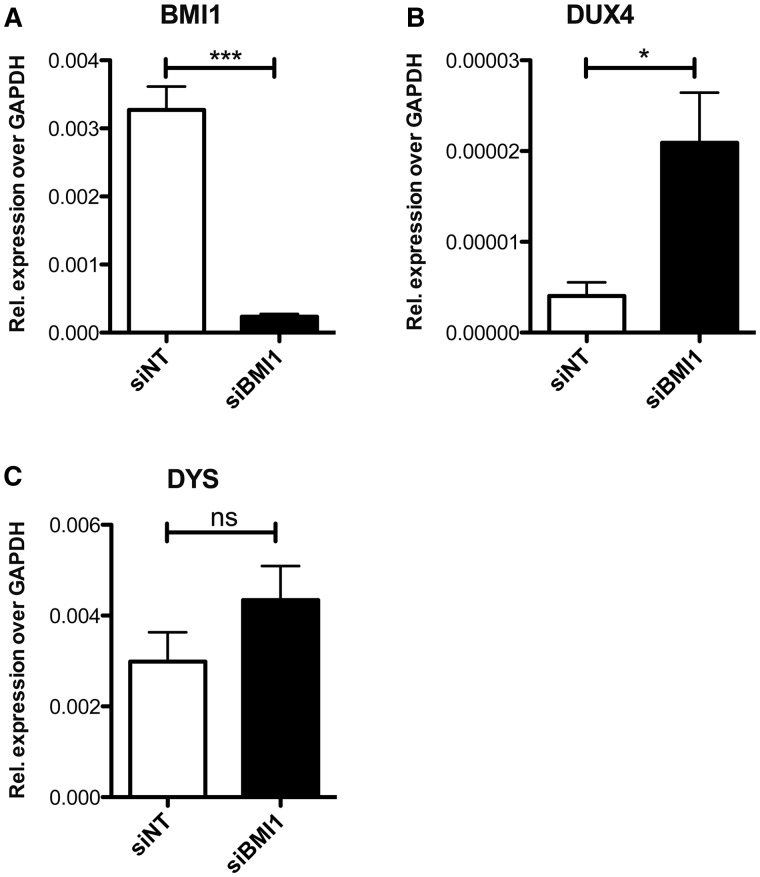
The above results indicate that PRC1 plays an important role in controlling the activity of the DUX4 promoter according to the D4Z4 copy number. To investigate the relevance of PRC1 in the physiological regulation of DUX4 expression, we performed BMI1 knockdown in human primary muscle cells obtained from healthy donors in which DUX4 is normally silent. As shown in Fig. 6, BMI1 depletion caused a significant de-repression of DUX4 compared with non-silencing controls. In conclusion, our data indicate that PRC1 plays a key role in the repression of DUX4 imposed by high D4Z4 copy number, which is lost in FSHD patients leading to disease.
Figure 6.
PRC1 is required to maintain the endogenous DUX4 gene repressed in human primary muscle cells. A. RT-qPCR of BMI1 expression in human primary myotubes from healthy donor, transfected with siRNA targeting BMI1 (siBMI1) or a non-targeting control (siNT). B. RT-qPCR showing selective DUX4 de-repression upon BMI1 depletion (siBMI1), in the same cells in (A). C. RT-qPCR of DYSTROPHIN (DYS) expression, showing no significant alteration of muscle differentiation, in the same cells in (A). BMI1, DUX4 and DYS levels are relative to GAPDH, used as normalizator. Error bars represent SEM of four independent experiments. Asterisks indicate statistical significance (P value) as evaluated by Student’s t-test. ***= P < 0.001; * P < 0.05; ns = non significant.
Discussion
Among genetic disorders, FSHD is one of the most peculiar for the major role played in its pathogenesis by epigenetic components. While a number of epigenetic alterations of the disease locus have been described (64), the molecular bases of D4Z4 copy-number dependency and incomplete penetrance of FSHD phenotype are not fully understood (65). Our results show how the size of DUX4 promoter tandem repeat arrays directly correlates with recruitment of and repression by the Polycomb Repression Complex 1, thus contributing to the comprehension of the molecular mechanisms responsible of the copy-number dependency in FSHD (65).
For practical reasons, we focused on a 180 bp region inside D4Z4 containing the DUX4 proximal promoter and displaying a CpG/GC density comparable to that of the entire D4Z4 repeat. We generated isogenic reporter muscle cell lines with DUX4p arrays of 12, 5 and 1 units, which would respectively approximate the status of the FSHD locus in healthy, mildly and severely affected patients. Remarkably, while with the 5 copies array we obtained only a partial repression, with the 12 copies array we obtained an almost complete shutdown of DUX4 promoter reporter expression, which is reminiscent of the threshold effect observed in DUX4 expression and FSHD pathogenesis (65,67).
D4Z4 is highly GC and CpG rich and it can be repeated in as many as 100 copies at each 4q35 allele. Since in mammals both features can harbor repressive machineries (88–91), it was important to address if the GC/CpG-dense or the repetitive nature of D4Z4 play a role in its ability to regulate gene expression. To dissect this, we generated reporter cell lines carrying either a high GC/CpG density, but in a single copy sequence, or a high number of repeats, but in a low GC/CpG context. By comparing gene expression in these reporters to 12 DUX4p array, we determined that the repetitive and GC/CpG-dense natures of DUX4p arrays are both important for establishing efficient repression of gene expression.
To determine the molecular mechanism responsible for the copy number-dependent repression by DUX4p, we analyzed the most relevant epigenetic repressive traits described on the FSHD locus (54,55,57,58). Despite the fact that DNA methylation, H3K9me3, PRC1 and PRC2 complexes are all enriched on DUX4p reporters at levels comparable to those of the endogenous FSHD locus in human cells, only PRC1 emerged as a good candidate to explain copy-number dependency in gene repression. In fact DNA methylation, H3K9me3 and the PRC2 core component Suz12 showed similar enrichments irrespectively of DUX4p copy number. On the contrary, the enrichment of the PRC1 core component Bmi1 is a function of DUX4p copy-number. Notably, we demonstrated that PRC1 is required for the silencing of the endogenous DUX4 in muscle cells of healthy subjects. Moreover, PRC1 is also required to maintain copy-number dependent gene silencing, since Bmi1 depletion caused a de-repression of the reporter that was significantly stronger with high DUX4p copy number. On the contrary, we found that repression by DUX4p arrays is PRC2-independent.
That PRC2 is enriched on but does not play a significant role in repression mediated by DUX4p arrays was somewhat unexpected. Nevertheless, at the FSHD locus, we previously showed that PRC2 and the associated H3K27me3 histone mark peak at the TSS of the lncRNA DBE-T to repress its transcription (58). Hence, it is tempting to speculate that PRC1 and PRC2 could play multiple and diverse roles at the FSHD locus.
In Drosophila, PcG proteins are recruited to genetic elements called Polycomb Response Elements (PREs). Much less is known about PcG recruitment to target genes in vertebrates (92). Cardinal features of Drosophila PREs are their ability to recruit PcG complexes and mediate PcG-dependent repression when inserted to ectopic sites. Accordingly, we found that DUX4p is capable to mediate robust ectopic recruitment of PRC1, PRC2 and mediate PcG-dependent gene silencing. Based on this, it is tempting to speculate that D4Z4 could be the first PRE involved in a human genetic disease. Besides our previous work on D4Z4 (58), there are several reports of Polycomb binding to repetitive elements (3,10,29,30,93–97) suggesting an important role of Polycomb proteins in genome stability through the stabilization of the repetitive genome (95). Moreover, recent data suggest that Polycomb might use repeats to achieve efficient chromatin compaction. Indeed, super-resolution images indicate that the chromatin packing density of Polycomb-repressed domains increases with domain length (98). Therefore, it has been suggested that a concatenation of PcG domains would lead to a more compact chromatin packaging and a more efficient repression of gene expression (98). Importantly, this function depends on PRC1 (98). This recent work (98) and the observation that chromatin compaction inversely correlates with FSHD disease (67,99) suggest that a copy number-dependent DUX4 promoter chromatin packing mediated by PRC1 contributes to DUX4 silencing in healthy subjects.
Recently, a correlation between DNA methylation levels and cumulative D4Z4 repeat copy number (including D4Z4 sequences from different human chromosomes) has been described (85). However, while an inverse correlation between D4Z4 copy number and DUX4 expression has been reported in FSHD1 (67), DNA methylation does not correlate with DUX4 expression (67), suggesting that additional epigenetic differences could account for DUX4 expression levels relative to D4Z4 copy number. Nevertheless, it is well established that DNA methylation of D4Z4 plays a protective role, since low levels of DNA methylation are required for the manifestation of FSHD symptoms independently from D4Z4 copy number (67,83,85).
To conclude, FSHD muscular dystrophy represents an extraordinary example of how copy-number variations in genomic elements can affect chromatin structure and gene expression (64). Our work, by exploring the role of repetitive elements in recruiting different heterochromatin regulators to repress gene expression, is relevant for understanding the contribution of repeats to organism complexity. Indeed, besides FSHD, there are a number of copy-number variations associated with diseases. In particular, similarly to FSHD, repeat expansion in neurological disorders leads to epigenetic changes and a correlation between repeat copy-number and disease phenotype has been documented (100–102). Since the molecular mechanism underlying these phenomena has remained elusive (101), the present work could shed new light on these disorders, especially when involving GC-rich regions like in Fragile X syndrome (102). In this context, the possibility to modulate PcG binding to repeats could be explored in the search of new epigenetic therapies.
Materials and Methods
Plasmid design and construction
For the generation of the 12-DUX4p-pMA construct, a sequence was designed in order to contain: an array of 11-DUX4p units with a size of 180 bp per unit, flanked by AvrII restriction sites; a terminal DUX4p unit prolonged to include DUX4 promoter until the ATG of DUX4 ORF; the EGFP gene sequence; the SV40 polyA site. This construct was synthesized by GeneArt (Life Technologies) according to the above plasmid design, in pMA backbone.
By digestion of the 12-DUX4p-pMA construct with AvrII (NEB), 11-DUX4p copies were excised, thus producing the 1-DUX4p-pMA construct.
By PCR amplification from 12-DUX4p-pMA construct, with primers DUX4p-F and DUX4p-R (in Supplementary Material, Table S1), a fragment containing 4-DUX4p units was isolated and subcloned into 1-DUX4p-pMA construct exploiting AvrII restriction site, thus generating the 5-DUX4p-pMA construct.
For the generation of FRT-constructs, the pcDNA5/FRT (Flp-In System, Life Technologies) backbone was modified as follows: the CMV promoter was excised by digestion with MfeI- NheI enzymes (NEB); a SV40 polyA site was inserted into the HindIII restriction site of the MCS of pcDNA5/FRT.
For the generation of 12-DUX4p-FRT, 5-DUX4p-FRT and 1-DUX4p-FRT constructs, SacI – NotI restriction fragments containing DUX4p arrays and EGFP reporter were excised respectively from 12-DUX4p-pMA, 5-DUX4p-pMA and 1-DUX4p-pMA constructs and subcloned into the modified backbone of pcDNA5/FRT.
For the generation of the CGI-B3gnt6-FRT construct, a 2 Kb fragment containing 110 CpGs flanked by SacI restriction sites was PCR amplified from C2C12 genomic (g) DNA with primers CGI-B3gnt6-F and CGI-B3gnt6-R (Supplementary Material, Table S1). The isolated fragment was subcloned into the 1-DUX4p-FRT plasmid in a 3’-5’ orientation, thus generating the CGI-B3gnt6-FRT construct carrying 110 non-repetitive CpGs, the DUX4 promoter unit and the EGFP reporter gene cassette.
For the generation of the 11-HbG-FRT construct, an array of 11 HbG copies of 180 pb per unit, flanked by 5’-SacI- 3'-AvrII restriction sites, was synthesized by GeneArt (Life Technologies) and subcloned into the 1-DUX4p-FRT construct, thus generating the 11-HbG-FRT construct carrying 11 HbG copies with 0 CpGs, the DUX4 promoter unit and the EGFP reporter gene cassette.
By digestion with BamHI (NEB) of the 11-HbG-FRT construct, 6-HbG copies were excised, thus producing the 5-HbG-FRT construct.
By digestion with HindIII (NEB) of the 11-HbG-FRT construct, 10-HbG copies were excised, thus producing the 1-HbG-FRT construct.
For screening and propagation purposes, Top10F’ (Life technologies) competent E.coli cells were employed. Bacteria were transformed by heat shock (42 °C – 1 min) with 1 ng of purified plasmids or 1-3 μl of ligation products. Cells were grown in LB medium plus ampicillin (100 μg/ml; Sigma) or kanamycin (50 μg/ml; Sigma) at 37 °C in a shaking incubator and plated in LB Agar + ampicillin (100 μg/ml) or + kanamycin (50 μg/ml) plates. When transformed with pGEM-T (Promega), cells were plated in LB + Amp plates supplemented with 20 μl of 50 mg/ml X-Gal (Sigma) to allow blue/white colony screening.
Mammalian cell lines culture
HEK293T cells were obtained by ATCC. The C2C12-frt cell line was derived using the Flp-In System (Life Technologies), where clones were screened for random integration of a single FRT site into the genome as previously described (103). Cells were maintained in DMEM-HIGH (Dulbecco’s Modified Eagle’s Medium, High Glucose with Sodium Pyruvate and L-Glutamine; EuroClone) supplemented with 10% FBS (Fetal Bovine Serum; EuroClone) and 1% Penicillin/Streptomycin (100 U/ml final concentration; EuroClone). Zeocine (100 µg/ml final concentration, Life Tecnologies) was added to the C2C12-frt medium.
For cell transfection, Lipofectamine LTX (Life Technologies) was used. For transient assays, HEK293T cells were transfected with equimolar quantities of either 1-DUX4p-pMA, or 5-DUX4p-pMA or 12-DUX4p-pMA, and co-transfected with pECFP-C1 (Clontech) in a ratio 5:1, where pMA constructs where 5 times more concentrated than pECFP-C1. Cells were collected after 48h for gene expression and flow cytometry analyses.
For the generation of 1-DUX4p-C2C12-frt, 5-DUX4p-C2C12-frt, 12-DUX4p-C2C12-frt, CGI-B3gnt6-C2C12-frt, 1-HbG-C2C12-frt, 5-HbG-C2C12-frt and 11-HbG-C2C12-frt stable cell lines, site-specific integration with corresponding constructs was performed via a Flp-FRT-mediated recombination (Flip In System, Life Technologies). C2C12-frt myoblasts were co-transfected with FRT constructs and the Flp recombinase expression vector, pOG44, in a ratio 9:1, where pOG44 vector was 9 times more concentrated than FRT construct. Positively transfected cells were selected with Hygromycin (100 µg/ml final concentration; InVivoGen). The Flp recombinase-mediated integration confers hygromycin resistance and zeocine sensitivity to correctly integrated cells. A minimal of two independent stable cell lines were generated for each construct.
For the generation of 1-DUX4p-C2C12-frt, 5-DUX4p-C2C12-frt and 12-DUX4p-C2C12-frt knock-down for Bmi1 or Suz12, cells were transfected with 25 nM siRNAs against Bmi1 (siRNA Mm_Bmi1_2, SI00167090 and Mm_Bmi1_7, SI02707642 in 1:1 ratio, Qiagen) or non- silencing control (All Stars Negative Control, 1027280, Qiagen) or Suz12 (L-040180-00-0005, ON-TARGETplus SMARTpool, Mouse, Thermo Scientific) or non-silencing control (D-001810-10, ONTARGETplus Non targeting pool, Thermo Scientific) following manufacturer’s instructions. Transfections were performed 72h prior to collection for gene expression and flow cytometry analyses.
For freezing, cell pellets were resuspended in FBS and transferred to cryovials. DMSO (dimethyl sulfoxide; Sigma) was added as cryoprotector at a final concentration of 10%. Cryovials were frozen into isopropanol-filled cryoboxes, stored at −80 °C overnight and, then, long-term stored in liquid nitrogen.
Culture and transfection of human primary muscle cells
All procedures involving human samples were approved by Fondazione San Raffaele del Monte Tabor Ethical Committee.
Human primary myoblasts were obtained from the University of Massachusetts Medical School Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center for FSHD (http://www.umassmed.edu/wellstone; date last accessed December 30, 2016) in Worcester, MA, USA.
Human primary myoblasts (07Ubic) were derived from the biceps of a healthy donor. They were cultured in a humidified atmosphere at 37 °C with 5% O2 and 5% CO2.
Myoblasts were seeded in 0.1% gelatin-coated dishes and cultured in Ham’s F-10 medium (Nutrient Mixture F10 Ham, with L-glutamine and sodium bicarbonate, Sigma) supplemented with 20% FBS (Fetal Bovine Serum; EuroClone), 0.5% chicken embryo extract, 1.2 mM CaCl2 (Sigma) and 1% penicillin/streptomycin (100 U/ml final concentration, EuroClone). When cells reached confluence of 90%, differentiation was induced by replacing growth medium with 4:1 D-MEM:199 Medium (Dulbecco’s Modified Eagle’s Medium, High Glucose with Sodium Pyruvate and L-Glutamine, EuroClone; Medium 199, Gibco), supplemented with 2% horse serum (Euroclone), 1% ITS (Insulin, transferrin and sodium selenite solution, Sigma) and 1% penicillin/streptomycin (Euroclone) for 4 days.
For siRNA transfection, Lipofectamine 3000 in Opti-MEM (Thermo Fisher Scientific) was used, following manufacturer’s instructions. siRNA for BMI1 and the non-silencing control were purchased from Thermo Fisher Scientific (SMARTpool: ON-TARGETplus Human BMI1 siRNA L-005230-01; non-targeting control D-001810-10) and used at a final concentration of 25nM. 48h after transfection, differentiation was induced, and after 4 days myotubes were collected.
Flow cytometry
For flow cytometry analysis of fluorescence intensity in HEK293T, cells transiently co-transfected with ECGP and either 1-DUX4p-pMA, 5-DUX4p-pMA, or 12-DUX4p-pMA constructs were collected with trypsinization followed by centrifugation 48h after transfection.
For flow cytometry analysis of fluorescence intensity in C2C12-frt cells, 1-DUX4p-C2C12-frt, 5-DUX4p-C2C12-frt, 12-DUX4p-C2C12-frt, CGI-B3gnt6-C2C12-frt, 1-HbG-C2C12-frt, 5-HbG-C2C12-frt or 11-HbG-C2C12-frt muscle cells expressing EGFP were collected with trypsinization followed by centrifugation at different passages from cell line generations.
For flow cytometry analysis of fluorescence intensity upon Bmi1 or Suz12 knockdown, 1-DUX4p-C2C12-frt, 5-DUX4p-C2C12-frt or 12-DUX4p-C2C12-frt myoblasts were collected with trypsinization followed by centrifugation 72h after siRNA transfection.
Aliquot of 0.5–1x106 cells were transferred into assay tubes, rinsed by centrifugation with growth medium and resuspended in 100 µl normal growth medium per assay tube. Analysis on flow cytometers was performed within 1h from trypsinization. LSR Fortessa (Becton Dickinson) was used for acquisition of EGFP, ECFP and morphological parameters, while Accuri C6 (BD Biosciences) for acquisition of EGFP signal and morphological parameters.
Fluorochrome spectral overlap of EGFP and ECFP signals was corrected with set up of automatic compensation with FACSDIVA software (BD Biosciences). Analyses were performed with FCSExpress 4.0 (De Novo), and EGFP intensity and percentage of EGFP positive cells were quantified in the subpopulation of ECFP positive events in the case of HEK293T co-transfected cells, or in the total population of living cells (singlet events) in the case of stable cell lines expressing EGFP.
RNA extraction, RT-PCR, and RT-qPCR analysis
Reporter gene expression and gene down-regulation upon siRNA treatment were evaluated by RT-qPCR analysis.
Total RNA from cultured cells was extracted with Trizol, treated with DNaseI and purified using PureLink RNA Mini Kit (Ambion).
Total RNA from human primary muscle cells was extracted with Lysis buffer supplemented with 0.01% β-mercaptoethanol and purified with RNA spin columns (PureLink RNA Mini kit, Ambion), including a DNAseI treatment step, following the manufacturer's instructions.
cDNA (using up to 1 µg of RNA) was synthesized using SuperScript III First-Strand Synthesis Super-Mix (Life Technologies). For DUX4 expression analyses in human primary cells, 2 µg of RNA were retrotranscribed using SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies). RNA was mixed with oligo(dT) and the RT-PCR reaction was performed following the manufacturer's protocol.
qPCRs were performed with SYBR GreenER qPCR SuperMix Universal (Life Technologies) using CFX96 or CFX384 Real-time System (Biorad). Validated primers are listed in Supplementary Material, Table S1. Expression analyses were performed using the ΔΔCT method (ΔCT method for human primary muscle cells). β-actin or GAPDH (for human primary muscle cells) were used as housekeeping genes for sample normalization.
Real-time PCR analysis
Biorad’s CFX96 or CFX384 Real-time System with CFX Manager Software V.1.6 were used. Real-time PCRs were conducted as follows: initial denaturation: 95 °C, 10 min; 40 cycles of denaturation (95 °C, 30 sec), annealing (temperature according to primers, 30 sec), amplification (72 °C, 30 sec). For Major Satellite analysis only, amplification time was 15 sec.
The specificity of the amplified products was monitored by performing melting curves at the end of each amplification reaction. The efficiency of each primer was assessed by performing primer validation qPCRs with serial dilutions of reference material (cDNA for qRT-PCR, gDNA for MeDIP, Input DNA for ChIP).
MeDIP
MeDIP analysis of 1-DUX4p-C2C12-frt, 5-DUX4p-C2C12-frt and 12-DUX4p-C2C12-frt muscle cells was carried out mainly as described in (104).
Briefly, cell pellets were resuspended in 300 μl of TE per 1x106 of cells and lysed with one volume of Lysis buffer (20 mM Tris pH 8.0, 4 mM EDTA, 20 mM NaCl and 1% SDS) containing 20 μl of proteinase K (10mg/ml, Promega). Samples were incubated at 55 °C for at least 5 h.
gDNA was extracted with phenol (Fluka) and chloroform (Fluka) extractions, followed by 10 min centrifugation at maximum speed at RT. Supernatant was precipitated using 2 volumes of 100% ethanol and 75 mM of Sodium Acetate, and DNA was resuspended in TE containing 20 μg/ml of RNAse A (Sigma) for 30 min. Then DNA was fragmented (200–400 bp) per 5 cycles 30 sec ON 30 second OFF at low intensity with Bioruptor sonication device (Diagenode). DNA was denatured for 10 min in boiling water and immediately cool down on ice for 10 min.
500-1000 ng of sonicated gDNA was precipitated using 5 μg of α-5mC Clone 33D3 (Diagenode) or normal mouse IgG (015-000-003, Jackson ImmunoResearch) in IP buffer (10 mM Na-Phospate pH 7.0, 140 mM NaCl, 0.005% Triton x-100) overnight at 4ºC with overhead incubation. Next day, 50 μl of Protein G Dynabeads (Life Technologies), previously washed with PBS-BSA 0.1%, were added to the mix and incubated for 2 h at 4ºC with overhead mixing. Sonicanted gDNA/antibody/beads complex were washed three times with IP buffer.
Immunoprecipitated DNA was extracted in proteinase K buffer (50 mM Tris pH 8.0, 10 mM EDTA and 0.5% SDS) with 7 μl of proteinase K (10 mg/ml, Promega) followed by phenol chloroform extraction. DNA was precipitated using 400 mM NaCl, glycogen (1 μl) and 2 volumes of 100% ethanol. Then DNA was resuspended in 60 μl of TE. 100% of the initial amount of DNA was used as Input.
DNA was analyzed by qPCR with SYBR GreenER qPCR SuperMix Universal (Life technologies) using Biorad’s CFX96 or CFX384 Real-time System with an amount of template equivalent to 1 µl of the original elution. For Major Satellite analysis only, the template was further diluted 1:1000. Validated primers are listed in Supplementary Material, Table S1. Analyses were performed using the ΔCT method, normalizing MeDIP-qPCR data for input DNA (reported as Relative enrichment to Input in the figures).
Chromatin immunoprecipitation
ChIP-qPCR was carried out mainly as described in (58) on 1-DUX4p-C2C12-frt, 5-DUX4p-C2C12-frt and 12-DUX4p-C2C12-frt muscle cells for ChIP evaluation of histone mark and protein enrichment, and on 1-DUX4p-C2C12-frt, 5-DUX4p-C2C12-frt and 12-DUX4p-C2C12-frt muscle cells treated with siBmi1 or siSuz12 for evaluation of protein downregulation on target sites.
Briefly, cells were washed once in PBS and fixed for 10 min in 1% formaldehyde in PBS (from a 37.5% formaldehyde/10% methanol stock, Sigma). After formaldehyde quenching with Glycine (final concentration 125 mM) for 5 min, cells were washed with PBS, harvested by scraping and pelleted at 1350 g for 5 min at 4 °C.
The pellet was lysed in a solution containing 50 mM Hepes-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40 and 0.25% Triton X100 for 10 min in ice. Nuclei were pelleted at 1350 g for 5 min at 4 °C and washed from detergents in a solution containing 10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA and 0.5 mM EGTA with gentle swirl for 10 min. Next, samples were centrifuged and the nuclei pellet was lysed in a solution with 10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-Deoxycholate and 0.5% N-laurylsarcosine. Chromatin was sheared by sonication using Bioruptor (Diagenode), typically 15 min (high intensity 30 sec ON 30 sec OFF) or as much as needed to reach an optimal size of the fragments between 200 bp and 700 bp. Triton X-100 was added to lysates at a final concentration of 1%, and a clarification step of 10 min at 16,360 g at 4 °C followed.
100 μg of chromatin were used for each immunoprecipitation (10 μg for ChIP to histones and histone modifications). Incubations were carried out at 4 °C overnight with 10 μg (5 μg for ChIP to histones and histone modifications) of the following antibodies: α-Bmi1 (ab85688, Abcam), α-Suz12 (ab12073, Abcam), α-H3K9me3 (Ab8898, Abcam), α-H3 (ab1791, Abcam) and whole molecule rabbit IgG (011-000-003, Jackson ImmunoResearch). Immunoprecipitation with 50 µl of Protein G Dynabeads (Life Technologies), previously washed with BSA 0.1% in PBS, was carried out for 3 h at 4 °C. Before starting the washes, 5% of the total ChIP volume was taken from the control IgG supernatant as Input fraction.
Immunoprecipitated chromatin was washed extensively with a solution containing 50 mM Hepes-KOH pH 7.6, 500 mM LiCl, 1 mM EDTA, 1% NP125 40 and 0.7% Na-Deoxycholate, and protein–DNA cross-links were reverted by heating at 65 °C overnight in TE buffer 2% SDS.
DNA was purified with QIAquick PCR Purification Kit (Qiagen) and eluted in 50 µl TE. qPCRs were performed with SYBR GreenER qPCR SuperMix Universal (Life Technologies), in CFX96 or CFX384 Real-time System (Biorad) with an amount of template equivalent to 1 µl of the original elution. For Major Satellite analysis only, the template was further diluted 1:1000. Validated primers are listed in Supplementary Material, Table S1. Analyses were performed using the ΔCT method, normalizing ChIP-qPCR data for input chromatin. For histone modification antibodies, the enrichment is expressed as percentage of total histone (reported in the figures as: Relative enrichment to H3). For non-histone antibodies, ChIP-qPCR data are shown relative to the bona fide PcG target gene Evx1 (reported in the figures as: Relative enrichment to Input – fold over Evx1), as described (79). ΔΔCT method was used for analyses versus non-silencing control in ChIP of RNAi samples.
SDS-PAGE and immunoblotting
Protein down-regulation was evaluated by immunoblotting with mouse α-Bmi1 (ab85688, Abcam) or α-Suz12 (ab12073, Abcam), using α-Vinculin (V9131, Sigma) as loading control. To this aim, SDS-polyacrylamide gels were prepared following the recipes described by Sambrook and Russel in Molecular cloning: a laboratory manual (105) for 10% or 12% gels.
Electrophoresis was performed at 100-120V at RT in 1X Tris-Glycine SDS (TGS) running buffer pH 8.3 [25mM Trizma (Sigma), 192 mM Glycine (Sigma), 0.1% SDS (National Diagnostics)] prepared in milliQ water.
For immunoblotting, transfers were performed using the iBlot dry blotting system (Life technologies), following manufacturer recommended conditions (Life technologies) with P3 program.
For immunohybridization, filters were saturated in 5% non-fat milk (Sigma) TBS-Tween 0.1% solution (Tris Buffered Saline: 10 mM TrisHCl pH 7.4, 140 mM NaCl, Tween-20; National Diagnostics) for 1h at RT or overnight at 4 °C. Incubation with the primary antibody was performed in 5% non-fat milk TBS-Tween 0.1% solution for 2h at RT or overnight at 4 °C. Filters were then washed 3 times (10 min each) with TBS-Tween 0.1% at RT. HRP-conjugated secondary antibodies α-mouse (715-035-150 Jackson ImmunoResearch) or α-rabbit (711-035-152, Jackson ImmunoResearch) were incubated 5% non-fat milk TBS-Tween 0.1% solution for 1-2 h at RT. Filters were washed again as described.
For detection, filters were incubated for 5 min with horseradish peroxidase (HRP) chemiluminescent substrate (SuperSignal West Pico or Dura Chemiluminescent Substrate; Thermo scientific).
Statistical analyses
All statistical analyses were performed using GraphPad Prism version 5.0a (GraphPad Software, San Diego, USA). The type of statistical test, P value, the number of independent experiments, mean and standard error of the mean are provided for each data set in the corresponding figure legends.
Supplementary Material
Supplementary Material are available at HMG online.
Supplementary Material
Acknowledgements
We thank the Flow cytometry Resource and Advanced Cytometry Technical Applications Laboratory. This work is a partial fulfillment of V.C. PhD in Neuroscience, Università Vita-Salute San Raffaele, Milan, Italy. D.G. is a Dulbecco Telethon Institute Senior Scientist.
Conflict of Interest statement. None declared.
Funding
This work was supported by the Italian Telethon Foundation [TCR13001 to D.G.]; the ERA-Net for Research on Rare Diseases [E-Rare-2 to D.G., E-Rare-2 through Canadian Institutes of Health Research and Muscular Dystrophy Canada to F.J.D.]; the Italian Epigenomics Flagship Project [to D.G.]; the Italian Ministry of Health [to D.G.]; and the FSHD Global Research Foundation [to D.G.]. Funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript should be declared. Funding to pay the Open Access publication charges for this article was provided by Fondazione Telethon.
References
- 1.Casa V., Gabellini D. (2012) A repetitive elements perspective in Polycomb epigenetics. Front. Genet., 3, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warburton P.E., Hasson D., Guillem F., Lescale C., Jin X., Abrusan G. (2008) Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genomics, 9, 533.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Day D.S., Luquette L.J., Park P.J., Kharchenko P.V. (2010) Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol., 11, R69.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. (2011) Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet., 7, e1002384.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norris J., Fan D., Aleman C., Marks J.R., Futreal P.A., Wiseman R.W., Iglehart J.D., Deininger P.L., McDonnell D.P. (1995) Identification of a new subclass of Alu DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J. Biol. Chem., 270, 22777–22782. [DOI] [PubMed] [Google Scholar]
- 6.Speek M. (2001) Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell Biol., 21, 1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen S., Lin L., Cai J.J., Jiang P., Kenkel E.J., Stroik M.R., Sato S., Davidson B.L., Xing Y. (2011) Widespread establishment and regulatory impact of Alu exons in human genes. Proc. Natl Acad. Sci. U S A, 108, 2837–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G., Kleinman M.E., Ponicsan S.L., Hauswirth W.W., Chiodo V.A., et al. (2011) DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature, 471, 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faulkner G.J., Carninci P. (2009) Altruistic functions for selfish DNA. Cell Cycle, 8, 2895–2900. [DOI] [PubMed] [Google Scholar]
- 10.Leeb M., Pasini D., Novatchkova M., Jaritz M., Helin K., Wutz A. (2010) Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev., 24, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters A.H., Kubicek S., Mechtler K., O'Sullivan R.J., Derijck A.A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., et al. (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell, 12, 1577–1589. [DOI] [PubMed] [Google Scholar]
- 12.Wang T., Zeng J., Lowe C.B., Sellers R.G., Salama S.R., Yang M., Burgess S.M., Brachmann R.K., Haussler D. (2007) Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl Acad. Sci. U S A, 104, 18613–18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourque G., Leong B., Vega V.B., Chen X., Lee Y.L., Srinivasan K.G., Chew J.L., Ruan Y., Wei C.L., Ng H.H., et al. (2008) Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res., 18, 1752–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chadwick B.P. (2008) DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res., 18, 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simeonova I., Lejour V., Bardot B., Bouarich-Bourimi R., Morin A., Fang M., Charbonnier L., Toledo F. (2012) Fuzzy tandem repeats containing p53 response elements may define species-specific p53 target genes. PLoS Genet., 8, e1002731.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faulkner G.J., Kimura Y., Daub C.O., Wani S., Plessy C., Irvine K.M., Schroder K., Cloonan N., Steptoe A.L., Lassmann T., et al. (2009) The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet., 41, 563–571. [DOI] [PubMed] [Google Scholar]
- 17.Conley A.B., Miller W.J., Jordan I.K. (2008) Human cis natural antisense transcripts initiated by transposable elements. Trends Genet., 24, 53–56. [DOI] [PubMed] [Google Scholar]
- 18.Vierstra J., Rynes E., Sandstrom R., Zhang M., Canfield T., Hansen R.S., Stehling-Sun S., Sabo P.J., Byron R., Humbert R., et al. (2014) Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science, 346, 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brahmachary M., Guilmatre A., Quilez J., Hasson D., Borel C., Warburton P., Sharp A.J. (2014) Digital genotyping of macrosatellites and multicopy genes reveals novel biological functions associated with copy number variation of large tandem repeats. PLoS Genet., 10, e1004418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mills R.E., Luttig C.T., Larkins C.E., Beauchamp A., Tsui C., Pittard W.S., Devine S.E. (2006) An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res., 16, 1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelfand Y., Rodriguez A., Benson G. (2007) TRDB–the Tandem Repeats Database. Nucleic Acids Res., 35, D80–D87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ames D., Murphy N., Helentjaris T., Sun N., Chandler V. (2008) Comparative analyses of human single- and multilocus tandem repeats. Genetics, 179, 1693–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blackburn E.H. (1984) The molecular structure of centromeres and telomeres. Annu. Rev. Biochem., 53, 163–194. [DOI] [PubMed] [Google Scholar]
- 24.Schueler M.G., Higgins A.W., Rudd M.K., Gustashaw K., Willard H.F. (2001) Genomic and genetic definition of a functional human centromere. Science, 294, 109–115. [DOI] [PubMed] [Google Scholar]
- 25.Tam R., Smith K.P., Lawrence J.B. (2004) The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatin unlike most human telomeres. J. Cell Biol., 167, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guenatri M., Bailly D., Maison C., Almouzni G. (2004) Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol., 166, 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNeil J.A., Smith K.P., Hall L.L., Lawrence J.B. (2006) Word frequency analysis reveals enrichment of dinucleotide repeats on the human X chromosome and [GATA]n in the X escape region. Genome Res., 16, 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris C.A., Moazed D. (2007) Centromere assembly and propagation. Cell, 128, 647–650. [DOI] [PubMed] [Google Scholar]
- 29.Pinter S.F., Sadreyev R.I., Yildirim E., Jeon Y., Ohsumi T.K., Borowsky M., Lee J.T. (2012) Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res., 22, 1864–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darrow E.M., Huntley M.H., Dudchenko O., Stamenova E.K., Durand N.C., Sun Z., Huang S.C., Sanborn A.L., Machol I., Shamim M., et al. (2016) Deletion ofDXZ4on the human inactive X chromosome alters higher-order genome architecture. Proceedings of the National Academy of Sciences, in press., 201609643. [DOI] [PMC free article] [PubMed]
- 31.Warburton P.E., Haaf T., Gosden J., Lawson D., Willard H.F. (1996) Characterization of a chromosome-specific chimpanzee alpha satellite subset: evolutionary relationship to subsets on human chromosomes. Genomics, 33, 220–228. [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin C.R., Chadwick B.P. (2011) Characterization of DXZ4 conservation in primates implies important functional roles for CTCF binding, array expression and tandem repeat organization on the X chromosome. Genome Biol., 12, R37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudd M.K., Wray G.A., Willard H.F. (2006) The evolutionary dynamics of alpha-satellite. Genome Res., 16, 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tremblay D.C., Alexander G. Jr., Moseley S., Chadwick B.P. (2010) Expression, tandem repeat copy number variation and stability of four macrosatellite arrays in the human genome. BMC Genomics, 11, 632.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruce H.A., Sachs N., Rudnicki D.D., Lin S.G., Willour V.L., Cowell J.K., Conroy J., McQuaid D.E., Rossi M., Gaile D.P., et al. (2009) Long tandem repeats as a form of genomic copy number variation: structure and length polymorphism of a chromosome 5p repeat in control and schizophrenia populations. Psychiatric Genet., 19, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez Castel A., Cleary J.D., Pearson C.E. (2010) Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol., 11, 165–170. [DOI] [PubMed] [Google Scholar]
- 37.Puschendorf M., Terranova R., Boutsma E., Mao X., Isono K., Brykczynska U., Kolb C., Otte A.P., Koseki H., Orkin S.H., et al. (2008) PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet., 40, 411–420. [DOI] [PubMed] [Google Scholar]
- 38.Saksouk N., Barth T.K., Ziegler-Birling C., Olova N., Nowak A., Rey E., Mateos-Langerak J., Urbach S., Reik W., Torres-Padilla M.E., et al. (2014) Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol. Cell, 56, 580–594. [DOI] [PubMed] [Google Scholar]
- 39.Oberdoerffer P., Sinclair D.A. (2007) The role of nuclear architecture in genomic instability and ageing. Nat. Rev. Mol. Cell Biol., 8, 692–702. [DOI] [PubMed] [Google Scholar]
- 40.Le H.Q., Ghatak S., Yeung C.C., Tellkamp F., Gunschmann C., Dieterich C., Yeroslaviz A., Habermann B., Pombo A., Niessen C.M., et al. (2016) Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol., in press. [DOI] [PubMed] [Google Scholar]
- 41.Deenen J.C., Arnts H., van der Maarel S.M., Padberg G.W., Verschuuren J.J., Bakker E., Weinreich S.S., Verbeek A.L., van Engelen B.G. (2014) Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology, 83, 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cabianca D.S., Gabellini D. (2010) The cell biology of disease: FSHD: copy number variations on the theme of muscular dystrophy. J. Cell Biol., 191, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., et al. (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet., 2, 26–30. [DOI] [PubMed] [Google Scholar]
- 44.Wijmenga C., Sandkuijl L.A., Moerer P., van der Boorn N., Bodrug S.E., Ray P.N., Brouwer O.F., Murray J.C., van Ommen G.J., Padberg G.W., et al. (1992) Genetic linkage map of facioscapulohumeral muscular dystrophy and five polymorphic loci on chromosome 4q35-qter. Am. J. Hum. Genet., 51, 411–415. [PMC free article] [PubMed] [Google Scholar]
- 45.van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R. (1993) FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet., 2, 2037–2042. [DOI] [PubMed] [Google Scholar]
- 46.Lunt P.W., Jardine P.E., Koch M.C., Maynard J., Osborn M., Williams M., Harper P.S., Upadhyaya M. (1995) Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35-facioscapulohumeral muscular dystrophy (FSHD). Hum. Mol. Genet., 4, 951–958. [DOI] [PubMed] [Google Scholar]
- 47.Ricci G., Scionti I., Sera F., Govi M., D'Amico R., Frambolli I., Mele F., Filosto M., Vercelli L., Ruggiero L., et al. (2013) Large scale genotype-phenotype analyses indicate that novel prognostic tools are required for families with facioscapulohumeral muscular dystrophy. Brain, 136, 3408–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ricci E., Galluzzi G., Deidda G., Cacurri S., Colantoni L., Merico B., Piazzo N., Servidei S., Vigneti E., Pasceri V., et al. (1999) Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann. Neurol., 45, 751–757. [DOI] [PubMed] [Google Scholar]
- 49.Tawil R., Forrester J., Griggs R.C., Mendell J., Kissel J., McDermott M., King W., Weiffenbach B., Figlewicz D. (1996) Evidence for anticipation and association of deletion size with severity in facioscapulohumeral muscular dystrophy. The FSH-DY Group. Ann. Neurol., 39, 744–748. [DOI] [PubMed] [Google Scholar]
- 50.Statland J.M., Donlin-Smith C.M., Tapscott S.J., Lemmers R.J., van der Maarel S.M., Tawil R. (2015) Milder phenotype in facioscapulohumeral dystrophy with 7-10 residual D4Z4 repeats. Neurology, 85, 2147–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin F., Wang Z.Q., Lin M.T., Murong S.X., Wang N. (2015) New Insights into Genotype-phenotype Correlations in Chinese Facioscapulohumeral Muscular Dystrophy: A Retrospective Analysis of 178 Patients. Chinese Med. J., 128, 1707–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goto K., Lee J.H., Matsuda C., Hirabayashi K., Kojo T., Nakamura A., Mitsunaga Y., Furukawa T., Sahashi K., Arahata K. (1995) DNA rearrangements in Japanese facioscapulohumeral muscular dystrophy patients: clinical correlations. Neuromuscul. Disord., 5, 201–208. [DOI] [PubMed] [Google Scholar]
- 53.Tonini M.M., Passos-Bueno M.R., Cerqueira A., Matioli S.R., Pavanello R., Zatz M. (2004) Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul. Disord., 14, 33–38. [DOI] [PubMed] [Google Scholar]
- 54.de Greef J.C., Wohlgemuth M., Chan O.A., Hansson K.B., Smeets D., Frants R.R., Weemaes C.M., Padberg G.W., van der Maarel S.M. (2007) Hypomethylation is restricted to the D4Z4 repeat array in phenotypic FSHD. Neurology, 69, 1018–1026. [DOI] [PubMed] [Google Scholar]
- 55.Zeng W., de Greef J.C., Chen Y.Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A., et al. (2009) Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet., 5, e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng W., Chen Y.Y., Newkirk D.A., Wu B., Balog J., Kong X., Ball A.R. Jr., Zanotti S., Tawil R., Hashimoto N., et al. (2014) Genetic and Epigenetic Characteristics of FSHD-Associated 4q and 10q D4Z4 that are Distinct from Non-4q/10q D4Z4 Homologs. Hum. Mutat., 35, 998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bodega B., Ramirez G.D., Grasser F., Cheli S., Brunelli S., Mora M., Meneveri R., Marozzi A., Mueller S., Battaglioli E., et al. (2009) Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol., 7, 41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cabianca D.S., Casa V., Bodega B., Xynos A., Ginelli E., Tanaka Y., Gabellini D. (2012) A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell, 149, 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gabellini D., Green M.R., Tupler R. (2002) Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell, 110, 339–348. [DOI] [PubMed] [Google Scholar]
- 60.Rijkers T., Deidda G., van Koningsbruggen S., van Geel M., Lemmers R.J., van Deutekom J.C., Figlewicz D., Hewitt J.E., Padberg G.W., Frants R.R., et al. (2004) FRG2, an FSHD candidate gene, is transcriptionally upregulated in differentiating primary myoblast cultures of FSHD patients. J. Med. Genet., 41, 826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Matteotti C., van Acker A.M., Leo O., et al. (2007) DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. U S A, 104, 18157–18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ansseau E., Laoudj-Chenivesse D., Marcowycz A., Tassin A., Vanderplanck C., Sauvage S., Barro M., Mahieu I., Leroy A., Leclercq I., et al. (2009) DUX4c is up-regulated in FSHD. It induces the MYF5 protein and human myoblast proliferation. PLoS One, 4, e7482.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J., et al. (2010) Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet., 6, e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daxinger L., Tapscott S.J., van der Maarel S.M. (2015) Genetic and epigenetic contributors to FSHD. Curr. Opin. Genet. Dev., 33, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L.H., Tawil R. (2016) Facioscapulohumeral Dystrophy. Curr. Neurol. Neurosci. Rep., 16, 66.. [DOI] [PubMed] [Google Scholar]
- 66.Neguembor M.V., Previtali S.C., Gabellini D.. Facioscapulohumeral dystrophy. OMMBID McGraw-Hill 2015. [Google Scholar]
- 67.Jones T.I., King O.D., Himeda C.L., Homma S., Chen J.C., Beermann M.L., Yan C., Emerson C.P. Jr., Miller J.B., Wagner K.R., et al. (2015) Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clin. Epigenet., 7, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dmitriev P., Petrov A., Ansseau E., Stankevicins L., Charron S., Kim E., Bos T.J., Robert T., Turki A., Coppee F., et al. (2011) The Kruppel-like factor 15 as a molecular link between myogenic factors and a chromosome 4q transcriptional enhancer implicated in facioscapulohumeral dystrophy. J. Biol. Chem., 286, 44620–44631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winokur S.T., Bengtsson U., Vargas J.C., Wasmuth J.J., Altherr M.R., Weiffenbach B., Jacobsen S.J. (1996) The evolutionary distribution and structural organization of the homeobox-containing repeat D4Z4 indicates a functional role for the ancestral copy in the FSHD region. Hum. Mol. Genet., 5, 1567–1575. [DOI] [PubMed] [Google Scholar]
- 70.Winokur S.T., Bengtsson U., Feddersen J., Mathews K.D., Weiffenbach B., Bailey H., Markovich R.P., Murray J.C., Wasmuth J.J., Altherr M.R., et al. (1994) The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Research: An International Journal on the Molecular, Supramolecular and Evolutionary Aspects of Chromosome Biology, 2, 225–234. [DOI] [PubMed] [Google Scholar]
- 71.Lyle R., Wright T.J., Clark L.N., Hewitt J.E. (1995) The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics, 28, 389–397. [DOI] [PubMed] [Google Scholar]
- 72.Clark L.N., Koehler U., Ward D.C., Wienberg J., Hewitt J.E. (1996) Analysis of the organisation and localisation of the FSHD-associated tandem array in primates: implications for the origin and evolution of the 3.3 kb repeat family. Chromosoma, 105, 180–189. [DOI] [PubMed] [Google Scholar]
- 73.Block G.J., Petek L.M., Narayanan D., Amell A.M., Moore J.M., Rabaia N.A., Tyler A., van der Maarel S.M., Tawil R., Filippova G.N., et al. (2012) Asymmetric bidirectional transcription from the FSHD-causing D4Z4 array modulates DUX4 production. PLoS One, 7, e35532.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Giacalone J., Friedes J., Francke U. (1992) A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat. Genet., 1, 137–143. [DOI] [PubMed] [Google Scholar]
- 75.Kogi M., Fukushige S., Lefevre C., Hadano S., Ikeda J.E. (1997) A novel tandem repeat sequence located on human chromosome 4p: isolation and characterization. Genomics, 42, 278–283. [DOI] [PubMed] [Google Scholar]
- 76.Chadwick B.P. (2009) Macrosatellite epigenetics: the two faces of DXZ4 and D4Z4. Chromosoma, 118, 675–681. [DOI] [PubMed] [Google Scholar]
- 77.Tsumagari K., Qi L., Jackson K., Shao C., Lacey M., Sowden J., Tawil R., Vedanarayanan V., Ehrlich M. (2008) Epigenetics of a tandem DNA repeat: chromatin DNaseI sensitivity and opposite methylation changes in cancers. Nucleic Acids Res., 36, 2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jabbari K., Bernardi G. (2004) Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene, 333, 143–149. [DOI] [PubMed] [Google Scholar]
- 79.Jermann P., Hoerner L., Burger L., Schubeler D. (2014) Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc. Natl Acad. Sci. U S A, 111, E3415–E3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ottaviani A., Rival-Gervier S., Boussouar A., Foerster A.M., Rondier D., Sacconi S., Desnuelle C., Gilson E., Magdinier F. (2009) The D4Z4 macrosatellite repeat acts as a CTCF and A-type lamins-dependent insulator in facio-scapulo-humeral dystrophy. PLoS Genet., 5, e1000394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ottaviani A., Schluth-Bolard C., Gilson E., Magdinier F. (2010) D4Z4 as a prototype of CTCF and lamins-dependent insulator in human cells. Nucleus, 1, 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ottaviani A., Schluth-Bolard C., Rival-Gervier S., Boussouar A., Rondier D., Foerster A.M., Morere J., Bauwens S., Gazzo S., Callet-Bauchu E., et al. (2009) Identification of a perinuclear positioning element in human subtelomeres that requires A-type lamins and CTCF. Embo J., 28, 2428–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gaillard M.C., Roche S., Dion C., Tasmadjian A., Bouget G., Salort-Campana E., Vovan C., Chaix C., Broucqsault N., Morere J., et al. (2014) Differential DNA methylation of the D4Z4 repeat in patients with FSHD and asymptomatic carriers. Neurology, 83, 733–742. [DOI] [PubMed] [Google Scholar]
- 84.Das S., Chadwick B.P. (2016) Influence of Repressive Histone and DNA Methylation upon D4Z4 Transcription in Non-Myogenic Cells. PLoS One, 11, e0160022.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lemmers R.J., Goeman J.J., van der Vliet P.J., van Nieuwenhuizen M.P., Balog J., Vos-Versteeg M., Camano P., Ramos Arroyo M.A., Jerico I., Rogers M.T., et al. (2015) Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum. Mol. Genet., 24, 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. (2007) Genome regulation by polycomb and trithorax proteins. Cell, 128, 735–745. [DOI] [PubMed] [Google Scholar]
- 87.Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D., et al. (2014) A comparative encyclopedia of DNA elements in the mouse genome. Nature, 515, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang J., Jia S.T., Jia S. (2016) New Insights into the Regulation of Heterochromatin. Trends Genet., 32, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lynch M.D., Smith A.J., De Gobbi M., Flenley M., Hughes J.R., Vernimmen D., Ayyub H., Sharpe J.A., Sloane-Stanley J.A., Sutherland L., et al. (2012) An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. Embo J., 31, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mendenhall E.M., Koche R.P., Truong T., Zhou V.W., Issac B., Chi A.S., Ku M., Bernstein B.E. (2010) GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet., 6, e1001244.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Deaton A.M., Bird A. (2011) CpG islands and the regulation of transcription. Genes Dev., 25, 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kassis J.A., Brown J.L. (2013) Polycomb group response elements in Drosophila and vertebrates. Adv. Genet., 81, 83–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bire S., Casteret S., Piegu B., Beauclair L., Moire N., Arensbuger P., Bigot Y. (2016) Mariner Transposons Contain a Silencer: Possible Role of the Polycomb Repressive Complex 2. PLoS Genet., 12, e1005902.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schoeftner S., Sengupta A.K., Kubicek S., Mechtler K., Spahn L., Koseki H., Jenuwein T., Wutz A. (2006) Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. Embo J., 25, 3110–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abdouh M., Hanna R., El Hajjar J., Flamier A., Bernier G. (2016) The Polycomb Repressive Complex 1 Protein BMI1 Is Required for Constitutive Heterochromatin Formation and Silencing in Mammalian Somatic Cells. J. Biol. Chem., 291, 182–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Walter M., Teissandier A., Perez-Palacios R., Bourc'his D. (2016) An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. eLife, 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sinclair D., Mottus R., Grigliatti T.A. (1983) Genes which suppress position-effect variegation in Drosophila melanogaster are clustered. Mol. Gen. Genet., 191, 326–333. [Google Scholar]
- 98.Boettiger A.N., Bintu B., Moffitt J.R., Wang S., Beliveau B.J., Fudenberg G., Imakaev M., Mirny L.A., Wu C.T., Zhuang X. (2016) Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature, 529, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Balog J., Thijssen P.E., de Greef J.C., Shah B., van Engelen B.G., Yokomori K., Tapscott S.J., Tawil R., van der Maarel S.M. (2012) Correlation analysis of clinical parameters with epigenetic modifications in the DUX4 promoter in FSHD. Epigenetics, 7, 579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Biagioli M., Ferrari F., Mendenhall E.M., Zhang Y., Erdin S., Vijayvargia R., Vallabh S.M., Solomos N., Manavalan P., Ragavendran A., et al. (2015) Htt CAG repeat expansion confers pleiotropic gains of mutant huntingtin function in chromatin regulation. Hum. Mol. Genet., 24, 2442–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nakamori M., Thornton C. (2010) Epigenetic changes and non-coding expanded repeats. Neurobiol. Dis., 39, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Usdin K., Kumari D. (2015) Repeat-mediated epigenetic dysregulation of the FMR1 gene in the fragile X-related disorders. Front. Genet., 6, 192.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sebastian S., Faralli H., Yao Z., Rakopoulos P., Palii C., Cao Y., Singh K., Liu Q.C., Chu A., Aziz A., et al. (2013) Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev., 27, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D. (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet., 37, 853–862. [DOI] [PubMed] [Google Scholar]
- 105.Sambrook J.F., Russel D.W. (2001) Molecular cloning: a laboratory manual Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.